Abstract
Thrombosis of small and large vessels is reported as a key player in COVID-19 severity. However, host genetic determinants of this susceptibility are still unclear. Congenital Thrombotic Thrombocytopenic Purpura is a severe autosomal recessive disorder characterized by uncleaved ultra-large vWF and thrombotic microangiopathy, frequently triggered by infections. Carriers are reported to be asymptomatic. Exome analysis of about 3000 SARS-CoV-2 infected subjects of different severities, belonging to the GEN-COVID cohort, revealed the specific role of vWF cleaving enzyme ADAMTS13 (A disintegrin-like and metalloprotease with thrombospondin type 1 motif, 13). We report here that ultra-rare variants in a heterozygous state lead to a rare form of COVID-19 characterized by hyper-inflammation signs, which segregates in families as an autosomal dominant disorder conditioned by SARS-CoV-2 infection, sex, and age. This has clinical relevance due to the availability of drugs such as Caplacizumab, which inhibits vWF–platelet interaction, and Crizanlizumab, which, by inhibiting P-selectin binding to its ligands, prevents leukocyte recruitment and platelet aggregation at the site of vascular damage.
1. Introduction
Congenital Thrombotic Thrombocytopenic Purpura (cTTP) is a severe autosomal recessive disorder due to ADAMTS13 (a disintegrin-like and metalloprotease with thrombospondin type 1 motif, 13) rare variants and is characterized by uncleaved ultra-large vWF and thrombotic microangiopathy, frequently triggered by infections. Common polymorphisms in the same enzyme are reported to be involved in thrombosis [1,2]. It is well-recognized that coagulation abnormalities with an increased risk of thrombosis are one of the complications of severe Coronavirus disease 2019 (COVID-19) disease, accompanied by a high level of IL-6 and D-dimer often together with a reduction in platelets [3,4,5,6]. ADAMTS13 gene encodes for a plasma glycoprotein with protease activity that plays a fundamental role in platelet adhesion and aggregation on vascular lesions, and the reduced activity of ADAMTS13 is already reported to be associated with a severe COVID-19 outcome [7]. Moreover, ADAMTS13 protein production is positively induced by estrogen, and this reflects the greater penetrance of acquired or congenital TTP in middle aged females (over 50 years), whose estrogen levels start to decrease in relation to males [8].
In our previous work, we modeled COVID-19 by the use of Artificial Intelligence (AI), and we identified variants related to COVID-19 severity [9]. Here, we specifically explore the role of one key genetic player: ultra-rare variants in ADAMTS13.
2. Materials and Methods
2.1. GEN-COVID Cohort
A cohort of 2988 SARS-CoV-2-positive subjects, collected within the GEN-COVID Multicenter study, was used in this work, including 48 from the Netherlands ConCOVID cohort. Among the 2988 subjects, 1781 were males and 1207 were females. The majority (2808 subjects, 94%) were European; the remaining 180 (6%) subjects were of African, Asian, American, and Hispanic ethnicity.
2.2. Whole Exome Sequencing Analysis (WES)
WES, with at least 97% coverage at 20×, was performed using the NovaSeq6000 System (Illumina, San Diego, CA, USA) as previously described [10]. WES data were represented in a binary mode on a gene-by-gene basis [9,10,11].
2.3. Phenotype Definition Adjusting by Age
An Ordered Logistic Regression (OLR) Model was applied, separately for males and females, using age to predict the clinical grading according to the WHO Outcome scale [12]. Each patient had a clinical classification equal to 0 (asymptomatic cases) if the actual patient grading was below the one predicted by the OLR or 1 (severely affected cases) if the grading was above the OLR prediction. The patients with a predicted gradient equal to the actual gradient were excluded from further analyses, by which we wanted to compare the “extreme ends” [9,10,11]. After the adjustment in females, there were 486 subjects with OLR equal to 1; 289 subjects with ORL equal to 0; and 432 subjects with a predicted gradient equal to the WHO gradient. In males, there were 672 subjects with OLR equal to 1; 552 subjects with ORL equal to 0; and 557 subjects with a predicted gradient equal to the WHO gradient.
