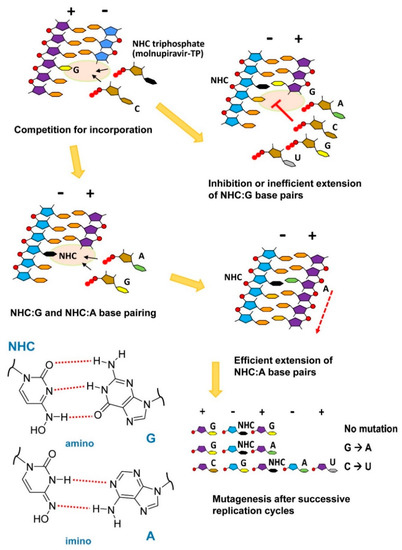
Abstract
In RNA viruses, a small increase in their mutation rates can be sufficient to exceed their threshold of viability. Lethal mutagenesis is a therapeutic strategy based on the use of mutagens, driving viral populations to extinction. Extinction catastrophe can be experimentally induced by promutagenic nucleosides in cell culture models. The loss of HIV infectivity has been observed after passage in 5-hydroxydeoxycytidine or 5,6-dihydro-5-aza-2′-deoxycytidine while producing a two-fold increase in the viral mutation frequency. Among approved nucleoside analogs, experiments with polioviruses and other RNA viruses suggested that ribavirin can be mutagenic, although its mechanism of action is not clear. Favipiravir and molnupiravir exert an antiviral effect through lethal mutagenesis. Both drugs are broad-spectrum antiviral agents active against RNA viruses. Favipiravir incorporates into viral RNA, affecting the G→A and C→U transition rates. Molnupiravir (a prodrug of β-d-N4-hydroxycytidine) has been recently approved for the treatment of SARS-CoV-2 infection. Its triphosphate derivative can be incorporated into viral RNA and extended by the coronavirus RNA polymerase. Incorrect base pairing and inefficient extension by the polymerase promote mutagenesis by increasing the G→A and C→U transition frequencies. Despite having remarkable antiviral action and resilience to drug resistance, carcinogenic risks and genotoxicity are important concerns limiting their extended use in antiviral therapy.
1. Quasispecies and Lethal Mutagenesis
Many viruses evolve rapidly [1]. Their compact genomes, high mutation rates, short replication times, and large population sizes are responsible for the generation of highly variable populations forming mutant swarms known as viral quasispecies [1]. A viral quasispecies refers to a population structure containing a large number of variant genomes with nonidentical, but closely related, mutant and recombinant genomes [2]. Also known as mutant spectra, mutant swarms, or mutant clouds, their composition and dynamics are continuously changing as viral replication and selection proceed. Mutant spectra containing related minority variants at low frequencies constitute phenotypic reservoirs and are well suited to respond to changing environments, a trait that is most relevant to viral populations.
The quasispecies concept was introduced by Eigen and Schuster in the 1970s to explain the self-organization and evolution of primitive RNA (or RNA-like) replicons when these molecules emerged from a mixture of prebiotic chemicals on the early Earth and began to replicate at the onset of life [3,4]. The existence of RNA virus quasispecies was independently demonstrated after studying the evolution of Escherichia coli bacteriophage Qβ RNA using a cell-free system [5]. The bacteriophage Qβ RNA is a small, single-stranded (ss)RNA that replicates through a double-stranded (ds)RNA intermediate. Qβ RNA can be replicated autocatalytically in vitro, while the sequence and composition of newly synthesized products depend on the fidelity of the Qβ replicase, nucleotide pools, metal cations, temperature, and speed of replication [6,7]. The mutation rates calculated for phage Qβ populations were estimated at around 10−4 per copied nucleotide, well above the rates calculated for DNA viruses and microorganisms [8,9].
Error catastrophe could be defined as a cumulative loss of genetic information in a lineage of organisms due to high mutation rates. The process leading to viral extinction through the accumulation of errors is known as lethal mutagenesis. Error catastrophe occurs when the mutation rate exceeds an error threshold. Viruses and bacteria have evolved to maintain a characteristic error rate. RNA viruses have very high mutation rates and replicate near the error threshold for the maintenance of genetic information [10]. A modest 1.1- to 2.8-fold increase in their mutation frequency can be sufficient to enter error catastrophe, as shown for vesicular stomatitis virus and poliovirus [11].
Lethal mutagenesis is an antiviral strategy that aims at extinguishing a virus by increasing the viral mutation rates during replication, usually through the use of mutagenic nucleoside analogs [12] (Figure 1).
