Abstract
Prion diseases are transmissible spongiform encephalopathies (TSEs) caused by pathogenic prion protein (PrPSc) originating from normal prion protein (PrPC) and have been reported in several types of livestock and pets. Recent studies have reported that the shadow of prion protein (Sho) encoded by the shadow of prion protein gene (SPRN) interacts with prion protein (PrP) and accelerates prion diseases. In addition, genetic polymorphisms in the SPRN gene are related to susceptibility to prion diseases. However, genetic polymorphisms in the feline SPRN gene and structural characteristics of the Sho have not been investigated in cats, a major host of feline spongiform encephalopathy (FSE). We performed amplicon sequencing to identify feline SPRN polymorphisms in the 623 bp encompassing the open reading frame (ORF) and a small part of the 3′ untranslated region (UTR) of the SPRN gene. We analyzed the impact of feline SPRN polymorphisms on the secondary structure of SPRN mRNA using RNAsnp. In addition, to find feline-specific amino acids, we carried out multiple sequence alignments using ClustalW. Furthermore, we analyzed the N-terminal signal peptide and glycosylphosphatidylinositol (GPI)-anchor using SignalP and PredGPI, respectively. We identified three novel SNPs in the feline SPRN gene and did not find strong linkage disequilibrium (LD) among the three SNPs. We found four major haplotypes of the SPRN polymorphisms. Strong LD was not observed between PRNP and SPRN polymorphisms. In addition, we found alterations in the secondary structure and minimum free energy of the mRNA according to the haplotypes in the SPRN polymorphisms. Furthermore, we found four feline-specific amino acids in the feline Sho using multiple sequence alignments among several species. Lastly, the N-terminal signal sequence and cutting site of the Sho protein of cats showed similarity with those of other species. However, the feline Sho protein exhibited the shortest signal sequence and a unique amino acid in the omega-site of the GPI anchor. To the best of our knowledge, this is the first report on genetic polymorphisms of the feline SPRN gene.
1. Introduction
Prion diseases are fatal and irreversible neurodegenerative disorders caused by a pathogenic isoform of prion protein (PrPSc) changed from normal prion protein (PrPC) and have been reported in various hosts, including Creutzfeldt–Jakob disease (CJD), fatal familial insomnia (FFI) and Gerstmann–Sträussler–Scheinker syndrome (GSS) in humans; scrapie in sheep and goats; bovine spongiform encephalopathy (BSE) in cattle; transmissible mink encephalopathy (TME) in mink; chronic wasting disease in elk and deer; and feline spongiform encephalopathy (FSE) in cats, cheetahs and pumas [1,2,3,4,5,6,7,8]. Prion diseases are classified into three major types according to etiology, including sporadic, iatrogenic and genetic forms [9].
In recent studies, several cofactors that interact with prion protein (PrP), a template of PrPSc, have been shown to accelerate the conversion of PrPC to PrPSc [10,11,12,13]. Among these cofactors, the shadow of prion protein (Sho) encoded by the shadow of prion protein gene (SPRN) plays a pivotal role in pathogenesis in several types of prion diseases [14]. The Sho is a member of the prion protein family and shows similar characteristics to PrP, including being a glycosylphosphatidylinositol (GPI) anchor protein and having tandem repeat sequences [15]. Since the Sho is an important interaction partner for PrP, genetic polymorphisms in the SPRN gene that affect conformational changes and/or the expression level of the Sho protein are related to susceptibility to prion diseases in several hosts. In humans, a frameshift polymorphism in the SPRN gene is observed only in variant CJD patients [16]. In addition, insertion/deletion polymorphisms in the SPRN gene are associated with vulnerability to scrapie-affected goats and atypical BSE-affected cattle [17]. Although the role of the Sho protein in prion diseases is apparent, genetic polymorphisms in the feline SPRN gene and structural characteristics of the Sho have not yet been investigated in cats, a major host of FSEs.