2.4. ADAMTS13 Assay
The TECHNOZYM® ADAMTS-13 Activity Enzyme-Linked Immunosorbent Assay (ELISA) was used for determination of the ADAMTS13 activity using the venous-drawn, frozen, citrated (3.2% sodium citrate), platelet-poor plasma of studied patients. Blood samples should be collected with minimal stasis and processed rapidly to avoid cellular and plasma activation. The assay is a chromogenic and quantitative test performed on microplate readers capable of reading wavelengths of 450 nm. The measured value is reported as a percentage of normal-pooled plasma, which has been calibrated and defined as 100% activity [13].
2.5. Statistical Analysis
Statistical analysis was performed using R version 4.1.3 (10 March 2022) and RStudio (2022.02.0-443) software. A p-value < 0.05 was considered statistically significant.
3. Results
3.1. ADAMTS13 Ultra-Rare Variants Associate with Severity in COVID-19
Exome analysis of 2988 SARS-CoV-2-infected subjects of different severities, belonging to GEN-COVID cohort, stratified by sex and adjusted by age, shows an association between ADAMTS13 ultra-rare variants (Minor Allele Frequency < 0.001) and severity in female patients with an OR = 3.32 (95% CI 1.37 to 8.05; p-value = 4.9 × 10–3) (Table 1a). No significant association was found in male patients (p-value = 0.252) (Table 1b). The adjustment by age was performed as explained in the paragraph “Phenotype definition adjusting by age” of the Section 2.

Table 1.
(a) ADAMTS13 ultra-rare variants correlation with COVID-19 severity in female cohort. (b) ADAMTS13 ultra-rare variants correlation with COVID-19 severity in male cohort.
3.2. Characterization of Ultra-Rare Variants
One of several heterozygous ADAMTS13 ultra-rare variants (Table 2), classified as either VUS or pathogenic, was identified in 124 SARS-CoV-2-infected patients (4.2%), including 49 females (39.5%) and 75 males (60.5%). Among these 124 subjects, 110 were of European ethnicity, and the remaining subjects were of African, Asian, and Hispanic ethnicities.

Table 2.
ADAMTS13 heterozygous mutations in the entire cohort of SARS-CoV-2 positive subjects.
Most of the subjects (106) had severe COVID-19 disease requiring hospitalization (85.5%). The remaining 18 subjects (14.5%) were not-hospitalized patients (Table 2). The majority of not-hospitalized patients (14 subjects) were either females under 50 years or males over 50 years of age. For the females under 50, a protective role of estrogen, which increases ADAMTS13 transcript, can be envisaged. Among the hospitalized patients, there were also three females younger than 10 months, as described below (Table 2).
3.3. Characterization of ADAMTS13 Activity of Ultra-Rare Variants
Eleven subjects (6 cases with ADAMTS13 mutations and 5 controls without mutations) had ad hoc blood drawn and successful ADAMTS13 activity assessed after SARS-CoV-2 infection. The ADAMTS13 assay results were (median (min–max)) 61% (48–84) and 85% (71–106) for Cases and Controls, respectively. Carriers of ultra-rare variants show a significant reduction of ADAMTS13 activity, p-value = 0.017 (Wilcoxon test), as expected for heterozygous subjects. The box plot (Figure 1) shows the distribution of the two groups.
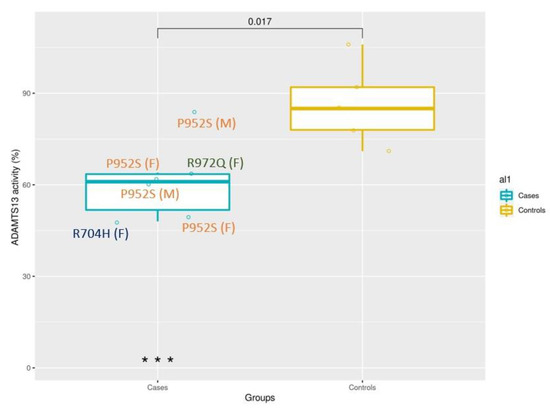
Figure 1.
Heterozygous ADAMTS13 ultra-rare variants are related to a reduction of protein detection. Box plot of patients with one ultra-rare variant (6 cases) and patients without ultra-rare variants (5 controls). The presence of ultra-rare variants is associated with a reduction of ADAMTS13 activity (p-value = 0.017 at Mann–Whitney U test).