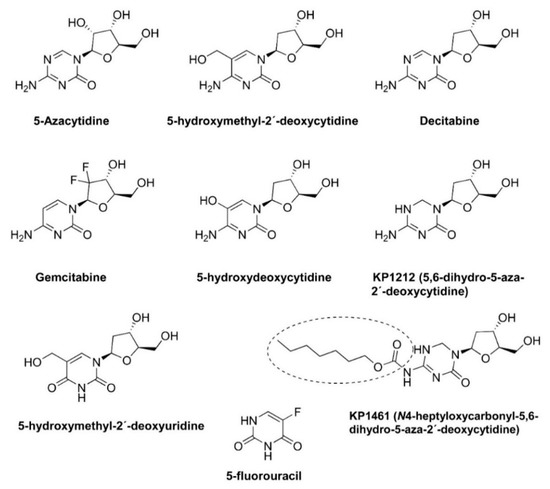
Figure 1.
Chemical structures of mutagenic base and nucleoside analogs and prodrugs used in lethal mutagenesis experiments. KP-1461 is a prodrug of KP-1212.
The term ‘lethal mutagenesis’ was coined in 1999 by Loeb and colleagues after showing that exposing the human immunodeficiency virus type 1 (HIV-1) LAI strain to 1 mM 5-hydroxydeoxycytidine (Figure 1) led to the loss of viral titer after 24 sequential passages in cell cultures [13]. The sequencing of part of the HIV-1 reverse transcriptase (RT) coding region of the penultimate passage prior to extinction revealed 2.6- and 5-fold increases in the frequency of A-to-G transitions (A→G) in two separate experiments [13]. These results stimulated further studies addressing the potential of mutagenic nucleoside analogs as antiviral agents driving viral populations to extinction. The other compounds shown in Figure 1 have been tested in HIV-1 and other RNA viruses, as well as in hepatitis B virus, with variable efficacies. In addition, virological and biochemical studies have demonstrated that the antiviral effect of approved drugs, such as ribavirin, favipiravir, and, more recently, molnupiravir, is due (at least in part) to their mutagenic action. Their chemical structures are shown in Figure 2. In this review, we discuss these studies and their implications for the further improvement of this antiviral strategy.
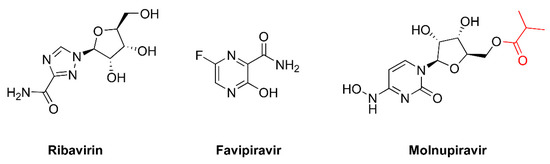
Figure 2.
Chemical structures of approved drugs inducing mutagenesis in viral replication assays. Molnupiravir is the isopropylester prodrug of β-d-N4-hydroxycytidine. The isopropylester moiety is shown in red.
2. Mutation Rates and Fidelity of Viral Polymerases
The mutation rates vary considerably among viruses [14,15]. While DNA viruses exhibit mutation rates ranging between 10−8 and 10−6 substitutions per nucleotide per cell infection, RNA viruses have mutation rates in the order of 10−6 to 10−4 [14,16]. A high replication rate combined with a high mutation rate allows RNA viruses to explore the sequence space and evade the immune system, even in situations where the majority of the viral progenies are nonviable [17].
Reverse-transcribing RNA viruses, such as retroviruses, use the viral RT to convert their ssRNA genomes into dsDNA, which is then integrated into the host genome and replicated along with it by eukaryotic DNA polymerases [18]. The mutation rates caused by the inactivation of a reporter gene range from 2 × 10−5 to 6 × 10−6 per nucleotide and replication cycle for many retroviruses, such as spleen necrosis virus, Rous sarcoma virus, murine leukemia virus (MLV), bovine leukemia virus, HIV-1, and human T-cell leukemia virus I (reviewed in [19]). O’Neil et al. [20] reported a mutation rate for HIV-1 of 8.5 × 10−5 mutations per base pair and replicative cycle, based on the variability observed at the long terminal repeats of the viral genome. In their study, the authors concluded that HIV-1 mutagenesis results from nucleotide misincorporation by the viral RT, although some contribution of the host RNA polymerase cannot be excluded.
Retroviral RTs lack a proofreading 3′–5′ exonuclease domain and have a relatively high propensity to extend mispaired 3′ ends when synthesizing viral DNA (reviewed in [21]). In addition, viral and host cells may also contribute to the observed variability in different retroviruses. Thus, feline immunodeficiency virus (FIV), equine infectious anemia virus (EIAV), mouse mammary tumor virus (MMTV), and Mason-Pfizer money virus (MPMV) encode a dUTP pyrophosphatase (dUTPase) within their genomes [22,23]. These enzymes reduce the mutation levels by preventing the incorporation of uracil into the viral genome, thereby safeguarding efficient reverse transcription [24]. In HIV-1, single-cycle replication assays using the lacZα gene as a mutational target showed that the deletion of the vpr gene produced a four-fold increase in the viral mutation rate [25]. On the other hand, apolipoprotein B mRNA-editing, catalytic polypeptide-like enzymes (APOBEC3) are cytidine deaminases of the host cell that cause hypermutations of nascent retroviral genomes by the deamination of cytidine residues [26,27]. APOBEC proteins are encapsidated within the virion, but the viral protein Vif suppresses their mutagenic effects by promoting APOBEC degradation in the ubiquitin-proteasome pathway (for a review, see [28]).