In the present study, we performed amplicon sequencing to identify feline SPRN polymorphisms and analyzed the impact of feline SPRN polymorphisms on the secondary structure of SPRN gene mRNA using RNAsnp [18]. In addition, we performed multiple sequence alignments among several prion-related species to identify unique characteristics of the feline Sho protein using ClustalW [19]. Furthermore, we analyzed the N-terminal signal peptide and GPI-anchor using SignalP and PredGPI, respectively [20,21].
2. Materials and Methods
2.1. Ethical Statements
All experiments were performed according to the Korea Experimental Animal Protection Act. All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Jeonbuk National University (IACUC number: CBNU 2019-00077).
2.2. Samples
A total of 193 cat samples were provided by Hemalgeun and Cool-Pet animal hospitals in the Republic of Korea. These samples were derived from regular health examinations and emasculations performed by qualified veterinary surgeons. The tissue and blood samples originated from 17 cat breeds, including Korean Domestic Shorthair (162), Persian (8), Turkish Angora (3), Abyssinian (2), Bengal (2), Norwegian Forest (2), Russian Blue (2), Scottish Fold (2), Siamese (2), American Curl (1), American Shorthair (1), British Shorthair (1), Malaysian Domestic Shorthair (1), Minuet (also called Napoleon) (1), Siberian (1), Sphynx (1) and Ragdoll (1). Since the official pedigree document was not available, and the sample size of each breed was very small, breed-specific analysis was not conducted.
2.3. Genomic DNA Isolation
Genomic DNA was isolated from tissue and blood samples using the Labopass Tissue Genomic DNA Isolation Kit Mini (Cosmogenetech, Seoul, Korea) and Bead Genomic DNA Prep Kit for Blood (Biofact, Daejeon, Korea) following the manufacturers’ protocols.
2.4. Genetic Analysis
The feline SPRN gene (Gene ID: 102902356) was amplified from genomic DNA by polymerase chain reaction (PCR) using gene-specific primers (F: 5′-GCTGCGGTCCTTTCTCCGTT-3′, R: 5′-GCAGCGTCTGTGGGTCAG-3′). The PCR mixture was composed of 2.5 µL 10× H-star Taq reaction buffer, 5 µL 5× band helper, 1 µL dNTP mix (10 mM each dNTP), 1 µL of each primer (10 µM), 0.2 µL H-star Taq DNA polymerase (BIOFACT, Daejeon, Korea) and nuclease-free water up to a total volume of 25 µL. The PCR conditions were as follows: 98 °C for 15 min for denaturation; 40 cycles of 98 °C for 20 s, 60 °C for 40 s and 72 °C for 1 min for annealing and extension; and 1 cycle of 72 °C for 5 min for the final extension. PCR was performed using a C1000 Touch Thermal Cycler (Bio–Rad, Hercules, CA, USA). The PCR products (623 bp) were sequenced by an ABI 3730 sequencer (ABI, Forster City, CA, USA). Electropherograms of the sequencing data were produced by Finch TV software (Geospiza Inc., Seattle, WA, USA). The genotyping was performed with DNA sequences of the PCR products except for the primer region (585 bp).
2.5. Statistical Analyses
The Hardy–Weinberg equilibrium (HWE), linkage disequilibrium (LD) and haplotype analyses of the SPRN polymorphisms were carried out using Haploview version 4.2 (Broad Institute, Cambridge, MA, USA). Haplotypes were identified by in silico analysis using a haplotype reconstruction method [22]. Information about the PRNP polymorphisms was obtained from a previous study [8]. LD analysis between PRNP and SPRN polymorphisms was performed using Haploview version 4.2 (Broad Institute, Cambridge, MA, USA).
2.6. Multiple Sequence Alignments
The amino acid sequences of the Sho protein were obtained from GenBank at the National Center for Biotechnology Information (NCBI), including those of human (Homo sapiens, NP_001012526.2), mouse (Mus musculus, AAH56484.1), cattle (Bos taurus, AAY83885.1), sheep (Ovis aries, NP_001156033.1), goat (Capra hircus, AGU17009.1), red deer (Cervus elaphus, ACF24724.1), dog (Canis lupus familiaris, XP_038296952.1), horse (Equus caballus, XP_023492126.1) and cat (Felis catus, XP_023096939.1). The amino acid sequences of the Sho protein were aligned using ClustalW based on progressive alignment methods.