Among heterozygous subjects, there is a large variability in the percentage of activity, likely due to different effects of each mutation and to additional genetic factors modulating the activity. The mutation c.2915G > A, p.R972Q has 64% of activity (normal value of up to 150%); the mutation c.2111G > A, p.R704H has 48% of activity; the mutation c.2854C > T, p.P952S has 63.8% of activity (mean of 4 subjects with a range of 49–85).
There is no data about low levels of ADAMTS13 in the other carriers, whose ADAMTS13 activity has not been measured.
3.4. Laboratory Values in Heterozygous Subjects
During hospitalization, both males and females with heterozygous ADAMTS13 variants have a tendency for hyper-inflammation (CRP mean 39, p = 0.005), higher D-dimer (mean 3024, p = 0.03), platelets consumption (platelet count mean 180, p = 0.07) and hemolysis (LDH mean 444, p = 0.009) (Table 3 and Table S1).

Table 3.
Correlations between ADAMTS13 ultra-rare variants and laboratory values in hospitalized patients.
The correlation is sustained mainly by females ≥50 years (CRP mean 55, p = 0.005; LDH mean 506, p = 0.006933) and males <50 years (platelet mean 153, p = 0.052) (Table 3 and Table S1). No significant correlation was observed between fibrinogen levels and carriers of ultra-rare variants (Table 3 and Table S1).
3.5. Autosomal Dominant Disorder Conditioned by SARS-CoV-2 Infection, Sex and Age
Complete clinical and molecular data were available for two families (Figure 2).
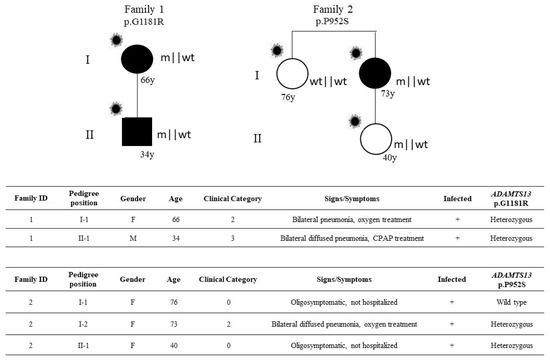
Figure 2.
Segregation analysis. Pedigree (upper panel) and respective segregation of ADAMTS13 variant and COVID-19 status (lower panel) are shown. Squares represent male family members; circles represent females. A virus cartoon close to the individual symbol indicates individuals infected by SARS-CoV-2 (  ). The inheritance pattern appears that of an autosomal dominant disorder conditioned by SARS-CoV-2 infection, sex, and age.
). The inheritance pattern appears that of an autosomal dominant disorder conditioned by SARS-CoV-2 infection, sex, and age.
 ). The inheritance pattern appears that of an autosomal dominant disorder conditioned by SARS-CoV-2 infection, sex, and age.
). The inheritance pattern appears that of an autosomal dominant disorder conditioned by SARS-CoV-2 infection, sex, and age.
Data of segregation analysis were able to demonstrate that the disorder segregates as autosomal dominant disorders conditioned by SARS-CoV-2 infection, sex, and age (Figure 2). In the first family, the 66-year-old female who required oxygen support transmitted the mutation to the 34-year-old son who required CPAP treatment. In the second family, the 73-year-old female treated by oxygen support transmitted the mutation to the 40-year-old daughter who was oligosymptomatic, likely due to the relatively young age; her sister, the 76-year-old without the mutation, was oligosymptomatic.
3.6. Pediatric Cases
Among the 127 patients with ADAMTS13 variants, three pediatric cases required hospitalization. Clinical and molecular characteristics are detailed below.
Case 1 (female) is the second child of a non-consanguineous couple of Filipino origin. Her pathological anamnesis is negative. The patient contracted SARS-CoV-2 infection when she was nine months old and her parents also turned out to be positive, but neither of them needed hospitalization, while her sister was negative. She had fever, cough, rhinitis, diuresis contraction and diarrhea (she tested negative for adenovirus and rotavirus); she did not have respiratory distress and her chest/lung ultrasound showed a B pattern. Among her blood tests, to be noticed: SGOT (serum glutamic oxaloacetic transaminase) 105 UI/L and SGPT (serum glutamic-pyruvic transaminase) 46 UI/l (maximum values during hospitalization) and D-dimer 1096 ng/mL. She has c.1016C > G, p.T339R ADAMTS13 heterozygous variant (MAF ExAC_NFE 0.0004; MAF ExAC_SAS 0.0012).