The HIV-1 RT is a heterodimer composed of subunits of 66 and 51 kDa. The large subunit contains an RNase H domain at its C-terminal end that is absent from p51. The RNase H activity of the HIV-1 RT degrades the RNA strand in the RNA/DNA duplexes formed during reverse transcription (for a review, see [18]). The DNA polymerase active site of the enzyme is located in the 66-kDa subunit and contains the catalytic residues Asp110, Asp185, and Asp186. The polymerase domain shares a subdomain arrangement found in many polymerases consisting of fingers, palm, and thumb subdomains, and a series of conserved motifs (A, B, C, D, and E) contributing key residues of the nucleotide-binding site [18]. In the fingers subdomain of p66, the incoming dNTP is tightly coordinated by the side-chains of Lys65 and Arg72, the main chain amido groups of Asp113 and Ala114, and two magnesium cations. Tyr115, Phe116, and Gln151 are additional residues delineating the dNTP binding pocket. Site-directed mutagenesis studies have shown that Lys65 has a major influence on the fidelity of HIV-1 RTs. Its substitution by Arg rendered enzymes with increased fidelity of DNA synthesis in HIV-1 M and O strains [29,30]. On the other hand, the substitution of Ala for Tyr115 conferred a mutator phenotype, as demonstrated by using enzymatic and cell-based assays [31,32,33].
HIV and other retroviruses evolve at rates similar to those of non-reverse transcribing RNA viruses (often referred to as riboviruses). Riboviral RNA-dependent RNA polymerases (RdRps) share the classical fingers–palm–thumb subdomain arrangement and lack 3′ exonuclease proofreading activity [34,35]. Notable exceptions are members of the Nidovirales order, including coronaviruses, a family of positive-strand RNA viruses encoding an RdRp complex (nsp7/(nsp8)2/nsp12) that associates with a protein subunit (nsp14) with 3′ exonuclease activity (for recent reviews, see [36,37]). The coronavirus genomes are among the largest known for RNA viruses, ranging from ∼26–32 kbp [38]. The amino-terminal half of SARS-CoV nsp14 (59 kDa) contains active site residues (Asp90, Glu92, Glu191, Asp273, and His268) also found in the cellular enzymes of the DEDD superfamily, including those that catalyze DNA proofreading. The substitution of Ala for Asp90 or Glu92 in SARS-CoV and the equivalent positions of murine hepatitis virus (MHV) rendered viable mutants that showed 15- to 20-fold increases in mutation rates, and were up to 18 times greater than those tolerated for fidelity mutants of other RNA viruses [39,40].
5. Mutagenic Effects of Ribavirin
Ribavirin [1-(β-d-ribofuranosyl)-1,2,4-triazole-3-carboxamide] (Figure 2) exhibits broad-spectrum antiviral activity against DNA- and RNA-based viruses. Despite being widely used in clinics for almost five decades, its efficacy has only been established for chronic hepatitis C virus infection, chronic hepatitis E virus infection in transplant recipients, and respiratory syncytial virus infection in infants and immunocompromised elderly patients. In addition, it is used to treat infections causing hemorrhagic fevers (e.g., Lassa fever, Crimean–Congo hemorrhagic fever, and Hantavirus infection), although it has very poor activity against filoviruses, such as Ebola or Marburg viruses [106,107].
The mechanism of action of ribavirin has remained largely elusive, probably due to the multiple targets of the drug [108,109]. These mechanisms include host-targeted effects, such as the inhibition of inosine monophosphate dehydrogenase (IMPDH) by ribavirin 5′-monophosphate (RMP), host immune response modulation (including the regulation of interferon-stimulated gene expression), and inhibition of translation initiation, through ribavirin binding to the translation initiation factor 4E (eIF4E) or enzymes responsible for RNA cap synthesis [110]. Interestingly, the lack of viral RNA capping triggers the antiviral host immune response by the recognition of a foreign viral RNA. IMPDH plays a key role in guanine nucleotide biosynthesis by catalyzing the conversion of inosine 5′-monophosphate into xanthine 5′-monophosphate, an intermediate in the de novo synthesis of guanosine. IMPDH regulates intracellular GTP pools necessary for RNA synthesis, and this could explain the activity of ribavirin against both DNA and RNA viruses [111]. However, nucleotide pool alterations due to IMPDH inhibition have a relatively small effect on the increased mutation frequencies observed during ribavirin treatments, as demonstrated for foot-and-mouth disease virus infections in cell cultures [75].