2.7. Secondary Structural Analysis of mRNA
‘Mode 1′ of RNAsnp (http://rth.dk/resources/rnasnp/) (accessed on 12 January 2022), which is specialized for short mRNA sequences (<1000 nt), was used to assess the effects of the feline SPRN polymorphisms on the mRNA structure according to RNA folding algorithms. The base-pair probabilities based on the global folding method were analyzed between wild-type and query RNA sequences. Structural differences between the wild-type and query RNA sequences were calculated using the Euclidean distance or Pearson correlation coefficient for all sequence intervals.
2.8. Prediction of the Signal Peptide of Feline Sho
The signal peptide and cleavage site of the Sho were predicted by SignalP 5.0 (https://services.healthtech.dtu.dk/service.php?SignalP-5.0) (accessed on 12 January 2022). The prediction of SignalP 5.0 was based on a deep neural network-based method combined with conditional random field classification and optimized transfer learning.
2.9. Prediction of Omega-Site and Signal Sequence of GPI-Anchor of Feline Sho
The omega-site and signal sequence of the GPI-anchor were predicted by PredGPI (http://gpcr.biocomp.unibo.it/predgpi/index.htm) (accessed on 12 January 2022). The anchoring signal was predicted based on a support vector machine (SVM). The omega-site was predicted based on a hidden Markov model (HMM).
3. Results
3.1. Identification of Novel Polymorphisms of the Feline SPRN Gene in 193 Cats
The feline SPRN gene is composed of two exons. To investigate feline SPRN gene polymorphisms, we performed PCR to amplify the open reading frame (ORF) region of the feline SPRN gene in Exon 2. The amplicons were composed of 623 bp, encompassing the ORF and a small part of the 3′ untranslated region (UTR) of the SPRN gene. We identified three novel single nucleotide polymorphisms (SNPs), including c.15C > A and c.430C > A in the ORF region and c.469C > T in the 3′ UTR (Figure 1). The genotype and allele frequencies of the SNPs in the feline SPRN gene are described in Table 1. The genotype frequencies of all polymorphisms were in HWE. We also investigated the LD values among the three feline SPRN polymorphisms with r2 values (Table 2). Notably, strong LD (r2 > 0.333) was not observed between any of the feline SPRN polymorphisms. In addition, we performed haplotype analysis of three polymorphisms in the feline SPRN gene (Table 3). The CCC haplotype was most frequently observed (63.5%), followed by the ACC (26.2%), CAC (10.1%) and CCT (3%) haplotypes in the feline SPRN gene (Table 3). Furthermore, we investigated LD values between feline PRNP and SPRN polymorphisms. Strong LD (r2 > 0.333) was not observed between feline PRNP and SPRN polymorphisms (Table 4).
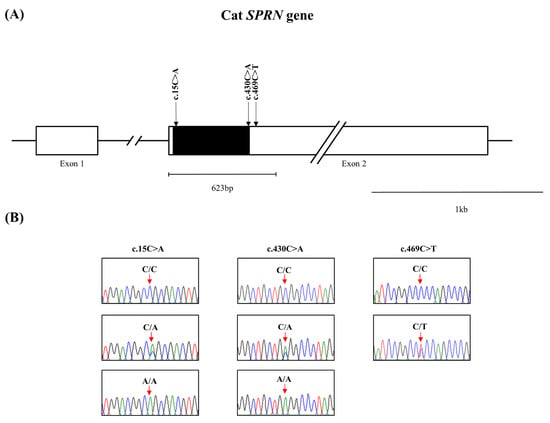
Figure 1.
Novel genetic polymorphisms in the feline shadow of prion protein gene (SPRN) in the present study. (A) Gene map of polymorphisms in the feline SPRN gene on chromosome D2. The open reading frame (ORF) is indicated by a shaded block, and the 5′ and 3′ untranslated regions (UTRs) are indicated by white blocks. The edged horizontal bar indicates the regions sequenced. (B) Electropherograms of 3 novel single nucleotide polymorphisms (SNPs) in the feline shadow of prion protein gene (SPRN) found in cats. The four colors of the peaks indicate each base of the DNA sequence (green: adenine; red: thymine; blue: cytosine; black: guanine).

