Abstract
Over the years, infectious diseases with high morbidity and mortality disrupted human healthcare systems and devastated economies globally. Respiratory viruses, especially emerging or re-emerging RNA viruses, including influenza and human coronavirus, are the main pathogens of acute respiratory diseases that cause epidemics or even global pandemics. Importantly, due to the rapid mutation of viruses, there are few effective drugs and vaccines for the treatment and prevention of these RNA virus infections. Of note, a class of antibodies derived from camelid and shark, named nanobody or single-domain antibody (sdAb), was characterized by smaller size, lower production costs, more accessible binding epitopes, and inhalable properties, which have advantages in the treatment of respiratory diseases compared to conventional antibodies. Currently, a number of sdAbs have been developed against various respiratory RNA viruses and demonstrated potent therapeutic efficacy in mouse models. Here, we review the current status of the development of antiviral sdAb and discuss their potential as therapeutics for respiratory RNA viral diseases.
1. Introduction
Respiratory diseases caused by various types of virus infections have been the focus of global health concerns and are one of the leading causes of death in developing countries [1]. According to the nucleic acid types, respiratory viruses can be divided into RNA and DNA viruses. However, the primary viruses causing the epidemics of respiratory infections in the last two decades were RNA viruses, such as the severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003, the influenza H1N1 virus in 2009, the Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012, and the SARS-CoV-2 in 2019 [2]. Therefore, the development of effective therapeutics for respiratory RNA viruses is rival to combat infectious diseases.
Respiratory RNA viruses include coronaviruses (SARS-CoV-1, MERS-CoV, and SARS-CoV-2), influenza viruses, respiratory syncytial virus (RSV), and others [3,4]. With the development of innovative recombinant DNA technologies, monoclonal antibodies (mAbs) have been proven effective in controlling respiratory RNA viral diseases. In 1998, the mAb palivizumab targeting RSV fusion (F) protein was approved by the FDA as prophylaxis against serious lower respiratory tract disease caused by RSV in children at high risk [5]. However, the high production costs of mAbs limit their commercial market, and the large size of mAbs leads to their low tissue accessibility and penetration, thus affecting their therapeutic efficacy. These features obstruct the development of mAbs [6]. Single-domain antibodies (sdAbs), consisting of only variable domains, have many advantages compared to mAbs. Their smaller size enables tissue penetration, so they could recognize epitopes that are normally not accessible for mAbs. In addition, the smaller size and higher stability of sdAbs make administration by inhalation possible, which is more suitable for treating respiratory diseases. It is also easy to express sdAbs in bacteria so that the production costs could be reduced. Therefore, sdAbs are a promising alternative to conventional mAbs [7,8]. In this review, we summarize the development of sdAb-based therapeutics for respiratory RNA virus infections and the strategies of antigen-specific sdAb screening.
2. Single-Domain Antibodies
In 1993, Hamers-Casterman et al. found that camelids could produce homodimeric heavy chain-only antibodies (HCAbs) devoid of light chains and the first constant domain (CH1) [9]. Two years later, Greenberg et al. reported that sharks and other cartilaginous fish could produce a type of HCAbs called Ig new antigen receptors (IgNARs) [10]. IgNARs compose of two identical heavy chains, each comprising five constant domains and a variable domain named VNAR that is responsible for antigen recognition [11]. The autonomous variable domains of HCAbs and IgNARs are called sdAbs, also known as nanobodies or VHHs if coming from the camelid family (including camels, llamas, and vicugna) [12]. Compared to cartilaginous fish, camelids are easier to access and can generate stronger sdAbs after antigen immunization, and VHHs share relatively higher homology with human immunoglobulin heavy chain variable region (IGHV) genes [11,13]. Therefore, VHHs attracted more interest than VNAR in the biological drug field.
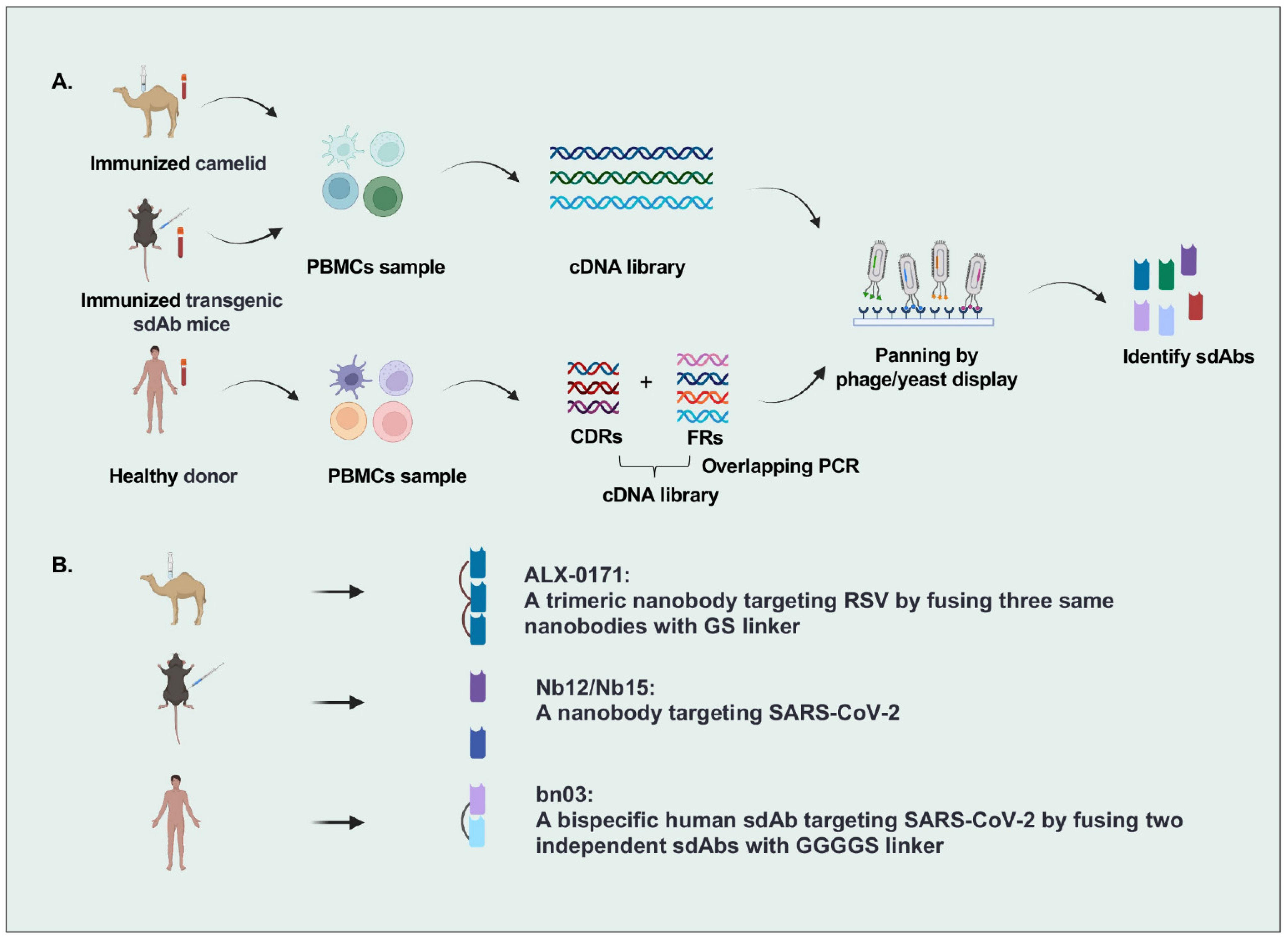
There are many strategies to identify sdAbs targeting specific antigens, including immunization of camelid and transgenic mice or panning by phage/yeast library with human sdAbs, camelid nanobodies, and IgNARs (Figure 1). The most popular strategy is that VHH genes were cloned from the peripheral blood lymphocytes of camelids immunized with specific antigens, followed by constructing a nanobody library by phage display and isolating nanobodies from this library. For example, Detalle et al. identified a monovalent RSV F protein-specific nanobody from llamas that received repetitive immunization with soluble recombinant F protein [14]. In another study, Xu et al. created mice (named nanomice) that could produce high-affinity nanobodies by inserting a VHH cassette instead of the VH locus in mouse embryonic stem cells. Then they immunized these nanomice with the receptor-binding domain (RBD) and the stabilized prefusion spike (S) protein of SARS-CoV-2 and isolated two nanobodies by phage display [15]. However, the sequences of VHHs with nonhuman origin may increase the immunogenicity risk in humans, leading to the need for humanization in VHH development. Wu et al. identified human VHs exhibiting biophysical properties very similar to VHHs from a fully human sdAb library constructed by using the germline 3-66*01 VH framework regions, indicating the potential of human sdAbs as alternatives to VHHs [16,17].