Viral replication is also a target of ribavirin. Thus, viral RNA-dependent RNA polymerases can be inhibited directly by ribavirin 5′-triphosphate (RTP) or can be incorporated (as RMP) into the viral genome, leading to viral mutagenesis [108]. Ribavirin does not have modifications in its sugar moiety and it is not clear how RMP incorporation inhibits RdRp. Ribavirin decreases viral RNA synthesis in infected cells, while in vitro studies have also demonstrated that the drug inhibits the polymerases of influenza A virus, hepatitis C virus, and vesicular stomatitis virus [112]. Studies carried out with poliovirus RdRp showed that RMP incorporation occurs opposite both template cytidine and uridine template residues [69,113]. Virological studies with poliovirus showed that ribavirin-mutagenized genomes had a 600-fold increase in G→A and C→U transition mutations [69]. C→U mutations would be a consequence of the incorporation of RTP as a GTP analog during negative-strand RNA synthesis. The effects of ribavirin as a lethal mutagen have been extensively studied in hepatitis C virus [74,75,76,77,78], but have also been demonstrated for other RNA viruses (Table 2).
The early studies with poliovirus suggested that the virus could develop resistance to ribavirin. However, the analysis of the polymerase-coding sequence of virus grown in the presence of 0.8 mM ribavirin showed that conserved motifs C–E remained unchanged, while some variability was observed at motifs A and B [114]. The passage of poliovirus in cell cultures in the presence of ribavirin led to the selection of G64S. This mutation is located at the fingers subdomain of the viral RdRp (Figure 3A). Ribavirin-resistant poliovirus displays increased fidelity of RNA synthesis in the absence of ribavirin, as well as increased survival in the presence of ribavirin and in the presence of 5-azacytidine [115].
Unexpectedly, the equivalent substitution in the RNA polymerase of foot-and-mouth disease virus (i.e., G62S) was never selected when the virus was passaged in the presence of ribavirin and was not detected as a minority variant in the mutant spectra of the virus that replicated in the absence or presence of ribavirin or other mutagenic agents [75,77]. Instead, P44S, P169S, and M296I in the RdRp of foot-and-mouth disease virus (serotype C) were shown to confer different levels of ribavirin resistance [77,78,116] (Figure 3B), while selection studies with the drug using hepatitis C virus replicons selected for P415Y in the thumb subdomain of the polymerase [117]. However, this substitution did not lead to treatment failure in infected patients treated with interferon and ribavirin, or to ribavirin resistance in cell culture assays [118,119]. More recently, Mejer et al. [120] showed, in a hepatitis C virus genotype 3a cell-culture-adapted strain, that certain combinations of mutations selected in patients treated with ribavirin (e.g., D148N/I363V, A150V/I363V, and T227S/S183P) conferred resistance, possibly by increasing the overall fidelity of the viral polymerase as a putative mechanism for ribavirin resistance. Although the structural basis of ribavirin resistance is still uncertain, many of the involved mutations are located at the periphery of the nucleotide entry site in the predicted polymerase structure.
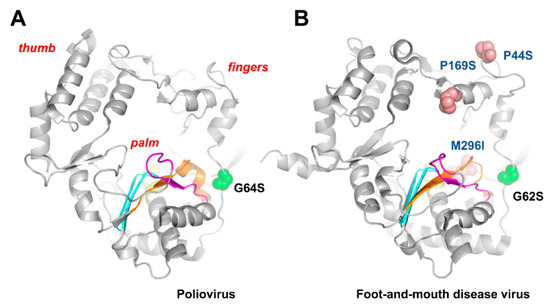

Figure 3.
Poliovirus (A) and foot-and-mouth disease virus (B) RdRp structures showing the location of motifs A–D and residues relevant for ribavirin binding and resistance. Locations of motifs A, B, C, and D are shown in orange, yellow, blue, and magenta, respectively [34,121]. Spheres are used to represent the location of relevant amino acid substitutions. Crystal structures were taken from PDB files 2ILY (poliovirus RdRp) and 1U09 (foot-and-mouth disease virus RdRp).
Apart from the structural constraints and mechanisms affecting ribavirin incorporation and nucleotide selectivity by the viral RdRp, it should be noted that viral fitness might be an important factor contributing to the viral response to lethal mutagenesis. Cell culture studies with hepatitis C virus showed that high-fitness viral quasispecies showed resistance to ribavirin and favipiravir without modifying the mutation-type bias typical of those mutagens, probably by limiting the expansion of their mutational spectra [122]. Although ribavirin has been extensively used for various decades, its interaction with the host cell machinery results in poor selectivity and toxicity, yielding undesirable side effects, including severe anemia.
6. Favipiravir as a Lethal Mutagenesis Agent
Favipiravir (6-fluoro-3-oxo-3,4-dihydropyrazine-2-carboxamide, T-705) (Figure 2) is an antiviral drug used in Japan to treat influenza. Favipiravir is a prodrug whose active form (favipiravir-ribofuranosyl-5′-triphosphate) mimics both guanosine and adenosine as substrates of viral RdRPs. It has a broad-spectrum antiviral effect, and, apart from influenza virus, favipiravir is also able to inhibit the replication of flavi-, alpha-, filo-, bunya-, arena-, noro-, and other RNA viruses, including neglected and (re)emerging viruses for which no antiviral therapy is currently available [123,124].