Case 1 (female) is the second child of a non-consanguineous couple of Filipino origin. Her pathological anamnesis is negative. The patient contracted SARS-CoV-2 infection when she was nine months old and her parents also turned out to be positive, but neither of them needed hospitalization, while her sister was negative. She had fever, cough, rhinitis, diuresis contraction and diarrhea (she tested negative for adenovirus and rotavirus); she did not have respiratory distress and her chest/lung ultrasound showed a B pattern. Among her blood tests, to be noticed: SGOT (serum glutamic oxaloacetic transaminase) 105 UI/L and SGPT (serum glutamic-pyruvic transaminase) 46 UI/l (maximum values during hospitalization) and D-dimer 1096 ng/mL. She has c.1016C > G, p.T339R ADAMTS13 heterozygous variant (MAF ExAC_NFE 0.0004; MAF ExAC_SAS 0.0012).
Case 2 (female) is the second child of a non-consanguineous European couple (Ukrainian mother, Italian father). She was born with a pulmonary CCAM (congenital cystic adenomatoid malformation) and a patent foramen ovale with a left-right shunt. The patient contracted SARS-CoV-2 infection when she was one month old and her sister and parents turned out to be positive but information on their clinical outcome is not available. She had fever, cough, rhinitis, diuresis contraction and diarrhea (she tested negative for adenovirus and rotavirus); she had slight respiratory distress, which did not require oxygen therapy, and her chest/lung ultrasound was negative. Among her blood tests, to be noticed: SGOT 61 UI/L and SGPT 36 UI/L at admission. She has c.1423C > T, p.P475S ADAMTS13 heterozygous variant (MAF ExAC_NFE 0.0006).
Case 3 (female) is the fourth child of a non-consanguineous couple of Nigerian origin. She was diagnosed with Rubinstein-Taybi syndrome (22q13.1q13.2 deletion) when she was a newborn. The patient contracted SARS-CoV-2 infection when she was seven months old and her mother turned out to be positive and she was not hospitalized. She had fever, cough, dyspnea, tachycardia, and respiratory distress (with oxygen saturation 88–89 and resuscitation); she also had a rhinovirus infection and part of her clinical picture is due to her genetic condition: in fact, she has a congenital heart defect. Her chest/lung ultrasound showed a B pattern. Among her blood tests, to be noticed: SGOT 52 UI/L and SGPT 71 UI/L at admission, which were normal when the child was discharged. She has c.2494G > A, p.V832M ADAMTS13 heterozygous variant (MAF ExAC_NFE 0.000017, MAF ExAC_AFR 0.013).
4. Discussion
Patients with severe COVID-19 can develop a wide range of complications, the most common of which is thrombosis of the large vessels. Thrombotic microangiopathy (TMA), whose pathophysiology is mostly due to endothelial dysfunction, has only been described in a few of these patients. TMAs include (i) congenital Thrombotic Thrombocytopenic Purpura (cTTP) characterized by no evidence of anti-ADAMTS-13 IgG antibodies and severe deficiency of ADAMTS13 activity and (ii) autoimmune TTP characterized by the presence of anti-ADAMTS-13 IgG antibodies. In primary autoimmune disease, no clear cause is identified, and instead in secondary autoimmune TTP a defined disorder or trigger can be identified, such as a viral infection [14,15]. The literature, recently reviewed by Singh B. et al. [16], contains few case reports of secondary autoimmune TTP in the course of COVID-19. In the descriptions of clinical cases, it is difficult to distinguish whether there is a cause-and-effect relationship between COVID-19 and TTP or whether SARS-CoV-2 infection is just present at the time of TTP diagnosis.