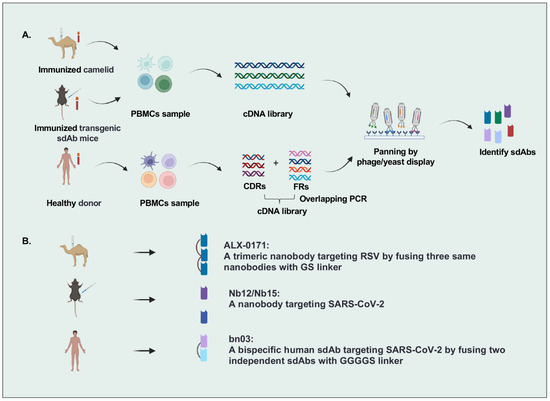
Figure 1.
Overview of single-domain antibodies screening strategies. (A). Strategies to identify single-domain antibodies. In brief, VHH or VH genes are cloned from peripheral blood lymphocytes of specific-antigen immunized camelids, transgenic sdAb mice, or healthy donors, and then antigen-specific sdAbs are identified by phage/yeast display panning. (B). Representative single-domain antibodies against respiratory RNA viruses are identified by different strategies. Figure generated with BioRender.
As the smallest antibodies, sdAbs have several beneficial characteristics, such as low molecular weight (12–15 kDa), small size (4 × 2.5 nm) [18], as well as high stability and solubility due to their longer CDR3 loops than conventional antibodies [19,20]. Because of the small size, sdAbs can recognize cavities or hidden epitopes that are not accessible to conventional antibodies [21]. For example, Wu et al. identified several fully human sdAbs that recognize a cryptic epitope located in the SARS-CoV-2 spike trimeric interface and have potent neutralization [16]. Moreover, sdAbs can be easily produced in bacteria, greatly reducing the production cost.
In addition, sdAbs can be easily engineered to be multivalent or multispecific, improving their binding affinity and breath [22] and prolonging half-life in vivo [23]. Lauren et al. reported four sdAbs, designed as SD36, SD38, SD83, and SD84, which could bind to highly conserved epitopes of the influenza A and B virus hemagglutinins (HAs), respectively. By fusing these sdAbs with peptide linkers, two multidomain antibodies (MDAbs) were generated and demonstrated the improved binding breadth and neutralizing potency of individual sdAbs [24]. In another study, a bispecific sdAbs bn03 was constructed by linking two sdAbs (n3113v and n3130v) that recognized two different conserved epitopes on SARS-CoV-2 RBD and exhibited potent neutralizing activity to all SARS-CoV-2 variants including Omicron [25]. Importantly, inhalation of bn03 significantly reduced viral titer in the lung in mild or severe SARS-CoV-2 infectious mice [25], indicating inhalation as a favorable route for delivering human sdAbs.
Currently, sdAbs have been developed to treat and diagnose various diseases [18]. In 2019, FDA approved Sanofi’s caplacizumab (an anti-von Willebrand factor nanobody and the first nanobody approved by FDA) for treating acquired thrombotic thrombocytopenic purpura (aTTP), a rare disease characterized by excessive blood clotting in small blood vessels [26]. A nanobody targeting human epidermal growth factor receptor 2 (HER2) is being evaluated in detecting breast-to-brain metastasis by Positron Emission Tomography (PET)/Computed Tomography (CT) imaging in phase II clinical trials (NCT03331601) [27,28]. Furthermore, due to the small size and favorable biophysical characteristics, some studies have reported that directly delivering nanobodies into the lung can block the viral invasion of airway epithelial cells in situ by inhalation [25,29,30]. Therefore, nanobodies can be administered by inhaled delivery and are particularly suitable for treating respiratory diseases caused by respiratory RNA virus infections.
3. The Mechanisms of Single-Domain Antibodies Inhibiting Respiratory RNA Virus Infections
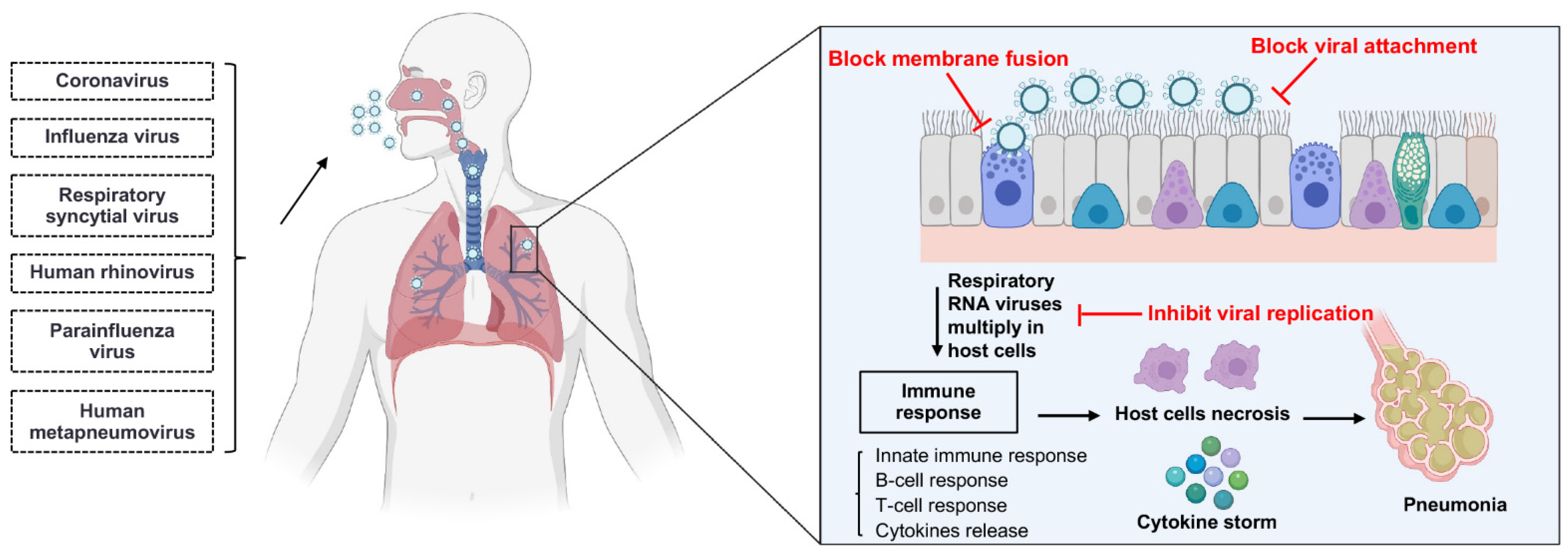
sdAbs can neutralize viruses by several different mechanisms in parallel with the virus life cycle (Figure 2) [31]. Respiratory RNA viruses can be classified into enveloped and non-enveloped viruses based on the presence or absence of a lipid membrane. Except for human rhinoviruses (HRVs), all other respiratory RNA viruses are enveloped viruses (Table 1) [32,33,34,35,36,37,38]. The main life-cycle steps of enveloped and non-enveloped viruses are similar, beginning with the entry into the host cell and ending with the release of virion progeny from the host cell. The majority of neutralizing sdAbs could serve as therapeutics for respiratory RNA virus infections by inhibiting the virion entry, while a few antiviral sdAbs can inhibit the viral genomic replication and the release of virion progeny [31].
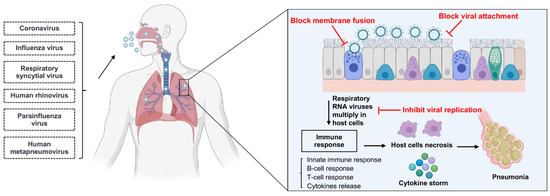
Figure 2.
The mechanisms of single-domain antibodies inhibiting respiratory RNA virus infections. Single-domain antibodies inhibit respiratory RNA virus infections by several different mechanisms in parallel with the virus life cycle. Respiratory RNA viruses primarily inoculate through the nose and then enter into host cells. After entry, respiratory RNA viruses multiply in the host cells, such as epithelium cells of the large and small airways, vascular endothelial cells, and alveolar macrophages. The infection of the respiratory tract can induce an immune response, and the immune response may lead to the cytokine storm; finally, the inflammation and necrosis of the epithelium cells may lead to pneumonia. The majority of neutralizing sdAbs inhibit the entry process by blocking the attachment of membrane fusion between virus and host cell. Moreover, some sdAbs neutralize the virus by inhibiting the viral genomic replication in host cells and the release of virion progeny from the host cells. Figure generated with BioRender.