Table 1.
Genotype and allele frequencies of shadow of prion protein gene (SPRN) polymorphisms in 193 domestic cats.

Table 2.
Linkage disequilibrium (LD) among genetic polymorphisms in the SPRN gene in cats.

Table 3.
Haplotype frequencies of 3 SPRN polymorphisms in cats.

Table 4.
LD between polymorphisms in the PRNP and SPRN genes with r2 values in cats.
3.2. mRNA Secondary Structure Analysis of the Feline SPRN Gene According to Polymorphisms
We investigated alterations of the mRNA secondary structure of the feline SPRN gene according to the haplotypes in the polymorphisms using the RNAsnp program (Figure 2). In the ACC haplotype in the feline SPRN polymorphisms, the graphical overview indicates a difference in the 1–215 nt region compared to the wild-type haplotype in the SPRN gene (CCC haplotype). The minimum free energies (CCC: −127.20 kcal/mol; ACC: −126.60 kcal/mol) and mRNA secondary structures showed differences between the CCC and ACC haplotypes (Figure 2A,B). In the CAC haplotype in the feline SPRN polymorphisms, the graphical overview indicates a difference in the 230–580 nt region compared to the wild-type haplotype in the SPRN gene (CCC haplotype). The minimum free energies (CCC: −206.00 kcal/mol; CAC: −205.90 kcal/mol) and mRNA secondary structures showed differences between the CCC and ACC haplotypes (Figure 2C,D). In the CCT haplotypes in the feline SPRN polymorphisms, the graphical overview indicates a difference in the 269–580 nt region compared to the wild-type haplotype in the SPRN gene (CCC haplotype). The minimum free energies (CCC: −176.10 kcal/mol; CCT: −175.20 kcal/mol) and mRNA secondary structures showed differences between the CCC and CCT haplotypes (Figure 2E,F).
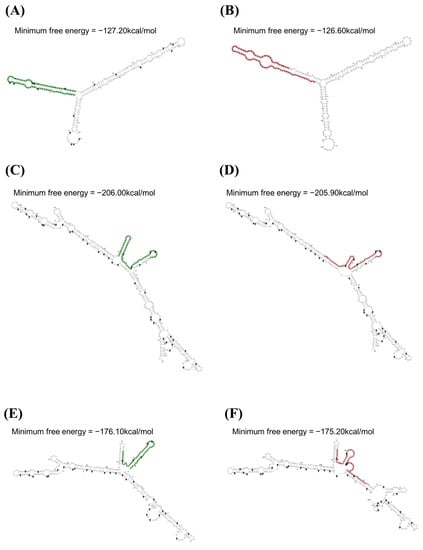
Figure 2.
Analysis of the mRNA secondary structure of the feline SPRN gene according to haplotypes in the feline SPRN polymorphisms. (A) Minimum free energy and mRNA secondary structure of the feline SPRN gene with the CCC haplotype (1–215 nt). (B) Minimum free energy and mRNA secondary structure of the feline SPRN gene with the ACC haplotype (1–215 nt). (C) Minimum free energy and mRNA secondary structure of the feline SPRN gene with the CCC haplotype (230–580 nt). (D) Minimum free energy and mRNA secondary structure of the feline SPRN gene with the CAC haplotype (230–580 nt). (E) Minimum free energy and mRNA secondary structure of the feline SPRN gene with the CCC haplotype (269–580 nt). (F) Minimum free energy and mRNA secondary structure of the feline SPRN gene with the CCT haplotype (269–580 nt).
3.3. Comparison of the Amino Acid Sequences of the Sho Protein among Species
We performed multiple sequence alignments of the amino acid sequences in the Sho protein among humans, mice, cattle, sheep, goats, red deer, dogs, horses and cats (Figure 3). The number of amino acids in the Sho protein was variable among species, and that of the human Sho protein was the longest (humans: 151; mice: 147; cattle: 143; sheep: 145; goats: 146; red deer: 143; dogs: 147; horses: 147; cats: 145). However, a low complexity Ala- and Gly-rich motif, in the interaction region of the Sho protein with PrP (red box), was conserved. In addition, although the NXT glycosylation motif (black box) was conserved among all species, cats have the unique amino acid sequence, NRT. The feline Sho protein showed a total of four feline-specific amino acids, including 21 I in the N-terminal domain, 106 R in the NXT glycosylation motif and 120 D and 138 V in the C-terminal domain.