All cases of TTP are due to reduced activity of ADAMTS13, the enzyme involved in the cleavage of ultra-large von Willebrand factor (vWF) multimers into smaller, less procoagulant multimers. The congenital or inherited form of TTP has autosomal recessive inheritance, a prevalence of 0.5–2 cases per million [17], and accounts for 2–10% of all TTP cases reported in international registries [18]. The diagnosis of secondary autoimmune TTP is possible in the presence of microangiopathic hemolytic anemia, thrombocytopenia, ADAMTS13 activity <10%, and demonstration of an anti-ADAMTS13 inhibitor [19]. TTP heterozygous, i.e., carriers of one mutated allele only, are reported to be healthy. Here, we show evidence that heterozygous subjects are at risk for severe COVID-19 through a micro-thrombotic mechanism. Furthermore, the disease segregates in families as an autosomal dominant disorder, conditioned by SARS-CoV-2 infection, sex, and age. It is also known that the TTP-recessive disease is more penetrant in females. Females have a lower basal level of ADAMTS13 than males. However, estrogens have the power to induce protein production. Indeed, we expect females from the puberal period until ovarian failure to be protected by the action of estrogens [8]. In line with this, we have identified that heterozygous females over 50 are at more risk. On the other hand, we have reported pediatric cases as well (all females), which also miss the beneficial effect of estrogens. In the other sex (male), the period with less estrogens is that from puberty to andropause and indeed, as shown by laboratory value, the tendency towards microangiopathy is more evident in males under 50.
The penetrance of the thrombotic disease triggered by SARS-CoV-2 infection in heterozygous ADAMTS13 subjects is incomplete. Other factors that may contribute to the imbalance in the vWF antigen (VWF:Ag):/ADAMTS13 ratio are age, as vWF levels increase with age [20], and the patient’s membership in a blood group other than 0, resulting in baseline vWF:Ag levels 25–30% higher than in group 0 patients [21]. Furthermore, common polymorphisms in thrombotic microangiopathy-associated genes such as the rs2230199 in C3, the rs800292 in CFH (26 patients), and the rs2301612 missense mutation (448E) in the gene itself ADAMTS13, reported in 60 patients with moderate-to-severe COVID-19 studied by Graviilaki E. et al., may contribute to penetrance modulation [22]. From a multistep pathogenetic perspective of TMA, the procoagulant environment that originates during SARS-CoV-2 infection could precipitate the clinical manifestation of TMA in patients with genetic variants of ADAMTS13. There is indeed a direct virus-induced endothelial damage and secondary inflammatory status to cytokine storms [23], which result in the release of vWF from endothelial storage sites and a further reduction in ADAMTS13 activity, creating an imbalance in the vWF: Ag/ADAMTS13 activity ratio [24,25,26,27].
5. Conclusions
In conclusion, data from the large multicenter GEN-COVID study allow us to define the prevalence of ADAMTS13 mutations in a SARS-CoV-2-positive population and to establish the severity of COVID-19 pathology in patients carrying the mutation. This finding has clinical relevance due to the availability of drugs such as Caplacizumab or Crizanlizumab that could be suggested to patients with ADAMTS13 variants exhibiting decreased enzymatic activity. Caplacizumab, an anti-vWF bivalent single-domain nanobody, inhibits vWF–platelet interaction and is already used to treat acquired thrombotic thrombocytopenic purpura. Besides, Crizanlizumab is a monoclonal antibody that prevents leukocyte recruitment and platelet aggregation at the site of vascular damage by inhibiting P-selectin binding to its ligands. These two drugs are likely to replace the reduced activity of the metalloproteinase due to certain mutations and therefore they could also be useful in decreasing hyper-inflammation signs in heterozygous ADAMTS13 patients.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v14061185/s1, Table S1: Correlation between ADAMTS13 ultra-rare variants and laboratory values.