Table 1.
The characteristics of respiratory RNA virus.
The process of viral entry usually starts from the attachment to a host cell; hence sdAbs can block viral entry by directly interfering with the interactions between the virus and host receptors, such as the RBD of SARS-CoV-1 and SARS-CoV-2 S proteins [39], and the receptor-binding site of influenza HA [40]. For example, a number of sdAbs targeting the RBD of SARS-CoV-2 S protein can block the interaction of RBD with the receptor angiotensin-converting enzyme 2 (ACE2) [41]. After attachment, the fusion of viral and host membranes is triggered and the viral genome is released into the cell cytoplasm from the endosome. Therefore, sdAbs can also block viral entry by binding to viral proteins that mediate membrane fusion and the release of the viral genome, such as the F protein of RSV [14], the HA and M2 of influenza viruses [42,43], and the S protein of SARS-CoV-2 [44]. Previous studies have reported that one sdAb targeting the non-ACE2 binding site of RBD can neutralize SARS-CoV-2 by inhibiting the conformational change of S protein that is essential for membrane fusion [45]. Moreover, some sdAbs can inhibit viral replication by binding to the viral proteins that are essential for virion release from the host cell, such as the neuraminidase (NA) of influenza viruses [46]. Apart from binding to the viral surface proteins, sdAbs can also inhibit the viral replication by binding to the viral intracellular proteins, such as the nucleoprotein of influenza viruses that mediates the viral nuclear trafficking and packaging [43,47].
5. Influenza Viruses
Influenza viruses are negative-sense single-stranded segmented enveloped RNA viruses belonging to the Orthomyxoviridae family and become the most common causes of human respiratory infections with high morbidity and mortality [94]. There are three genera that could infect humans: influenza A virus (IAV), influenza B virus (IBV), and influenza C virus (ICV) (Table 1). The majority of seasonal influenza viruses are IAVs and IBVs [95], which contain eight RNA segments that encode eight proteins, including two major targets of neutralizing antibodies, HA (a homotrimer that mediates the viral entry process) and NA (critical for viral release) [96]. IAVs are classified into subtypes based on the combination of HA and NA. Furthermore, these subtypes are classified into group 1 (H1, H2, H5, H6, H8, H9, H11, H12, H13, H16, H17) and group 2 (H3, H4, H7, H10, H14, H15), according to the phylogenetics of HA [97]. Due to the high genome mutation rate and the existence of antigenic drift and shift, new IAV types constantly appear and escape from the currently available neutralizing antibodies of influenza viruses [98]. Therefore, it is necessary to develop universal antibodies recognizing the conserved sites of HA or NA.
The monomer HA contains two subunits, HA1 and HA2. The head domain of the HA1 subunit is antigenic and variable, resulting in specific neutralizing of one influenza virus type for several sdAbs recognizing this region [40,77,78]. However, the receptor-binding site (RBS) is relatively conserved and has been recognized as the main target of broadly neutralizing antibodies. In contrast, the HA2 mediated membrane fusion shows more conserved [99], and, and the majority of broadly neutralizing sdAbs recognize the stem region [24,80]. Laursen et al. reported four sdAbs isolated from the immunized llamas, among which SD36, SD38, and SD83 bound to the conserved stem region of HA, while SD84 bound to the relatively conserved region of the HA head domain. SD36 and SD38 exhibited exquisite neutralizing breadth of IAVs, while SD83 and SD84 potently neutralized IBVs (Table 2). Moreover, the multidomain antibody was engineered by genetically fusing individual sdAbs with peptide linkers and then linking to human IgG1-Fc, named MD3606, exhibited enhanced broadly neutralizing activity against IAVs and IBVs and superior protective activity in mice from influenza B infection [24]. Further, humanized MD3606 expressed using an adeno-associated virus vector showed potent prophylactic efficacy in mice challenged with H1N1, H3N2, or IBV at a dose of 4 × 107 to 5 × 109 genome copies per mouse by intranasal administration [24]. In another study, R1a-B6 binding to the stem region of HA with high affinity was developed from an immunized alpaca by phage display and demonstrated neutralizing breadth against H1N1, H5N1, and H9N2 viruses. The bivalent R1a-B6 showed more neutralizing strength, including the H2N2 virus [80,82] (Table 2). Moreover, R1a-B6-Fc fusion protein delivered by AAV could be sustained in sera for more than 6 months and protected mice challenged with lethal H1N1 or H5N1 viruses [81]. In contrast, some sdAbs have been reported that bound to the HA stem region; however, they were only effective for a specific strain of influenza virus [78,79].
NA is a tetramer with each monomer consisting of four distinct domains: the catalytic head, the stalk region, the transmembrane region, and the cytoplasmic tail [100]. Due to the low mutation rate of HA, some universal mAbs targeting NA have been isolated from infected or vaccinated people, demonstrating NA as a promising target for broad protection against various influenza viruses [101]. N1-3-VHHb, a bivalent nanobody isolated from immunized alpaca and binding to H5N1 NA, showed potent NA-inhibiting activity and antiviral potential in vitro, as well as prophylactic efficacy in mice, challenged with lethal H5N1 virus administered intranasally at 60 μg [46].
M2 is an ion channel protein activated by the low pH of the endosome and plays essential roles in releasing the viral genome from the endosome to the cytoplasm [43]. The 1–9, His37, and Trp41 residues of M2 in all IAVs are extremely conserved. Therefore, sdAbs targeting these residues of M2 showed universal protection against IAVs [102]. Wei et al. reported a sdAb M2-7A that bound to the recombinant M2 protein and the native M2 on the virion, as well as the M2, expressed on the cell surface, but not to the synthetic ectodomain of M2 (M2e, residues 1–24) (Table 2). M2-7A broadly inhibited the replication of H3N2 and H1N1 in vitro and protected mice challenged with lethal H1N1 virus in vivo [42]. In addition, some VHHs targeting M2e showed broad binding breadth to most H2N2, H1N1 and H3N2 avian and swine influenza viruses. One of these VHHs, named M2e-VHH-23m, was fused with FcγRIV by a (GGGGS)3 linker and exhibited potently prophylactic efficacy in mice challenged with lethal H3N2 virus at 50 μg [83]. Furthermore, an mRNA treatment that encoded this bispecific sdAb also showed protection against the same H3N2 virus strain [84] (Table 2).
Because of the relatively high mutation rate of HA, NA, and M2, the development of truly universal antibodies remains challenging. To overcome this challenge, researchers focus on the less variable proteins, such as the nucleoprotein coating the viral RNA that is critical to transporting viral ribonucleoproteins (vRNPs) into the nucleus. More than 20 sdAbs targeting the nucleoprotein have been identified [47,103,104]. One of them, named αNP-VHH1, exhibited potent antiviral activity by blocking vRNP nuclear import and subsequent viral transcription and replication [47,105] (Table 2). In addition, Schmidt et al. reported a lentiviral screening strategy that developed several nucleoprotein-specific VHHs showing IAV-inhibiting activity by blocking the nuclear import of vRNPs and viral mRNA transcription [104].
6. Respiratory Syncytial Virus
Respiratory syncytial virus (RSV) is the main cause of respiratory virus infections in young children and elderly people. Apart from a humanized mAb that is licensed for high-risk infants as passive immunoprophylaxis, there are no effective RSV vaccines [38]. RSV is a non-segmented, negative-sense, single-stranded RNA virus belonging to the Pneumoviridae family (Table 1). Based on the antigenic subgroups, RSV is classified into two types: RSV-A and RSV-B. RSV is a filamentous enveloped virus and its envelope contains three proteins on the surface: the glycoprotein that plays an important role in host cell attachment and is the most variable structural protein, the fusion protein (F) that mediates the process of fusion and cell entry, and the small hydrophobic protein that is not required for the entry process [38,106]. The sequences of the F ectodomains share a high similarity (~90%) between RSV-A and RSV-B. Therefore, most sdAbs were developed for RSV F protein. ALX-0171, the most intensively studied nanobody targeting RSV F, is a trimeric nanobody developed by Ablynx with two GS linkers among each subunit (Table 2). ALX-0171 demonstrated high binding affinity with the antigenic site II epitope of F and broadly neutralizing activity against different clinical RSV isolates in vitro, as well as therapeutic efficacy in cotton rats challenged with RSV Tracy by nebulization administration [14]. In the preclinical study, ALX-0171 also exhibited robust antiviral effects and safety in newborn lambs infected with hRSV-M37 [29]. The phase I/IIa clinical trial showed that ALX-0171 administrated by inhalation at 1.2 mg/kg in infants infected with RSV could reduce the nasal viral titters with no safety concerns (https://www.clinicaltrialsregister.eu/ctr-search/trial/2014-002841-23/results, accessed on 18 August 2016). However, ALX-0171 showed no significant effect in children who were hospitalized with lower respiratory RSV infection in the phase IIb clinical trial [107]. With extensive research on the structure and function of RSV surface F glycoproteins, a number of sdAbs specifically targeting the prefusion conformation of F protein (PreF) exhibited higher binding affinity and inhibitory activity against RSV in vitro [85,86,87]. For example, F-VHH-4 and F-VHH-L66 could bind to PreF with <18 pM and 154 pM, respectively, and broadly neutralized RSV in vitro (Table 2). The crystal structures of F-VHH-4 and -L66 showed that they were both bound to a cavity formed by the boundary of two F protomers. In addition, F-VHH-4 demonstrated better prophylactic efficacy than palivizumab in mice challenged with RSV A2 at 10 μg [86]. These PreF-binding sdAbs with picomolar affinity are potential therapeutics for RSV infections.