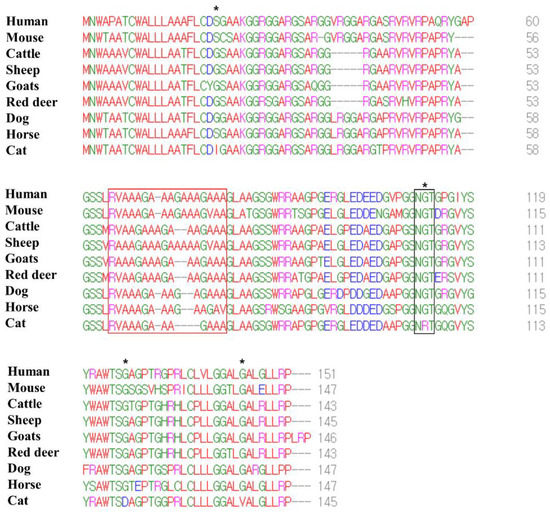
Figure 3.
Multiple sequence alignments of the amino acid sequences in the shadow of prion protein (Sho) in humans, mice, cattle, sheep, goats, red deer, dogs, horses and cats. Colors indicate the chemical properties of the amino acids; blue: acidic; red: small and hydrophobic; magenta: basic; green: hydroxyl, sulfhydryl, amine and glycine. Asterisks indicate cat-specific residues. The red box indicates the interaction region of the Sho protein with prion protein (PrP). The black box indicates the NXT glycosylation motif of the Sho protein.
3.4. Identification of Differences in the N-Terminal Signal Peptide of the Sho Protein among Species
We analyzed the N-terminal signal peptide of the Sho protein among humans, mice, cattle, sheep, goats, red deer, dogs, horses and cats. Detailed information on the amino acid sequences of the signal peptide is presented in Figure 4. The murine Sho has been reported to have an N-terminal signal sequence of 1–24 aa [23,24], and the prediction results in the present study were consistent with the data of the previous study. In brief, the length of the signal peptide of mice and dogs (24 aa) was longer than that of humans, cattle, sheep, goats, red deer, horses and cats (23 aa). Notably, the amino acid of the cutting site was alanine in all species.
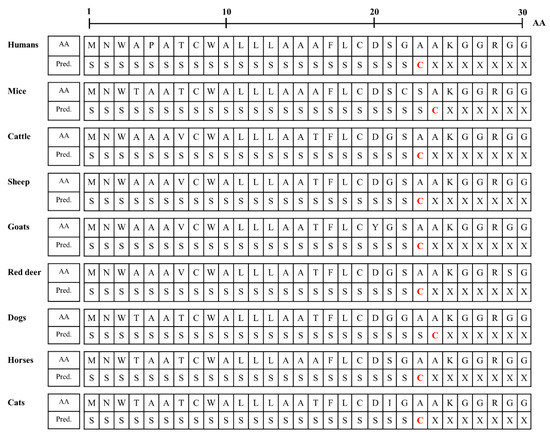
Figure 4.
Signal peptide predictions of amino acid sequences of Sho protein in human, mouse, cattle, sheep, goats, red deer, dog, horse and cat. AA: amino acids; Pred.: prediction; S: signal peptide; C: cutting site; X: non-signal peptide.
3.5. Investigation of the Omega-Site and Signal Sequence of the GPl-Anchor of the Sho Protein
We analyzed the omega-site and signal sequences of the GPl anchor among humans, mice, cattle, sheep, goats, red deer, dogs, horses and cats (Table 5). The murine Sho has been reported to have a GPI signal sequence of 123–147 aa [23,25], and the prediction results (116–147 aa) of the present study showed a slight difference from the previous study. Mice and cattle had the longest signal sequences of the GPI anchor of the Sho protein (32 aa). In contrast, cats showed the shortest signal sequence of the GPI anchor of the Sho protein (27 aa). The amino acid of the omega-site of humans, sheep, goats, red deer, dogs and horses was serine. However, that of the mice and cattle was tyrosine. Notably, the amino acid of the omega-site of cats was aspartic acid.