Enzyme kinetic analysis with influenza A virus RdRp demonstrated that favipiravir-ribofuranosyl-5′-triphosphate inhibited the incorporation of ATP and GTP in a competitive manner [125,126]. In addition, the incorporation of favipiravir into the nascent RNA strand as a purine nucleotide analog inhibited its further extension [125]. Cell culture studies with influenza A H1N1 viruses showed that favipiravir treatment produced an increased number of G→A and C→T mutations, suggesting that the favipiravir-ribofuranosyl-5′-triphosphate base pairs with either cytosine or uracil [127]. The increased mutation frequency is also dose-dependent, demonstrating that favipiravir is also a lethal mutagen. Therefore, the combination of ambiguous base-pairing with low discrimination of favipiravir-ribofuranosyl-5′-triphosphate is a key factor contributing to the mutagenic effect of favipiravir.
Evidence of the mutagenic effects of favipiravir in infected animals was reported after studying norovirus infections in a mouse model [80]. Viral RNA isolated from treated animals showed reduced infectivity, while a five- to six-fold increase in mutation frequency was obtained after the sequence analysis of individual molecular clones isolated from populations subjected to four passages in the presence of a drug concentration of 200 μM. Interestingly, these increases were higher than those obtained with ribavirin in parallel experiments (estimated at around three-fold). Favipiravir treatments led to a slight increase in the A→G and U→C transition rates in these assays. The study by Arias et al. [80] constitutes a proof of concept for lethal mutagenesis in vivo, supporting antiviral therapies based on mutagenic compounds at the clinical level. Table 2 shows several examples of viruses where favipiravir has been successfully used to extinguish the virus through lethal mutagenesis, with a concomitant excess of G→A and C→U transitions in the mutant spectrum of preextinction virus populations [91,92,95,103], although these preferences were not always predominant, as reported for favipiravir extinction experiments with foot-and-mouth disease virus [79] and West Nile virus [84].
The heterotrimeric influenza virus polymerase, containing the PA, PB1, and PB2 proteins, catalyzes viral RNA replication and transcription in the nucleus of infected cells. Studies with H1N1, H3N2, and H7N9 strains of influenza A virus showed that K229R in the catalytic PB1 subunit confers favipiravir resistance while impairing viral replication [128]. K229R reduced the mutagenic effect of favipiravir at a cost to growth, but this effect could be alleviated by P653L in the PA subunit. The combination of both mutations led to a virus that was 30-fold less susceptible to favipiravir relative to the wild-type virus and not impaired in replication kinetics [128]. Although the clinical relevance of these mutations is unclear, studies with ferrets have shown the transmissibility of the favipiravir-resistant strains in vivo [129].
Favipiravir resistance has also been mapped in the RNA polymerases of chikungunya virus (i.e., K291R) [130] and enterovirus 71 (i.e., S121N) [131], although the antiviral effects were assessed in replication assays and the mutation frequencies were not determined. Favipiravir has been approved for COVID-19 treatment in Russia, but there is no evidence of resistance so far. Based on the structural information of SARS-CoV-2 RdRp bound to favipiravir-ribofuranosyl-5′-triphosphate [132] and the variability observed in circulating viruses, a number of residues have been predicted as potentially involved in favipiravir resistance [133]. However, the phenotypic effects on resistance caused by the proposed substitutions are still being investigated.
One of the major limitations to the use of favipiravir is its relatively low bioavailability, resulting in relatively low plasma concentrations of the drug. This is apparently due to its short half-life caused by rapid renal elimination. Strategies to increase its potency are needed, particularly considering its strong antiviral effect against different viruses and in different animal models (most notably, mice, Guinea pigs, and non-human primates) (reviewed in [112]). Despite these limitations, Clinicaltrials.gov (accessed on 10 April 2022) currently includes more than one hundred clinical trials testing the efficacy of favipiravir alone or in combination with other drugs (https://clinicaltrials.gov/, accessed on 10 April 2022). Most of these trials (>85%) evaluate the efficacy of favipiravir as a drug against SARS-CoV-2, although there are trials where the compound has been tested against influenza, Ebola, or Lassa virus infections.