7. Other Respiratory RNA Viruses
In addition to CoVs, influenza viruses, and RSV, there are other respiratory RNA viruses, such as HRVs, human metapneumoviruses (hMPVs), and parainfluenza viruses (PIVs). HRVs, usually associated with upper respiratory tract infection, are with positive-sense, single-stranded RNA genome and belong to the Picornaviridae family (Table 1). They are genetically classified into three types (RV-A, RV-B, and RV-C) [33]. The capsid of HRVs consists of four proteins, VP1, VP2, VP3, and VP4. Except for VP4, VP1, VP2, and VP3 are exposed at the viral surface and therefore are the main targets for neutralizing antibodies. The canyon in VP1 is the attachment site for host cell receptors. For most known HRV serotypes (>90%), the intercellular adhesion molecule 1 (ICAM-1) acts as their receptor for viral entry, whereas a small portion of HRVs enter the host cell via the low-density lipoprotein receptor (LDLR) [33,108]. A number of mAbs and Fabs targeting the VP1 [109], VP3 [110], or ICAM-1 [111] have been identified with neutralizing activity, while there are no available sdAbs targeting HRVs. PIVs are the second leading cause of acute respiratory tract infections in children. They are negative-sense single-stranded enveloped RNA viruses belonging to the Paramyxoviridae family [35,112] (Table 1). Based on the genetic diversity, PIVs can be divided into four serotypes, PIV-1, PIV-2, PIV-3, and PIV-4. There are two glycoproteins studded in the viral envelope. One of them is the HA-NA (HN) protein, which serves as the attachment protein by binding to the sialic acid-containing molecules of the host cells. The other one is the F protein responsible for the fusion. These two proteins are exposed on the viral surface and are the main targets for neutralizing antibodies and prophylactic vaccines [112]. There are no effective vaccines or therapeutics for PIV infections currently, and only several antibodies targeting the apex of F protein were developed and exhibited protective activity in cotton rats [113]. hMPVs, a common cause of viral pneumonia among infants and children, are enveloped viruses with negative-sense single-stranded RNA genomes and belong to the Pneumoviridae family [34] (Table 1). Similar to RSVs, there are three proteins exposed on the hMPV surface, the glycoprotein, the F protein, and the small hydrophobic protein. Among them, the F protein could induce neutralizing antibodies and hence is the target for developing neutralizing antibodies or vaccines [114].
8. Conclusions and Perspectives
In the past twenty years, respiratory RNA virus infections caused several outbreaks, with high morbidity and mortality [115,116,117]. The main characteristics of respiratory RNA viruses are strong transmission capacity and a high mutation rate. For instance, the current pandemic of SARS-CoV-2 has lasted for more than two years and five variants of concern (VOCs), including Alpha, Beta, Gamma, Delta, and Omicron, have been identified. The latest mutant strain Omicron has been reported to be resistant to most neutralizing antibodies and currently available vaccines [118,119]. Because of these characteristics, the developments of universal vaccines and effective therapeutics specific for respiratory RNA viruses remain a great challenge.
mAb-based therapeutics for respiratory RNA virus infections have been demonstrated to be effective [120]. However, the development of mAbs against viral infections has been hampered due to expensive production costs and limited commercial markets. Since the end of the last century, sdAbs with smaller size, higher stability and solubility, and lower immunogenicity and production costs have attracted more and more attention [18]. Compared to mAbs, sdAbs have advantages that have been confirmed by many studies. Firstly, sdAb can penetrate deep inside the “sterically hidden” interface of the virus and neutralize the virus. As the enveloped virus with flexible viral states, the large size of mAbs might bind the cryptic epitope in one state; however, it exhibited neglectable neutralization. Importantly, these cryptic domains are usually highly conserved and induce a low immune response in humans. Secondly, sdAb can be easily engineered to be multivalent to improve neutralizing potency and breadth [22]. Notably, by inhaled delivery, sdAbs can be directly delivered to the lung as the main infectious tissue in respiratory diseases [25]. Therefore, these sdAbs are promising to develop universal antiviral therapeutics for respiratory RNA virus infections and provide insights into the rational design of effective vaccines.
Despite the superior properties of sdAbs, mAbs are still dominant in the treatment of viral infections. For instance, 6 mAbs have been FDA approved for COVID-19 therapy and more than 50 ongoing clinical trials. The short half-life in vivo may be the limitation of sdAb developments. Currently, many strategies have been used to extend the half-life of sdAbs, such as fusing antiviral sdAbs with anti-human serum albumin nanobodies or IgG1 Fc fragments [121,122,123]. So far, only one sdAb has been approved for treating aTTP, and more than 30 sdAb-based drug candidates are in clinical trials. We expect recent technological advances in the fields of half-life extension, nebulized delivery, and production processes to advance more sdAbs into the clinic for the treatment of respiratory diseases.
Author Contributions
K.H., T.Y. and Y.W. conceived and wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the National Key R & D Program of China (2019YFA0904400), National Natural Science Foundation of China (81902108), Shanghai Municipal Education Commission “Chenguang program” (C620297), and Shanghai Municipal Health Commission (GWV-10.2-XD01, GWV-10.2-YQ06).