Table 5.
Prediction of the omega-site and signal sequences of the glycosylphosphatidylinositol (GPI)-anchor of the Sho protein by PredGPI.
4. Discussion
In the present study, we identified three novel SNPs in the feline SPRN gene (Figure 1, Table 1). We analyzed the effect of the three SNPs on the secondary structure of the mRNA of the SPRN gene using an in silico program. Notably, the secondary structure and minimum free energy of mRNAs were variable according to the haplotypes in the SNPs in the feline SPRN gene (Figure 2). In addition, SNPs can affect the transcription efficiency of genes according to how they affect the secondary structure of the mRNA. Thus, further analysis of the transcription level of the SPRN gene according to the haplotypes in the three SNPs is needed to validate the effects of the three SNPs. Among the three novel SNPs, c.469C > T was found in the 3′ UTR (Figure 1). The 3′ UTR is the transcription regulatory region that acts via the binding of microRNA [26,27]. Furthermore, a previous study reported that an insertion/deletion polymorphism in the caprine SPRN gene located on the 3′ UTR was associated with susceptibility to scrapie [28]. Since the expression level of the SPRN gene is important to the pathomechanism of prion diseases, further association analysis using FSE-infected animals and a reporter assay according to the genotype of c.469C > T is highly desirable in the future to validate the effect of c.469C > T.
To identify specific characteristics of the feline Sho protein, we compared the amino acid sequences of the Sho from several species. Notably, a low complexity Ala- and Gly-rich motif of the Sho was conserved among species. In addition, the feline Sho has a unique amino acid sequence: NRT in the NXT glycosylation motif (Figure 3). In addition, four feline-specific amino acids were found (Figure 3). One feline-specific amino acid, 106 R, was located in the NXT glycosylation motif, and the other three feline-specific amino acids were located on the signal sequences of the N-terminal signal peptide and GPI-anchor. Thus, we investigated the effect of feline-specific amino acids on the N-terminal signal peptide and the signal sequence and omega-site of the GPI-anchor. In the N-terminal signal peptide, the cutting site and length of the signal sequence of cats showed similarity with those of humans, cattle, sheep, goats, red deer and horses (Figure 4). This in silico result may suggest that the feline-specific amino acid 21I did not significantly affect the interspecific conserved signal sequence or cutting site of the N-terminal signal peptide. However, the feline Sho protein showed the shortest signal sequence and a unique amino acid in the omega-site of the GPI anchor (Table 5). This in silico result may suggest that the feline-specific amino acids 120 D and 138 V significantly affect the interspecific conserved signal sequence and omega-site of the GPI-anchor. However, since these in silico analyses were the prediction, further validation of the N-terminal signal sequence and omega site of GPI-anchor is highly desirable using biochemical analyses. In recent studies, the Sho protein has been shown to have a stress-resistant function and to be essential for early mouse embryogenesis and mammary development and differentiation [6,7,29]. Future analysis of the effect of GPI-anchor-related differences on the native function and physiology of the Sho protein is greatly desired.
5. Conclusions
In conclusion, we identified three novel SNPs in the feline SPRN gene. Strong LD was not observed among the three SNPs. We also found four major haplotypes in the SPRN polymorphisms. Strong LD was not observed between PRNP and SPRN polymorphisms. In addition, we found different secondary structures and minimum free energies of mRNA according to the haplotypes in the SPRN polymorphisms. Furthermore, we found four feline-specific amino acids in the feline Sho using multiple sequence alignments among several species. Lastly, the signal sequence and the cutting site of the N-terminal signal peptide of the Sho protein in cats showed similarity with those in humans, cattle, sheep, goats, red deer and horses. However, the feline Sho protein showed the shortest signal sequence and a unique amino acid in the omega-site of the GPI anchor. To the best of our knowledge, this is the first report on genetic polymorphisms in the feline SPRN gene.