8. Future Directions and Challenges
The development of lethal mutagens faces several challenges. Most notably, the cell toxicity needs to be very low, particularly in relation to their carcinogenic potential. Similar to the case of other antivirals, drug resistance is a fundamental issue, not only because it could lead to drug inactivation, but also because a mutagen could spur the emergence of novel virus variants with potentially increased pathogenicity and transmissibility. Attempts to design and develop bona fide lethal mutagens (such as KP1212, for example) have not been successful and issues related to their efficacy and selectivity need to be addressed. However, lethal mutagens, such as ribavirin or favipiravir, were approved for clinical use based on their antiviral efficacy before the demonstration of their mutagenic effect. In the case of ribavirin, the drug has been used for decades to treat chronic hepatitis C, although with limited efficacy and adverse side effects. The mutagenic effect of ribavirin has been demonstrated with several viruses, including hepatitis C virus (Table 2). However, ribavirin has multiple targets, including enzymes and processes occurring in the host cell, and its side effects seem to be unrelated to mutagenicity.
Interestingly, favipiravir and molnupiravir act mainly by driving viruses to error catastrophe. Both compounds have broad-spectrum activity and show some resilience to drug resistance. Molnupiravir shows a very high genetic barrier to resistance and the selection of mutants conferring drug resistance has not been successful. However, learning from experience with HIV and other viruses [52,152], combination therapies should be the choice for a most efficient treatment and, in the case of COVID-19, further studies will be necessary to test the efficacy of molnupiravir combined with remdesivir (Veklury) or the protease inhibitor nirmatrelvir (Paxlovid). Future research efforts should also concentrate on finding or designing novel, more specific mutagenic agents acting on viral polymerases, suitable for combination therapies in order to avoid the selection of escape mutants.
Concerns about the potential genotoxicity of lethal mutagens are justified and have to be carefully addressed. However, in acute infections leading to COVID-19 or flu, the relatively short incubation time limits the extent of the medication and, therefore, the exposure to the mutagenic compounds. Reliable assessment of mutagenicity and carcinogenicity is still complex, since the results obtained in current tests (usually carried out in mice or rats) are difficult to confirm in primate models or humans. Another issue that is not always considered in genotoxicity assays is the impact of the drug exposure time. This could be short in acute viral infections, but long and probably hazardous in chronic infections. Finally, the prescription of mutagenic nucleosides should take into account long-term pharmacovigilance. A registry of patients treated with those drugs would be very helpful for a long-term follow-up of potential adverse effects, including genetic, carcinogenic, teratogenic, and embryotoxic damage.
Author Contributions
Writing—original draft preparation, I.H.H. and L.M.-A.; writing—review and editing, I.H.H., M.B.M. and L.M.-A.; funding acquisition, L.M.-A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Spanish Ministry of Science and Innovation (grant PID2019-104176RB-I00/AEI/10.13039/501100011033), and an institutional grant of Fundación Ramón Areces (Madrid, Spain) to the CBMSO.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Sanjuán, R.; Nebot, M.R.; Chirico, N.; Mansky, L.M.; Belshaw, R. Viral mutation rates. J. Virol. 2010, 84, 9733–9748. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E.; García-Crespo, C.; Perales, C. Historical perspective on the discovery of the quasispecies concept. Annu. Rev. Virol. 2021, 8, 51–72. [Google Scholar] [CrossRef] [PubMed]
- Eigen, M. Error catastrophe and antiviral strategy. Proc. Natl. Acad. Sci. USA 2002, 99, 13374–13376. [Google Scholar] [CrossRef] [PubMed]
- Eigen, M. From Strange Simplicity to Complex Familiarity: A Treatise on Matter, Information, Life and Thought; Oxford University Press: Cary, NC, USA, 2013. [Google Scholar]
- Weissmann, C.; Billeter, M.A.; Goodman, H.M.; Hindley, J.; Weber, H. Structure and function of phage RNA. Annu. Rev. Biochem. 1973, 42, 303–328. [Google Scholar] [CrossRef] [PubMed]