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Jordan, P.C.; Stevens, S.K.; Deval, J. Nucleosides for the treatment of respiratory RNA virus infections. Antivir. Chem. Chemother. 2018, 26, 2040206618764483. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, S.J.; Jacobson, R.M.; Dowdle, W.R.; Poland, G.A. 2009 H1N1 influenza. Mayo Clin. Proc. 2010, 85, 64–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kikkert, M. Innate Immune Evasion by Human Respiratory RNA Viruses. J. Innate Immun. 2020, 12, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, Z.; Li, M.; Wang, X. Comparative Review of SARS-CoV-2, SARS-CoV, MERS-CoV, and Influenza A Respiratory Viruses. Front. Immunol. 2020, 11, 552909. [Google Scholar] [CrossRef] [PubMed]
- Shadman, K.A.; Wald, E.R. A review of palivizumab and emerging therapies for respiratory syncytial virus. Expert Opin. Biol. 2011, 11, 1455–1467. [Google Scholar] [CrossRef] [PubMed]
- Samaranayake, H.; Wirth, T.; Schenkwein, D.; Raty, J.K.; Yla-Herttuala, S. Challenges in monoclonal antibody-based therapies. Ann. Med. 2009, 41, 322–331. [Google Scholar] [CrossRef]
- Respaud, R.; Vecellio, L.; Diot, P.; Heuze-Vourc’h, N. Nebulization as a delivery method for mAbs in respiratory diseases. Expert Opin. Drug Deliv. 2015, 12, 1027–1039. [Google Scholar] [CrossRef]
- Voss, J.E. Engineered single-domain antibodies tackle COVID variants. Nature 2021, 595, 176–178. [Google Scholar] [CrossRef]
- Hamers-Casterman, C.; Atarhouch, T.; Muyldermans, S.; Robinson, G.; Hamers, C.; Songa, E.B.; Bendahman, N.; Hamers, R. Naturally occurring antibodies devoid of light chains. Nature 1993, 363, 446–448. [Google Scholar] [CrossRef]
- Greenberg, A.S.; Avila, D.; Hughes, M.; Hughes, A.; McKinney, E.C.; Flajnik, M.F. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 1995, 374, 168–173. [Google Scholar] [CrossRef]
- Rossotti, M.A.; Belanger, K.; Henry, K.A.; Tanha, J. Immunogenicity and humanization of single-domain antibodies. FEBS J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Wolfson, W. Ablynx makes nanobodies from llama bodies. Chem. Biol. 2006, 13, 1243–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klarenbeek, A.; El Mazouari, K.; Desmyter, A.; Blanchetot, C.; Hultberg, A.; de Jonge, N.; Roovers, R.C.; Cambillau, C.; Spinelli, S.; Del-Favero, J.; et al. Camelid Ig V genes reveal significant human homology not seen in therapeutic target genes, providing for a powerful therapeutic antibody platform. MAbs 2015, 7, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Detalle, L.; Stohr, T.; Palomo, C.; Piedra, P.A.; Gilbert, B.E.; Mas, V.; Millar, A.; Power, U.F.; Stortelers, C.; Allosery, K.; et al. Generation and Characterization of ALX-0171, a Potent Novel Therapeutic Nanobody for the Treatment of Respiratory Syncytial Virus Infection. Antimicrob. Agents Chemother. 2016, 60, 6–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Xu, K.; Jung, S.; Conte, A.; Lieberman, J.; Muecksch, F.; Lorenzi, J.C.C.; Park, S.; Schmidt, F.; Wang, Z.; et al. Nanobodies from camelid mice and llamas neutralize SARS-CoV-2 variants. Nature 2021, 595, 278–282. [Google Scholar] [CrossRef]
- Wu, Y.; Li, C.; Xia, S.; Tian, X.; Kong, Y.; Wang, Z.; Gu, C.; Zhang, R.; Tu, C.; Xie, Y.; et al. Identification of Human Single-Domain Antibodies against SARS-CoV-2. Cell Host Microbe 2020, 27, 891–898.e5. [Google Scholar] [CrossRef]
- Wu, Y.; Li, Q.; Kong, Y.; Wang, Z.; Lei, C.; Li, J.; Ding, L.; Wang, C.; Cheng, Y.; Wei, Y.; et al. A highly stable human single-domain antibody-drug conjugate exhibits superior penetration and treatment of solid tumors. Mol. Ther. 2022. [Google Scholar] [CrossRef]
- Khodabakhsh, F.; Behdani, M.; Rami, A.; Kazemi-Lomedasht, F. Single-Domain Antibodies or Nanobodies: A Class of Next-Generation Antibodies. Int. Rev. Immunol. 2018, 37, 316–322. [Google Scholar] [CrossRef]
- Muyldermans, S. Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Jiang, S.; Ying, T. Single-Domain Antibodies as Therapeutics against Human Viral Diseases. Front. Immunol. 2017, 8, 1802. [Google Scholar] [CrossRef]
- Bao, G.; Tang, M.; Zhao, J.; Zhu, X. Nanobody: A promising toolkit for molecular imaging and disease therapy. EJNMMI Res. 2021, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Kang, G.; Yuan, H.; Cao, X.; Huang, H.; de Marco, A. Research Progress and Applications of Multivalent, Multispecific and Modified Nanobodies for Disease Treatment. Front. Immunol. 2021, 12, 838082. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cheng, L.; Fu, M.; Huang, B.; Zhu, L.; Xu, S.; Shi, H.; Zhang, D.; Yuan, H.; Nawaz, W.; et al. A potent bispecific nanobody protects hACE2 mice against SARS-CoV-2 infection via intranasal administration. Cell Rep. 2021, 37, 109869. [Google Scholar] [CrossRef] [PubMed]
- Laursen, N.S.; Friesen, R.H.E.; Zhu, X.; Jongeneelen, M.; Blokland, S.; Vermond, J.; van Eijgen, A.; Tang, C.; van Diepen, H.; Obmolova, G.; et al. Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science 2018, 362, 598–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Zhan, W.; Yang, Z.; Tu, C.; Hu, G.; Zhang, X.; Song, W.; Du, S.; Zhu, Y.; Huang, K.; et al. Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody. Cell 2022, 185, 1389–1401.e18. [Google Scholar] [CrossRef]
- Duggan, S. Caplacizumab: First Global Approval. Drugs 2018, 78, 1639–1642. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Lopez, E.; Schuhmacher, A.J. Transportation of Single-Domain Antibodies through the Blood-Brain Barrier. Biomolecules 2021, 11, 1131. [Google Scholar] [CrossRef]
- Keyaerts, M.; Xavier, C.; Heemskerk, J.; Devoogdt, N.; Everaert, H.; Ackaert, C.; Vanhoeij, M.; Duhoux, F.P.; Gevaert, T.; Simon, P.; et al. Phase I Study of 68Ga-HER2-Nanobody for PET/CT Assessment of HER2 Expression in Breast Carcinoma. J. Nucl. Med. 2016, 57, 27–33. [Google Scholar] [CrossRef] [Green Version]
- Larios Mora, A.; Detalle, L.; Gallup, J.M.; Van Geelen, A.; Stohr, T.; Duprez, L.; Ackermann, M.R. Delivery of ALX-0171 by inhalation greatly reduces respiratory syncytial virus disease in newborn lambs. MAbs 2018, 10, 778–795. [Google Scholar] [CrossRef] [Green Version]
- Nambulli, S.; Xiang, Y.; Tilston-Lunel, N.L.; Rennick, L.J.; Sang, Z.; Klimstra, W.B.; Reed, D.S.; Crossland, N.A.; Shi, Y.; Duprex, W.P. Inhalable Nanobody (PiN-21) prevents and treats SARS-CoV-2 infections in Syrian hamsters at ultra-low doses. Sci. Adv. 2021, 7, eabh0319. [Google Scholar] [CrossRef]
- Marasco, W.A.; Sui, J. The growth and potential of human antiviral monoclonal antibody therapeutics. Nat. Biotechnol. 2007, 25, 1421–1434. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov, D.S. Virus entry: Molecular mechanisms and biomedical applications. Nat. Rev. Microbiol. 2004, 2, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, S.E.; Lamson, D.M.; St. George, K.; Walsh, T.J. Human rhinoviruses. Clin. Microbiol. Rev. 2013, 26, 135–162. [Google Scholar] [CrossRef] [Green Version]
- Ballegeer, M.; Saelens, X. Cell-Mediated Responses to Human Metapneumovirus Infection. Viruses 2020, 12, 542. [Google Scholar] [CrossRef] [PubMed]
- Russell, E.; Ison, M.G. Parainfluenza Virus in the Hospitalized Adult. Clin. Infect. Dis. 2017, 65, 1570–1576. [Google Scholar] [CrossRef]