Author Contributions
Y.-C.K. and B.-H.J. conceived and designed the experiment. H.-H.K., Y.-C.K., K.K. and A.-D.K. performed the experiments. Y.-C.K. and B.-H.J. analyzed the data. Y.-C.K. and B.-H.J. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant, funded by the Korean government (MSIT) (2021R1A2C1013213, 2022R1C1C2004792). This research was supported by the Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education (2017R1A6A1A03015876, 2021R1A6A3A010864).
Institutional Review Board Statement
All experiments were performed according to the Korea Experimental Animal Protection Act. All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Jeonbuk National University (IACUC number: CBNU 2019-00077).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
Y.-C.K. and H.-H.K. were supported by the BK21 plus program in the Department of Bioactive Material Sciences.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Prusiner, S.B. The prion diseases. Brain Pathol. 1998, 8, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Houston, F.; Andreoletti, O. Animal prion diseases: The risks to human health. Brain Pathol. 2019, 29, 248–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.C.; Park, K.J.; Hwang, J.Y.; Park, H.C.; Kang, H.E.; Sohn, H.J.; Jeong, B.H. In-depth examination of PrP(Sc) in Holstein cattle carrying the E211K somatic mutation of the bovine prion protein gene (PRNP). Transbound. Emerg. Dis. 2021. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.C.; Won, S.Y.; Jeong, B.H. Altered expression of glymphatic system-related proteins in prion diseases: Implications for the role of the glymphatic system in prion diseases. Cell Mol. Immunol. 2021, 18, 2281–2283. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.J.; Samuel, M.D.; O’Rourke, K.I.; Johnson, C.J. The role of genetics in chronic wasting disease of North American cervids. Prion 2012, 6, 153–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roh, I.S.; Kim, Y.C.; Won, S.Y.; Park, K.J.; Park, H.C.; Hwang, J.Y.; Kang, H.E.; Sohn, H.J.; Jeong, B.H. Association Study of the M132L Single Nucleotide Polymorphism with Susceptibility to Chronic Wasting Disease in Korean Elk: A Meta-Analysis. Front. Vet. Sci. 2021, 8, 804325. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.C.; Jeong, B.H. The First Meta-Analysis of the M129V Single-Nucleotide Polymorphism (SNP) of the Prion Protein Gene (PRNP) with Sporadic Creutzfeldt-Jakob Disease. Cells 2021, 10, 3132. [Google Scholar] [CrossRef]
- Kim, H.H.; Kim, Y.C.; Kim, K.; Kim, A.D.; Jeong, B.H. Novel Polymorphisms and Genetic Features of the Prion Protein Gene (PRNP) in Cats, Hosts of Feline Spongiform Encephalopathy. Genes 2020, 12, 13. [Google Scholar] [CrossRef]
- Baldwin, K.J.; Correll, C.M. Prion Disease. Semin. Neurol. 2019, 39, 428–439. [Google Scholar] [CrossRef]
- Taylor, D.R.; Whitehouse, I.J.; Hooper, N.M. Glypican-1 mediates both prion protein lipid raft association and disease isoform formation. PLoS Pathog. 2009, 5, e1000666. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.C.; Lee, J.; Lee, D.W.; Jeong, B.H. Large-scale lipidomic profiling identifies novel potential biomarkers for prion diseases and highlights lipid raft-related pathways. Vet. Res. 2021, 52, 105. [Google Scholar] [CrossRef]
- Deleault, N.R.; Piro, J.R.; Walsh, D.J.; Wang, F.; Ma, J.; Geoghegan, J.C.; Supattapone, S. Isolation of phosphatidylethanolamine as a solitary cofactor for prion formation in the absence of nucleic acids. Proc. Natl. Acad. Sci. USA 2012, 109, 8546–8551. [Google Scholar] [CrossRef] [Green Version]