- Haruna, I.; Spiegelman, S. Recognition of size and sequence by an RNA replicase. Proc. Natl. Acad. Sci. USA 1965, 54, 1189–1193. [Google Scholar] [CrossRef] [PubMed]
- Haruna, I.; Spiegelman, S. Specific template requirments of RNA replicases. Proc. Natl. Acad. Sci. USA 1965, 54, 579–587. [Google Scholar] [CrossRef]
- Drake, J.W.; Charlesworth, B.; Charlesworth, D.; Crow, J.F. Rates of spontaneous mutation. Genetics 1998, 148, 1667–1686. [Google Scholar] [CrossRef]
- Drake, J.W.; Holland, J.J. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 1999, 96, 13910–13913. [Google Scholar] [CrossRef]
- Perales, C.; Domingo, E. Antiviral strategies based on lethal mutagenesis and error threshold. Curr. Top. Microbiol. Immunol. 2016, 392, 323–339. [Google Scholar] [CrossRef]
- Holland, J.J.; Domingo, E.; de la Torre, J.C.; Steinhauer, D.A. Mutation frequencies at defined single codon sites in vesicular stomatitis virus and poliovirus can be increased only slightly by chemical mutagenesis. J. Virol. 1990, 64, 3960–3962. [Google Scholar] [CrossRef]
- Anderson, J.P.; Daifuku, R.; Loeb, L.A. Viral error catastrophe by mutagenic nucleosides. Annu. Rev. Microbiol. 2004, 58, 183–205. [Google Scholar] [CrossRef] [PubMed]
- Loeb, L.A.; Essigmann, J.M.; Kazazi, F.; Zhang, J.; Rose, K.D.; Mullins, J.I. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc. Natl. Acad. Sci. USA 1999, 96, 1492–1497. [Google Scholar] [CrossRef] [PubMed]
- Sanjuán, R.; Domingo-Calap, P. Mechanisms of viral mutation. Cell. Mol. Life Sci. 2016, 73, 4433–4448. [Google Scholar] [CrossRef] [PubMed]
- Combe, M.; Sanjuán, R. Variation in RNA virus mutation rates across host cells. PLoS Pathog. 2014, 10, e1003855. [Google Scholar] [CrossRef]
- Peck, K.M.; Lauring, A.S. Complexities of viral mutation rates. J. Virol. 2018, 92, e01031-17. [Google Scholar] [CrossRef]
- Coffin, J.M. HIV population dynamics in vivo: Implications for genetic variation, pathogenesis, and therapy. Science 1995, 267, 483–489. [Google Scholar] [CrossRef]
- Menéndez-Arias, L.; Sebastián-Martín, A.; Álvarez, M. Viral reverse transcriptases. Virus Res. 2017, 234, 153–176. [Google Scholar] [CrossRef]
- Menéndez-Arias, L. Mutation rates and intrinsic fidelity of retroviral reverse transcriptases. Viruses 2009, 1, 1137–1165. [Google Scholar] [CrossRef]
- O’Neil, P.K.; Sun, G.; Yu, H.; Ron, Y.; Dougherty, J.P.; Preston, B.D. Mutational analysis of HIV-1 long terminal repeats to explore the relative contribution of reverse transcriptase and RNA polymerase II to viral mutagenesis. J. Biol. Chem. 2002, 277, 38053–38061. [Google Scholar] [CrossRef]
- Menéndez-Arias, L. Molecular basis of fidelity of DNA synthesis and nucleotide specificity of retroviral reverse transcriptases. Prog. Nucleic Acid Res. Mol. Biol. 2002, 71, 91–147. [Google Scholar] [CrossRef]
- Elder, J.H.; Lerner, D.L.; Hasselkus-Light, C.S.; Fontenot, D.J.; Hunter, E.; Luciw, P.A.; Montelaro, R.C.; Phillips, T.R. Distinct subsets of retroviruses encode dUTPase. J. Virol. 1992, 66, 1791–1794. [Google Scholar] [CrossRef] [PubMed]
- Köppe, B.; Menéndez-Arias, L.; Oroszlan, S. Expression and purification of the mouse mammary tumor virus gag-pro transframe protein p30 and characterization of its dUTPase activity. J. Virol. 1994, 68, 2313–2319. [Google Scholar] [CrossRef] [PubMed]
- Lerner, D.L.; Wagaman, P.C.; Phillips, T.R.; Prospero-García, O.; Henriksen, S.J.; Fox, H.S.; Bloom, F.E.; Elder, J.H. Increased mutation frequency of feline immunodeficiency virus lacking a functional deoxyuridine-triphosphatase. Proc. Natl. Acad. Sci. USA 1995, 92, 7480–7484. [Google Scholar] [CrossRef] [PubMed]
- Mansky, L.M. The mutation rate of human immunodeficiency virus type 1 is influenced by the vpr gene. Virology 1996, 222, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Sheehy, A.M.; Gaddis, N.C.; Choi, J.D.; Malim, M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 2002, 418, 646–650. [Google Scholar] [CrossRef]
- Mangeat, B.; Turelli, P.; Caron, G.; Friedli, M.; Perrin, L.; Trono, D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 2003, 424, 99–103. [Google Scholar] [CrossRef]
- Takaori-Kondo, A.; Shindo, K. HIV-1 Vif: A guardian of the virus that opens up a new era in the research field of restriction factors. Front Microbiol. 2013, 4, 34. [Google Scholar] [CrossRef]