- Miller, K.; McGrath, M.E.; Hu, Z.; Ariannejad, S.; Weston, S.; Frieman, M.; Jackson, W.T. Coronavirus interactions with the cellular autophagy machinery. Autophagy 2020, 16, 2131–2139. [Google Scholar] [CrossRef]
- Te Velthuis, A.J.; Fodor, E. Influenza virus RNA polymerase: Insights into the mechanisms of viral RNA synthesis. Nat. Rev. Microbiol. 2016, 14, 479–493. [Google Scholar] [CrossRef] [Green Version]
- Battles, M.B.; McLellan, J.S. Respiratory syncytial virus entry and how to block it. Nat. Rev. Microbiol. 2019, 17, 233–245. [Google Scholar] [CrossRef]
- Wrapp, D.; De Vlieger, D.; Corbett, K.S.; Torres, G.M.; Wang, N.; Van Breedam, W.; Roose, K.; van Schie, L.; Team, V.-C.C.-R.; Hoffmann, M.; et al. Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies. Cell 2020, 181, 1004–1015.e15. [Google Scholar] [CrossRef]
- Gaiotto, T.; Ramage, W.; Ball, C.; Risley, P.; Carnell, G.W.; Temperton, N.; Engelhardt, O.G.; Hufton, S.E. Nanobodies mapped to cross-reactive and divergent epitopes on A(H7N9) influenza hemagglutinin using yeast display. Sci. Rep. 2021, 11, 3126. [Google Scholar] [CrossRef]
- Huo, J.; Le Bas, A.; Ruza, R.R.; Duyvesteyn, H.M.E.; Mikolajek, H.; Malinauskas, T.; Tan, T.K.; Rijal, P.; Dumoux, M.; Ward, P.N.; et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020, 27, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Meng, W.; Guo, H.; Pan, W.; Liu, J.; Peng, T.; Chen, L.; Chen, C.Y. Potent neutralization of influenza A virus by a single-domain antibody blocking M2 ion channel protein. PLoS ONE 2011, 6, e28309. [Google Scholar] [CrossRef] [PubMed]
- Vanderlinden, E.; Naesens, L. Emerging antiviral strategies to interfere with influenza virus entry. Med. Res. Rev. 2014, 34, 301–339. [Google Scholar] [CrossRef] [Green Version]
- Koenig, P.A.; Das, H.; Liu, H.; Kummerer, B.M.; Gohr, F.N.; Jenster, L.M.; Schiffelers, L.D.J.; Tesfamariam, Y.M.; Uchima, M.; Wuerth, J.D.; et al. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science 2021, 371, eabe6230. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, Y.; Jin, Y.; Zhu, Y.; Wu, Y.; Li, C.; Kong, Y.; Song, W.; Tian, X.; Zhan, W.; et al. A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants. Signal Transduct. Target. Ther. 2021, 6, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, F.M.; Ibanez, L.I.; Van den Hoecke, S.; De Baets, S.; Smet, A.; Roose, K.; Schepens, B.; Descamps, F.J.; Fiers, W.; Muyldermans, S.; et al. Single-domain antibodies targeting neuraminidase protect against an H5N1 influenza virus challenge. J. Virol. 2014, 88, 8278–8296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashour, J.; Schmidt, F.I.; Hanke, L.; Cragnolini, J.; Cavallari, M.; Altenburg, A.; Brewer, R.; Ingram, J.; Shoemaker, C.; Ploegh, H.L. Intracellular expression of camelid single-domain antibodies specific for influenza virus nucleoprotein uncovers distinct features of its nuclear localization. J. Virol. 2015, 89, 2792–2800. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [Green Version]
- Tortorici, M.A.; Veesler, D. Structural insights into coronavirus entry. Adv. Virus Res. 2019, 105, 93–116. [Google Scholar] [CrossRef]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.; Muller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.; Zaki, A.; Fouchier, R.A.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef] [Green Version]
- Hartenian, E.; Nandakumar, D.; Lari, A.; Ly, M.; Tucker, J.M.; Glaunsinger, B.A. The molecular virology of coronaviruses. J. Biol. Chem. 2020, 295, 12910–12934. [Google Scholar] [CrossRef] [PubMed]
- Chi, X.; Liu, X.; Wang, C.; Zhang, X.; Li, X.; Hou, J.; Ren, L.; Jin, Q.; Wang, J.; Yang, W. Humanized single domain antibodies neutralize SARS-CoV-2 by targeting the spike receptor binding domain. Nat. Commun. 2020, 11, 4528. [Google Scholar] [CrossRef] [PubMed]
- Hanke, L.; Das, H.; Sheward, D.J.; Perez Vidakovics, L.; Urgard, E.; Moliner-Morro, A.; Kim, C.; Karl, V.; Pankow, A.; Smith, N.L.; et al. A bispecific monomeric nanobody induces spike trimer dimers and neutralizes SARS-CoV-2 in vivo. Nat. Commun. 2022, 13, 155. [Google Scholar] [CrossRef] [PubMed]
- Mast, F.D.; Fridy, P.C.; Ketaren, N.E.; Wang, J.; Jacobs, E.Y.; Olivier, J.P.; Sanyal, T.; Molloy, K.R.; Schmidt, F.; Rutkowska, M.; et al. Highly synergistic combinations of nanobodies that target SARS-CoV-2 and are resistant to escape. ELife 2021, 10, e73027. [Google Scholar] [CrossRef]
- Li, J.-F.; He, L.; Deng, Y.-Q.; Qi, S.-H.; Chen, Y.-H.; Zhang, X.-L.; Hu, S.-X.; Fan, R.-W.; Zhao, G.-Y.; Qin, C.-F. Generation and Characterization of a Nanobody Against SARS-CoV. Virol. Sin. 2021, 36, 1484–1491. [Google Scholar] [CrossRef]
- Stalin Raj, V.; Okba, N.M.A.; Gutierrez-Alvarez, J.; Drabek, D.; van Dieren, B.; Widagdo, W.; Lamers, M.M.; Widjaja, I.; Fernandez-Delgado, R.; Sola, I.; et al. Chimeric camel/human heavy-chain antibodies protect against MERS-CoV infection. Sci. Adv. 2018, 4, eaas9667. [Google Scholar] [CrossRef] [Green Version]
- Zhao, G.; He, L.; Sun, S.; Qiu, H.; Tai, W.; Chen, J.; Li, J.; Chen, Y.; Guo, Y.; Wang, Y.; et al. A Novel Nanobody Targeting Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Receptor-Binding Domain Has Potent Cross-Neutralizing Activity and Protective Efficacy against MERS-CoV. J. Virol. 2018, 92, e00837-18. [Google Scholar] [CrossRef] [Green Version]
- Sun, D.; Sang, Z.; Kim, Y.J.; Xiang, Y.; Cohen, T.; Belford, A.K.; Huet, A.; Conway, J.F.; Sun, J.; Taylor, D.J.; et al. Potent neutralizing nanobodies resist convergent circulating variants of SARS-CoV-2 by targeting diverse and conserved epitopes. Nat. Commun. 2021, 12, 4676. [Google Scholar] [CrossRef]
- Esparza, T.J.; Martin, N.P.; Anderson, G.P.; Goldman, E.R.; Brody, D.L. High affinity nanobodies block SARS-CoV-2 spike receptor binding domain interaction with human angiotensin converting enzyme. Sci. Rep. 2020, 10, 22370. [Google Scholar] [CrossRef]
- Esparza, T.J.; Chen, Y.; Martin, N.P.; Bielefeldt-Ohmann, H.; Bowen, R.A.; Tolbert, W.D.; Pazgier, M.; Brody, D.L. Nebulized delivery of a broadly neutralizing SARS-CoV-2 RBD-specific nanobody prevents clinical, virological and pathological disease in a Syrian hamster model of COVID-19. bioRxiv 2021, 14, 1. [Google Scholar] [CrossRef]
- Guttler, T.; Aksu, M.; Dickmanns, A.; Stegmann, K.M.; Gregor, K.; Rees, R.; Taxer, W.; Rymarenko, O.; Schunemann, J.; Dienemann, C.; et al. Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies. EMBO J. 2021, 40, e107985. [Google Scholar] [CrossRef] [PubMed]
- Gai, J.; Ma, L.; Li, G.; Zhu, M.; Qiao, P.; Li, X.; Zhang, H.; Zhang, Y.; Chen, Y.; Ji, W.; et al. A potent neutralizing nanobody against SARS-CoV-2 with inhaled delivery potential. MedComm 2021, 2, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Cai, H.; Yao, H.; Zhou, B.; Zhang, N.; van Vlissingen, M.F.; Kuiken, T.; Han, W.; GeurtsvanKessel, C.H.; Gong, Y.; et al. A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection. Nat. Commun. 2021, 12, 4635. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Kwon, H.J.; Cachau, R.; Chen, C.Z.; Butay, K.J.; Duan, Z.; Li, D.; Ren, H.; Liang, T.; Zhu, J.; et al. Camel nanobodies broadly neutralize SARS-CoV-2 variants. BioRxiv Prepr. Serv. Biol. 2021. [Google Scholar] [CrossRef]
- Haga, K.; Takai-Todaka, R.; Matsumura, Y.; Song, C.; Takano, T.; Tojo, T.; Nagami, A.; Ishida, Y.; Masaki, H.; Tsuchiya, M.; et al. Nasal delivery of single-domain antibody improves symptoms of SARS-CoV-2 infection in an animal model. PLoS Pathog. 2021, 17, e1009542. [Google Scholar] [CrossRef]