- Lewis, V.; Hooper, N.M. The role of lipid rafts in prion protein biology. Front. Biosci. 2011, 16, 151–168. [Google Scholar] [CrossRef] [Green Version]
- Ciric, D.; Richard, C.A.; Moudjou, M.; Chapuis, J.; Sibille, P.; Daude, N.; Westaway, D.; Adrover, M.; Beringue, V.; Martin, D.; et al. Interaction between Shadoo and PrP Affects the PrP-Folding Pathway. J. Virol. 2015, 89, 6287–6293. [Google Scholar] [CrossRef] [Green Version]
- Daude, N.; Westaway, D. Biological properties of the PrP-like Shadoo protein. Front. Biosci. 2011, 16, 1505–1516. [Google Scholar] [CrossRef]
- Beck, J.A.; Campbell, T.A.; Adamson, G.; Poulter, M.; Uphill, J.B.; Molou, E.; Collinge, J.; Mead, S. Association of a null allele of SPRN with variant Creutzfeldt-Jakob disease. J. Med. Genet. 2008, 45, 813–817. [Google Scholar] [CrossRef]
- Gurgul, A.; Polak, M.P.; Larska, M.; Slota, E. PRNP and SPRN genes polymorphism in atypical bovine spongiform encephalopathy cases diagnosed in Polish cattle. J. Appl. Genet. 2012, 53, 337–342. [Google Scholar] [CrossRef]
- Sabarinathan, R.; Tafer, H.; Seemann, S.E.; Hofacker, I.L.; Stadler, P.F.; Gorodkin, J. The RNAsnp web server: Predicting SNP effects on local RNA secondary structure. Nucleic Acids Res. 2013, 41, W475–W479. [Google Scholar] [CrossRef] [Green Version]
- Sievers, F.; Higgins, D.G. Clustal omega. Curr. Protoc. Bioinf. 2014, 48, 3–13. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gislason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022. [Google Scholar] [CrossRef]
- Pierleoni, A.; Martelli, P.L.; Casadio, R. PredGPI: A GPI-anchor predictor. BMC Bioinf. 2008, 9, 392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, J.E.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watts, J.C.; Drisaldi, B.; Ng, V.; Yang, J.; Strome, B.; Horne, P.; Sy, M.S.; Yoong, L.; Young, R.; Mastrangelo, P.; et al. The CNS glycoprotein Shadoo has PrP(C)-like protective properties and displays reduced levels in prion infections. EMBO J. 2007, 26, 4038–4050. [Google Scholar] [CrossRef] [PubMed]
- Ciric, D.; Rezaei, H. Biochemical insight into the prion protein family. Front. Cell Dev. Biol. 2015, 3, 5. [Google Scholar] [CrossRef] [Green Version]
- Dondapati, D.T.; Cingaram, P.R.; Ayaydin, F.; Nyeste, A.; Kanyó, A.; Welker, E.; Fodor, E. Membrane Domain Localization and Interaction of the Prion-Family Proteins, Prion and Shadoo with Calnexin. Membranes 2021, 11, 978. [Google Scholar] [CrossRef]
- Fang, Z.; Rajewsky, N. The impact of miRNA target sites in coding sequences and in 3′UTRs. PLoS ONE 2011, 6, e18067. [Google Scholar] [CrossRef]
- Didiano, D.; Hobert, O. Molecular architecture of a miRNA-regulated 3′ UTR. RNA 2008, 14, 1297–1317. [Google Scholar] [CrossRef] [Green Version]
- Peletto, S.; Bertolini, S.; Maniaci, M.G.; Colussi, S.; Modesto, P.; Biolatti, C.; Bertuzzi, S.; Caramelli, M.; Maurella, C.; Acutis, P.L. Association of an indel polymorphism in the 3′UTR of the caprine SPRN gene with scrapie positivity in the central nervous system. J. Gen. Virol. 2012, 93, 1620–1623. [Google Scholar] [CrossRef]
- Passet, B.; Castille, J.; Makhzami, S.; Truchet, S.; Vaiman, A.; Floriot, S.; Moazami-Goudarzi, K.; Vilotte, M.; Gaillard, A.L.; Helary, L.; et al. The Prion-like protein Shadoo is involved in mouse embryonic and mammary development and differentiation. Sci. Rep. 2020, 10, 6765. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).