Author Contributions
Conceptualization: K.Z., M.B. (Margherita Baldassarri), C.F., S.F. and A.R. Methodology: C.F., F.F., M.B. (Margherita Baldassarri), G.B., R.L., L.G., M.C. (Michele Cirianni), P.C., M.L., A.O., A.B., G.M. (Giuseppe Marotta) and M.T. Software: K.Z. and S.F.; Validation: C.F., S.F. and A.R. Formal analysis: K.Z., S.D., C.F., G.B. and S.F. Investigation: K.Z., M.B. (Margherita Baldassarri), F.F., G.B. and A.R. Resources: A.R., S.R., M.S., D.F., E.S., E.M., M.G., S.B., S.G.P., S.P., C.P., M.C. (Mario Capasso), D.T., C.S.R., S.L., G.M. (Giovanni Morelli), A.G., G.G. and P.D.K. Data curation: M.B. (Margherita Baldassarri), F.F., C.F., A.R. and GEN-COVID Multicenter Study. Writing—original draft preparation: K.Z., C.F., M.B. (Monica Bocchia), F.F., A.G., G.G. and A.R. Writing—Review & Editing: K.Z., M.B. (Margherita Baldassarri), F.F., S.R., P.D.K., M.B. (Monica Bocchia), C.F., S.F. and A.R. Visualization: S.D., K.Z., A.O., C.F. and A.R. Supervision: C.F., S.F. and A.R. Project Administration: A.R. Funding acquisition: A.R. All authors have reviewed and approved the paper. All authors have read and agreed to the published version of the manuscript.
Funding
MIUR project “Dipartimenti di Eccellenza 2018–2020” to Department of Medical Biotechnologies University of Siena, Italy (Italian D.L. n.18 17 March 2020). Private donors for COVID-19 research. “Bando Ricerca COVID-19 Toscana” project to Azienda Ospedaliero-Universitaria Senese. Charity fund 2020 from Intesa San Paolo dedicated to the project N. B/2020/0119 “Identificazione delle basi genetiche determinanti la variabilità clinica della risposta a COVID-19 nella popolazione italiana”. The Italian Ministry of University and Research for funding within the “Bando FISR 2020” in COVID-19 and the Istituto Buddista Italiano Soka Gakkai for funding the project “PAT-COVID: Host genetics and pathogenetic mechanisms of COVID-19” (ID n. 2020–2016_RIC_3). EU project H2020-SC1-FA-DTS-2018-2020, entitled “International consortium for integrative genomics prediction (INTERVENE)”—Grant Agreement No. 101016775. Health Holland LSHM20056 grant (PDK).
Institutional Review Board Statement
The GEN-COVID was approved by the University Hospital (Azienda Ospedaliero-Universitaria Senese) Ethical Review Board, Siena, Italy (Prot n. 16917, dated 16 March 2020). The ConCOVID study was reviewed and approved by the institutional review board of the Erasmus University Medical Center.
Informed Consent Statement
Informed consent was obtained from all subjects or legally authorized representatives involved in the study.
Data Availability Statement
The data are available for sharing through the COVID-19 dedicated section (http://nigdb.cineca.it (accessed on 1 March 2022)), within the Network for Italian Genome (http://www.nig.cineca.it (accessed on 1 March 2022)). The data and samples referenced here are housed in the GEN-COVID Patient Registry and the GEN-COVID Biobank and are available for consultation. You may contact the corresponding author, Prof. Alessandra Renieri (e-mail: alessandra.renieri@unisi.it).
Acknowledgments
This study is part of the GEN-COVID Multicenter Study, https://sites.google.com/dbm.unisi.it/gen-covid (accessed on 1 March 2022), the Italian multicenter study aimed at identifying the COVID-19 host genetic bases. Specimens were provided by the COVID-19 Biobank of Siena, which is part of the Genetic Biobank of Siena, member of BBMRI-IT, of Telethon Network of Genetic Biobanks (project no. GTB18001), of EuroBioBank, and of RD-Connect. We thank Yvonne M. Mueller, Arvind Gharbharan, Casper Rokx and Bart J.A. Rijnders from Erasmus University Medical Center for providing ConCOVID samples. We thank the CINECA consortium for providing computational resources