- Custodio, T.F.; Das, H.; Sheward, D.J.; Hanke, L.; Pazicky, S.; Pieprzyk, J.; Sorgenfrei, M.; Schroer, M.A.; Gruzinov, A.Y.; Jeffries, C.M.; et al. Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2. Nat. Commun. 2020, 11, 5588. [Google Scholar] [CrossRef]
- Chen, X.; Gentili, M.; Hacohen, N.; Regev, A. A cell-free nanobody engineering platform rapidly generates SARS-CoV-2 neutralizing nanobodies. Nat. Commun. 2021, 12, 5506. [Google Scholar] [CrossRef]
- Ma, H.; Zeng, W.; Meng, X.; Huang, X.; Yang, Y.; Zhao, D.; Zhou, P.; Wang, X.; Zhao, C.; Sun, Y.; et al. Potent Neutralization of SARS-CoV-2 by Hetero-bivalent Alpaca Nanobodies Targeting the Spike Receptor-Binding Domain. J. Virol. 2021, 95, e02438-20. [Google Scholar] [CrossRef]
- Lu, Q.; Zhang, Z.; Li, H.; Zhong, K.; Zhao, Q.; Wang, Z.; Wu, Z.; Yang, D.; Sun, S.; Yang, N.; et al. Development of multivalent nanobodies blocking SARS-CoV-2 infection by targeting RBD of spike protein. J. Nanobiotechnol. 2021, 19, 33. [Google Scholar] [CrossRef]
- Ye, G.; Gallant, J.P.; Massey, C.; Shi, K.; Tai, W.; Zheng, J.; Odle, A.E.; Vickers, M.A.; Shang, J.; Wan, Y.; et al. The Development of a Novel Nanobody Therapeutic for SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zupancic, J.M.; Desai, A.A.; Schardt, J.S.; Pornnoppadol, G.; Makowski, E.K.; Smith, M.D.; Kennedy, A.A.; de Mattos Barbosa, M.G.; Cascalho, M.; Lanigan, T.M.; et al. Directed evolution of potent neutralizing nanobodies against SARS-CoV-2 using CDR-swapping mutagenesis. Cell Chem. Biol. 2021, 28, 1379–1388.e7. [Google Scholar] [CrossRef] [PubMed]
- Huo, J.; Mikolajek, H.; Le Bas, A.; Clark, J.J.; Sharma, P.; Kipar, A.; Dormon, J.; Norman, C.; Weckener, M.; Clare, D.K.; et al. A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19. Nat. Commun. 2021, 12, 5469. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Nambulli, S.; Xiao, Z.; Liu, H.; Sang, Z.; Duprex, W.P.; Schneidman-Duhovny, D.; Zhang, C.; Shi, Y. Versatile, Multivalent Nanobody Cocktails Efficiently Neutralize SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Dong, J.; Huang, B.; Wang, B.; Titong, A.; Gallolu Kankanamalage, S.; Jia, Z.; Wright, M.; Parthasarathy, P.; Liu, Y. Development of humanized tri-specific nanobodies with potent neutralization for SARS-CoV-2. Sci. Rep. 2020, 10, 17806. [Google Scholar] [CrossRef]
- Dong, J.; Huang, B.; Jia, Z.; Wang, B.; Gallolu Kankanamalage, S.; Titong, A.; Liu, Y. Development of multi-specific humanized llama antibodies blocking SARS-CoV-2/ACE2 interaction with high affinity and avidity. Emerg. Microbes Infect. 2020, 9, 1034–1036. [Google Scholar] [CrossRef]
- Schoof, M.; Faust, B.; Saunders, R.A.; Sangwan, S.; Rezelj, V.; Hoppe, N.; Boone, M.; Billesbølle, C.B.; Puchades, C.; Azumaya, C.M.; et al. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science 2020, 370, 1473–1479. [Google Scholar] [CrossRef]
- Tillib, S.V.; Ivanova, T.I.; Vasilev, L.A.; Rutovskaya, M.V.; Saakyan, S.A.; Gribova, I.Y.; Tutykhina, I.L.; Sedova, E.S.; Lysenko, A.A.; Shmarov, M.M.; et al. Formatted single-domain antibodies can protect mice against infection with influenza virus (H5N2). Antivir. Res. 2013, 97, 245–254. [Google Scholar] [CrossRef]
- Ramage, W.; Gaiotto, T.; Ball, C.; Risley, P.; Carnell, G.W.; Temperton, N.; Cheung, C.Y.; Engelhardt, O.G.; Hufton, S.E. Cross-Reactive and Lineage-Specific Single Domain Antibodies against Influenza B Hemagglutinin. Antibodies 2019, 8, 14. [Google Scholar] [CrossRef] [Green Version]
- Voronina, D.V.; Shcheblyakov, D.V.; Esmagambetov, I.B.; Derkaev, A.A.; Popova, O.; Shcherbinin, D.N. Development of Neutralizing Nanobodies to the Hemagglutinin Stem Domain of Influenza A Viruses. Acta Nat. 2021, 13, 33–41. [Google Scholar] [CrossRef]
- Hufton, S.E.; Risley, P.; Ball, C.R.; Major, D.; Engelhardt, O.G.; Poole, S. The breadth of cross sub-type neutralisation activity of a single domain antibody to influenza hemagglutinin can be increased by antibody valency. PLoS ONE 2014, 9, e103294. [Google Scholar] [CrossRef] [Green Version]
- Del Rosario, J.M.M.; Smith, M.; Zaki, K.; Risley, P.; Temperton, N.; Engelhardt, O.G.; Collins, M.; Takeuchi, Y.; Hufton, S.E. Protection from Influenza by Intramuscular Gene Vector Delivery of a Broadly Neutralizing Nanobody Does Not Depend on Antibody Dependent Cellular Cytotoxicity. Front. Immunol. 2020, 11, 627. [Google Scholar] [CrossRef] [PubMed]
- Gaiotto, T.; Hufton, S.E. Cross-Neutralising Nanobodies Bind to a Conserved Pocket in the Hemagglutinin Stem Region Identified Using Yeast Display and Deep Mutational Scanning. PLoS ONE 2016, 11, e0164296. [Google Scholar] [CrossRef]
- De Vlieger, D.; Hoffmann, K.; Van Molle, I.; Nerinckx, W.; Van Hoecke, L.; Ballegeer, M.; Creytens, S.; Remaut, H.; Hengel, H.; Schepens, B.; et al. Selective Engagement of FcgammaRIV by a M2e-Specific Single Domain Antibody Construct Protects Against Influenza A Virus Infection. Front. Immunol. 2019, 10, 2920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Hoecke, L.; Verbeke, R.; De Vlieger, D.; Dewitte, H.; Roose, K.; Van Nevel, S.; Krysko, O.; Bachert, C.; Schepens, B.; Lentacker, I.; et al. mRNA Encoding a Bispecific Single Domain Antibody Construct Protects against Influenza A Virus Infection in Mice. Mol. Nucleic Acids 2020, 20, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Xun, G.; Song, X.; Hu, J.; Zhang, H.; Liu, L.; Zhang, Z.; Gong, R. Potent Human Single-Domain Antibodies Specific for a Novel Prefusion Epitope of Respiratory Syncytial Virus F Glycoprotein. J. Virol. 2021, 95, e0048521. [Google Scholar] [CrossRef]
- Rossey, I.; Gilman, M.S.; Kabeche, S.C.; Sedeyn, K.; Wrapp, D.; Kanekiyo, M.; Chen, M.; Mas, V.; Spitaels, J.; Melero, J.A.; et al. Potent single-domain antibodies that arrest respiratory syncytial virus fusion protein in its prefusion state. Nat. Commun. 2017, 8, 14158. [Google Scholar] [CrossRef]
- Rossey, I.; Hsieh, C.L.; Sedeyn, K.; Ballegeer, M.; Schepens, B.; McLellan, J.S.; Saelens, X. A vulnerable, membrane-proximal site in human respiratory syncytial virus F revealed by a prefusion-specific single-domain antibody. J. Virol. 2021, 95, e02279-20. [Google Scholar] [CrossRef]
- Hultberg, A.; Temperton, N.J.; Rosseels, V.; Koenders, M.; Gonzalez-Pajuelo, M.; Schepens, B.; Ibanez, L.I.; Vanlandschoot, P.; Schillemans, J.; Saunders, M.; et al. Llama-derived single domain antibodies to build multivalent, superpotent and broadened neutralizing anti-viral molecules. PLoS ONE 2011, 6, e17665. [Google Scholar] [CrossRef] [Green Version]
- Schepens, B.; Ibanez, L.I.; De Baets, S.; Hultberg, A.; Bogaert, P.; De Bleser, P.; Vervalle, F.; Verrips, T.; Melero, J.; Vandevelde, W.; et al. Nanobodies(R) specific for respiratory syncytial virus fusion protein protect against infection by inhibition of fusion. J. Infect. Dis. 2011, 204, 1692–1701. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Tai, W.; Li, J.; Chen, Y.; Gao, Y.; Li, J.; Sun, S.; Zhou, Y.; Du, L.; Zhao, G. Enhanced Ability of Oligomeric Nanobodies Targeting MERS Coronavirus Receptor-Binding Domain. Viruses 2019, 11, 166. [Google Scholar] [CrossRef] [Green Version]