and the Network for Italian Genomes (NIG; http://www.nig.cineca.it (accessed on 1 March 2022)) for its support. We thank private donors for the support provided to AR (Department of Medical Biotechnologies, University of Siena) for the COVID-19 host genetics research project (D.L n.18 of 17 March 2020). We also thank the COVID-19 Host Genetics Initiative (https://www.covid19hg.org/ (accessed on 1 March 2022), MIUR project ‘Dipartimenti di Eccellenza 2018–2020′ to the Department of Medical Biotechnologies University of Siena, Italy, and ‘Bando Ricerca COVID-19 Toscana’ project to Azienda Ospedaliero-Universitaria Senese. We thank Intesa San Paolo for the 2020 charity fund dedicated to the project N B/2020/0119 ‘Identificazione delle basi genetiche determinanti la variabilità clinica della risposta a COVID-19 nella popolazione italiana’. The Italian Ministry of University and Research for funding within the “Bando FISR 2020” in COVID-19 fo the project “Editing dell’RNA contro il Sars-CoV-2: hackerare il virus per identificare bersagli molecolari e attenuare l’infezione—HACKTHECOV) and the Istituto Buddista Italiano Soka Gakkai for funding the project “PAT-COVID: Host genetics and pathogenetic mechanisms of COVID-19” (ID n. 2020–2016_RIC_3). We thank EU project H2020-SC1-FA-DTS-2018–2020, entitled “International consortium for integrative genomics prediction (INTERVENE)”—Grant Agreement No. 101016775.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pagliari, M.T.; Cairo, A.; Boscarino, M.; Mancini, I.; Pappalardo, E.; Bucciarelli, P.; Martinelli, I.; Rosendaal, F.R.; Peyvandi, F. Role of ADAMTS13, VWF and F8 genes in deep vein thrombosis. PLoS ONE 2021, 16, e0258675. [Google Scholar] [CrossRef]
- Haybar, H.; Khodadi, E.; Kavianpour, M.; Saki, N. Mutations and Common Polymorphisms in ADAMTS13 and vWF Genes Responsible for Increasing Risk of Thrombosis. Cardiovasc. Hematol. Disord. Drug Targets 2018, 18, 176–181. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Tang, N.; Li, D.; Wang, X.; Sun, Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020, 18, 1233–1234. [Google Scholar] [CrossRef] [Green Version]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Qu, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Cao, B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Hafez, W.; Ziade, M.A.; Arya, A.; Saleh, H.; Ali, S.; Rao, S.R.; Alla, O.F.; Ali, M.; Al Zouhbi, M.; Abdelrahman, A. Reduced ADAMTS13 Activity in Correlation with Pathophysiology, Severity, and Outcome of COVID-19: A Retrospective Observational Study. Int. J. Infect. Dis. 2022, 117, 334–344. [Google Scholar] [CrossRef]
- Powazniak, Y.; Kempfer, A.C.; Pereyra, J.C.; Palomino, J.P.; Lazzari, M.A. VWF and ADAMTS13 behavior in estradiol-treated HUVEC. Eur. J. Haematol. 2011, 86, 140–147. [Google Scholar] [CrossRef]
- Fallerini, C.; Picchiotti, N.; Baldassarri, M.; Zguro, K.; Daga, S.; Fava, F.; Benetti, E.; Amitrano, S.; Bruttini, M.; Palmieri, M.; et al. Common, low-frequency, rare, and ultra-rare coding variants contribute to COVID-19 severity. Hum. Genet. 2022, 141, 147–173. [Google Scholar] [CrossRef]
- Daga, S.; Fallerini, C.; Baldassarri, M.; Fava, F.; Valentino, F.; Doddato, G.; Benetti, E.; Furini, S.; Giliberti, A. Employing a systematic approach to biobanking and analyzing clinical and genetic data for advancing COVID-19 research. Eur. J. Hum. Genet. 2021, 29, 745–759. [Google Scholar] [CrossRef]
- Picchiotti, N.; Benetti, E.; Fallerini, C.; Daga, S.; Baldassarri, M.; Fava, F.; Zguro, K.; Valentino, F.; Doddato, G.; Giliberti, A.; et al. Post-Mendelian genetic model in COVID-19. Cardiol. Cardiovasc. Med. 2021, 5, 673–694. [Google Scholar] [CrossRef]
- WHO. COVID-19 Therapeutic Trial Synopsis; WHO R & D Blueprint Novel Coronavirus; COVID-19 Therapeutic Trial Synopsis; R & D Blueprint; WHO: New York, NY, USA, 2020. [Google Scholar]
- Chiasakul, T.; Cuker, A. Clinical and laboratory diagnosis of TTP: An integrated approach. Hematol. Am. Soc. Hematol. Educ. Program. 2018, 2018, 530–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, H.M. Pathophysiology of thrombotic thrombocytopenic purpura. Int. J. Hematol. 2010, 91, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Scully, M.; Cataland, S.; Coppo, P.; De La Rubia, J.; Friedman, K.D.; Hovinga, J.A.K.; Lämmle, B.; Matsumoto, M.; Pavenski, K.; Sadler, E.; et al. Consensus on the standardization of terminology in thrombotic thrombocytopenic purpura and related thrombotic microangiopathies. J. Thromb. Haemost. 2017, 15, 312–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, B.; Kaur, P.; Mekheal, E.; Fasulo, S.; Maroules, M. COVID-19 and thrombotic thrombocytopenic purpura: A review of literature. Hematol. Transfus. Cell Ther. 2021, 43, 529–531. [Google Scholar] [CrossRef] [PubMed]
- Kremer Hovinga, J.A.; George, J.N. Hereditary Thrombotic Thrombocytopenic Purpura. N. Engl. J. Med. 2019, 381, 1653–1662. [Google Scholar] [CrossRef] [PubMed]
- Alwan, F.; Vendramin, C.; Liesner, R.; Clark, A.; Lester, W.; Dutt, T.; Thomas, W.; Gooding, R.; Biss, T.; Watson, H.G.; et al. Characterization and treatment of congenital thrombotic thrombocytopenic purpura. Blood 2019, 133, 1644–1651. [Google Scholar] [CrossRef] [Green Version]
- Page, E.E.; Kremer Hovinga, J.A.; Terrell, D.R.; Vesely, S.K.; George, J.N. Thrombotic thrombocytopenic purpura: Diagnostic criteria, clinical features, and long-term outcomes from 1995 through 2015. Blood Adv. 2017, 1, 590–600. [Google Scholar] [CrossRef]
- Laffan, M. Can you grow out of von Willebrand disease? Haemoph. Off. J. World Fed. Hemoph. 2017, 23, 807–809. [Google Scholar] [CrossRef] [Green Version]
- Ward, S.E.; O’Sullivan, J.M.; O’Donnell, J.S. The relationship between ABO blood group, von Willebrand factor, and primary hemostasis. Blood 2020, 136, 2864–2874. [Google Scholar] [CrossRef]
- Graviilaki, E.; Kokoris, S.; Touloumenidouet, T.; Koravou, E.; Koutra, M.; Karali, V.; Anagnostopoulos, A. Thrombotic microangiopathy variants are independently associated with critical disease in COVID-19 patients. Blood 2020, 136 (Suppl. S1), 21–22. [Google Scholar]
- Panigada, M.; Bottino, N.; Tagliabue, P.; Grasselli, G.; Novembrino, C.; Chantarangkul, V.; Pesenti, A.; Peyvandi, F.; Tripodi, A. Hypercoagulability of COVID-19 patients in intensive care unit: A report of thromboelastography findings and other parameters of hemostasis. J. Thromb. Haemost. 2020, 18, 1738–1742. [Google Scholar] [CrossRef] [PubMed]
- Joly, B.S.; Darmon, M.; Dekimpe, C. Imbalance of von Willebrand factor and ADAMTS13 axis is rather a biomarker of strong inflammation and endothelial damage than a cause of thrombotic process in critically ill COVID-19 patients. J. Thromb. Haemost. 2021, 19, 2193–2198. [Google Scholar] [CrossRef] [PubMed]
- Pascreau, T.; Zia-Chahabi, S.; Zuber, B.; Tcherakian, C.; Farfour, E.; Vasse, M. ADAMTS 13 deficiency is associated with abnormal distribution of von Willebrand factor multimers in patients with COVID-19. Thromb. Res. 2021, 204, 138–140. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, G.; Maji, M.; Mandal, S.; Vaniyath, S.; Negi, G.; Nath, U.K. COVID 19 infection associated with thrombotic thrombocytopenic purpura. J. Thromb. Thrombolysis 2021, 52, 504–507. [Google Scholar] [CrossRef]
- Mancini, I.; Baronciani, L.; Artoni, A.; Colpani, P.; Biganzoli, M.; Cozzi, G.; Peyvandi, F. The ADAMTS13-von Willebrand factor axis in COVID-19 patients. J. Thromb. Haemost. 2021, 19, 513–521. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).