- Chouchane, L.; Grivel, J.C.; Farag, E.; Pavlovski, I.; Maacha, S.; Sathappan, A.; Al-Romaihi, H.E.; Abuaqel, S.W.; Ata, M.M.A.; Chouchane, A.I.; et al. Dromedary camels as a natural source of neutralizing nanobodies against SARS-CoV-2. JCI Insight 2021, 6, e145785. [Google Scholar] [CrossRef] [PubMed]
- Hanke, L.; Vidakovics Perez, L.; Sheward, D.J.; Das, H.; Schulte, T.; Moliner-Morro, A.; Corcoran, M.; Achour, A.; Karlsson Hedestam, G.B.; Hallberg, B.M.; et al. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat. Commun. 2020, 11, 4420. [Google Scholar] [CrossRef] [PubMed]
- Pymm, P.; Adair, A.; Chan, L.J.; Cooney, J.P.; Mordant, F.L.; Allison, C.C.; Lopez, E.; Haycroft, E.R.; O’Neill, M.T.; Tan, L.L.; et al. Nanobody cocktails potently neutralize SARS-CoV-2 D614G N501Y variant and protect mice. Proc. Natl. Acad. Sci. USA 2021, 118, e2101918118. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K.; Kash, J.C. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 2010, 7, 440–451. [Google Scholar] [CrossRef] [Green Version]
- Petrova, V.N.; Russell, C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018, 16, 47–60. [Google Scholar] [CrossRef]
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K.; Doherty, P.C.; Palese, P.; Shaw, M.L.; Treanor, J.; Webster, R.G.; et al. Influenza. Nat. Rev. Dis. Primers 2018, 4, 3. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, D.; Wu, Y.; Ying, T. Recent advances in “universal” influenza virus antibodies: The rise of a hidden trimeric interface in hemagglutinin globular head. Front. Med. 2020, 14, 149–159. [Google Scholar] [CrossRef] [Green Version]
- Uyeki, T.M. Influenza. Ann. Intern. Med. 2017, 167, ITC33–ITC48. [Google Scholar] [CrossRef]
- Sedeyn, K.; Saelens, X. New antibody-based prevention and treatment options for influenza. Antivir. Res. 2019, 170, 104562. [Google Scholar] [CrossRef]
- McAuley, J.L.; Gilbertson, B.P.; Trifkovic, S.; Brown, L.E.; McKimm-Breschkin, J.L. Influenza Virus Neuraminidase Structure and Functions. Front. Microbiol. 2019, 10, 39. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.-Q.; Wohlbold, T.J.; Zheng, N.-Y.; Huang, M.; Huang, Y.; Neu, K.E.; Lee, J.; Wan, H.; Rojas, K.T.; Kirkpatrick, E.; et al. Influenza Infection in Humans Induces Broadly Cross-Reactive and Protective Neuraminidase-Reactive Antibodies. Cell 2018, 173, 417–429.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manzoor, R.; Igarashi, M.; Takada, A. Influenza A Virus M2 Protein: Roles from Ingress to Egress. Int. J. Mol. Sci. 2017, 18, 2649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, C.J.; Crank, M.C.; Shiver, J.; Graham, B.S.; Mascola, J.R.; Nabel, G.J. Next-generation influenza vaccines: Opportunities and challenges. Nat. Rev. Drug. Discov. 2020, 19, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, F.I.; Hanke, L.; Morin, B.; Brewer, R.; Brusic, V.; Whelan, S.P.; Ploegh, H.L. Phenotypic lentivirus screens to identify functional single domain antibodies. Nat. Microbiol. 2016, 1, 16080. [Google Scholar] [CrossRef] [Green Version]
- Hanke, L.; Knockenhauer, K.E.; Brewer, R.C.; van Diest, E.; Schmidt, F.I.; Schwartz, T.U.; Ploegh, H.L. The Antiviral Mechanism of an Influenza A Virus Nucleoprotein-Specific Single-Domain Antibody Fragment. mBio 2016, 7, e01569-16. [Google Scholar] [CrossRef] [Green Version]
- Borchers, A.T.; Chang, C.; Gershwin, M.E.; Gershwin, L.J. Respiratory syncytial virus—A comprehensive review. Clin. Rev. Allergy Immunol. 2013, 45, 331–379. [Google Scholar] [CrossRef]
- Cunningham, S.; Piedra, P.A.; Martinon-Torres, F.; Szymanski, H.; Brackeva, B.; Dombrecht, E.; Detalle, L.; Fleurinck, C.; Cunningham, S.; Piedra, P.A.; et al. Nebulised ALX-0171 for respiratory syncytial virus lower respiratory tract infection in hospitalised children: A double-blind, randomised, placebo-controlled, phase 2b trial. Lancet Respir. Med. 2021, 9, 21–32. [Google Scholar] [CrossRef]
- Touabi, L.; Aflatouni, F.; McLean, G.R. Mechanisms of Rhinovirus Neutralisation by Antibodies. Viruses 2021, 13, 360. [Google Scholar] [CrossRef]
- Smith, T.J.; Olson, N.H.; Cheng, R.H.; Chase, E.S.; Baker, T.S. Structure of a human rhinovirus-bivalently bound antibody complex: Implications for viral neutralization and antibody flexibility. Proc. Natl. Acad. Sci. USA 1993, 90, 7015–7018. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Liu, Y.; Jiang, W.; Smith, T.J.; Xu, Z.; Rossmann, M.G. Antibody-induced uncoating of human rhinovirus B14. Proc. Natl. Acad. Sci. USA 2017, 114, 8017–8022. [Google Scholar] [CrossRef] [Green Version]
- Traub, S.; Nikonova, A.; Carruthers, A.; Dunmore, R.; Vousden, K.A.; Gogsadze, L.; Hao, W.; Zhu, Q.; Bernard, K.; Zhu, J.; et al. An anti-human ICAM-1 antibody inhibits rhinovirus-induced exacerbations of lung inflammation. PLoS Pathog. 2013, 9, e1003520. [Google Scholar] [CrossRef] [Green Version]
- Pawelczyk, M.; Kowalski, M.L. The Role of Human Parainfluenza Virus Infections in the Immunopathology of the Respiratory Tract. Curr. Allergy Asthma Rep. 2017, 17, 16. [Google Scholar] [CrossRef] [PubMed]
- Boonyaratanakornkit, J.; Singh, S.; Weidle, C.; Rodarte, J.; Bakthavatsalam, R.; Perkins, J.; Stewart-Jones, G.B.E.; Kwong, P.D.; McGuire, A.T.; Pancera, M.; et al. Protective antibodies against human parainfluenza virus type 3 infection. MAbs 2021, 13, 1912884. [Google Scholar] [CrossRef] [PubMed]
- Skiadopoulos, M.H.; Biacchesi, S.; Buchholz, U.J.; Amaro-Carambot, E.; Surman, S.R.; Collins, P.L.; Murphy, B.R. Individual contributions of the human metapneumovirus F, G, and SH surface glycoproteins to the induction of neutralizing antibodies and protective immunity. Virology 2006, 345, 492–501. [Google Scholar] [CrossRef] [Green Version]
- Cherry, J.D.; Krogstad, P. SARS: The first pandemic of the 21st century. Pediatr. Res. 2004, 56, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fineberg, H.V. Pandemic preparedness and response--lessons from the H1N1 influenza of 2009. N. Engl. J. Med. 2014, 370, 1335–1342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zumla, A.; Hui, D.S.; Perlman, S. Middle East respiratory syndrome. Lancet 2015, 386, 995–1007. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R.; et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022, 602, 657–663. [Google Scholar] [CrossRef]
- Cele, S.; Jackson, L.; Khoury, D.S.; Khan, K.; Moyo-Gwete, T.; Tegally, H.; San, J.E.; Cromer, D.; Scheepers, C.; Amoako, D.G.; et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 2022, 602, 654–656. [Google Scholar] [CrossRef]
- Jin, Y.; Lei, C.; Hu, D.; Dimitrov, D.S.; Ying, T. Human monoclonal antibodies as candidate therapeutics against emerging viruses. Front. Med. 2017, 11, 462–470. [Google Scholar] [CrossRef] [Green Version]
- Ying, T.; Gong, R.; Ju, T.W.; Prabakaran, P.; Dimitrov, D.S. Engineered Fc based antibody domains and fragments as novel scaffolds. Biochim. Biophys. Acta 2014, 1844, 1977–1982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ying, T.; Ju, T.W.; Wang, Y.; Prabakaran, P.; Dimitrov, D.S. Interactions of IgG1 CH2 and CH3 Domains with FcRn. Front. Immunol. 2014, 5, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rondon, A.; Mahri, S.; Morales-Yanez, F.; Dumoulin, M.; Vanbever, R. Protein Engineering Strategies for Improved Pharmacokinetics. Adv. Funct. Mater. 2021, 31, 2101633. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).