- Shah, F.S.; Curr, K.A.; Hamburgh, M.E.; Parniak, M.; Mitsuya, H.; Arnez, J.G.; Prasad, V.R. Differential influence of nucleoside analog-resistance mutations K65R and L74V on the overall mutation rate and error specificity of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 2000, 275, 27037–27044. [Google Scholar] [CrossRef]
- Barrioluengo, V.; Alvarez, M.; Barbieri, D.; Menéndez-Arias, L. Thermostable HIV-1 group O reverse transcriptase variants with the same fidelity as murine leukaemia virus reverse transcriptase. Biochem. J. 2011, 436, 599–607. [Google Scholar] [CrossRef]
- Martín-Hernández, A.M.; Domingo, E.; Menéndez-Arias, L. Human immunodeficiency virus type 1 reverse transcriptase: Role of Tyr115 in deoxynucleotide binding and misinsertion fidelity of DNA synthesis. EMBO J. 1996, 15, 4434–4442. [Google Scholar] [CrossRef]
- Cases-González, C.E.; Gutiérrez-Rivas, M.; Menéndez-Arias, L. Coupling ribose selection to fidelity of DNA synthesis: The role of Tyr-115 of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 2000, 275, 19759–19767. [Google Scholar] [CrossRef] [PubMed]
- Mansky, L.M.; Le Rouzic, E.; Benichou, S.; Gajary, L.C. Influence of reverse transcriptase variants, drugs, and Vpr on human immunodeficiency virus type 1 mutant frequencies. J. Virol. 2003, 77, 2071–2080. [Google Scholar] [CrossRef] [PubMed]
- Peersen, O.B. Picornaviral polymerase structure, function, and fidelity modulation. Virus Res. 2017, 234, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Gong, P. A structural view of the RNA-dependent RNA polymerases from the Flavivirus genus. Virus Res. 2017, 234, 34–43. [Google Scholar] [CrossRef]
- Hillen, H.S. Structure and function of SARS-CoV-2 polymerase. Curr. Opin. Virol. 2021, 48, 82–90. [Google Scholar] [CrossRef]
- Robson, F.; Khan, K.S.; Le, T.K.; Paris, C.; Demirbag, S.; Barfuss, P.; Rocchi, P.; Ng, W.-L. Coronavirus RNA proofreading: Molecular basis and therapeutic targeting. Mol. Cell 2020, 79, 710–727. [Google Scholar] [CrossRef]
- Masters, P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006, 66, 193–292. [Google Scholar] [CrossRef]
- Eckerle, L.D.; Lu, X.; Sperry, S.M.; Choi, L.; Denison, M.R. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 2007, 81, 12135–12144. [Google Scholar] [CrossRef]
- Eckerle, L.D.; Becker, M.M.; Halpin, R.A.; Li, K.; Venter, E.; Lu, X.; Scherbakova, S.; Graham, R.L.; Baric, R.S.; Stockwell, T.B.; et al. Infidelity of SARS-CoV nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010, 6, e1000896. [Google Scholar] [CrossRef]
- Pathak, V.K.; Temin, H.M. 5-Azacytidine and RNA secondary structure increase the retrovirus mutation rate. J. Virol. 1992, 66, 3093–3100. [Google Scholar] [CrossRef]
- LaCasse, R.A.; Remington, K.M.; North, T.W. The mutation frequency of feline immunodeficiency virus enhanced by 3′-azido-3′-deoxythymidine. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1996, 12, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Julias, J.G.; Kim, T.; Arnold, G.; Pathak, V.K. The antiretrovirus drug 3′-azido-3′-deoxythymidine increases the retrovirus mutation rate. J. Virol. 1997, 71, 4254–4263. [Google Scholar] [CrossRef] [PubMed]
- Mansky, L.M.; Bernard, L.C. 3′-Azido-3′-deoxythymidine (AZT) and AZT-resistant reverse transcriptase can increase the in vivo mutation rate of human immunodeficiency virus type 1. J. Virol. 2000, 74, 9532–9539. [Google Scholar] [CrossRef]
- Dapp, M.J.; Clouser, C.L.; Patterson, S.; Mansky, L.M. 5-Azacytidine can induce lethal mutagenesis in human immunodeficiency virus type 1. J. Virol. 2009, 83, 11950–11958. [Google Scholar] [CrossRef] [PubMed]
- Roth, M.; McDaniel, Y.Z.; Daly, M.B.; Talledge, N.; Greggs, W.M., 3rd; Patterson, S.E.; Kim, B.; Mansky, L.M. Distinct antiretroviral mechanisms elicited by a viral mutagen. J. Mol. Biol. 2021, 433, 167111. [Google Scholar] [CrossRef] [PubMed]
- Rawson, J.M.O.; Daly, M.B.; Xie, J.; Clouser, C.L.; Landman, S.R.; Reilly, C.S.; Bonnac, L.; Kim, B.; Patterson, S.E.; Mansky, L.M. 5-Azacytidine enhances the mutagenesis of HIV-1 by reduction to 5-aza-2′-deoxycytidine. Antimicrob. Agents Chemother. 2016, 60, 2318–2325. [Google Scholar] [CrossRef]
- Sánchez-Jiménez, C.; Olivares, I.; de Ávila Lucas, A.I.; Toledano, V.; Gutiérrez-Rivas, M.; Lorenzo-Redondo, R.; Grande-Pérez, A.; Domingo, E.; López-Galíndez, C. Mutagen-mediated enhancement of HIV-1 replication in persistently infected cells. Virology 2012, 424, 147–153. [Google Scholar] [CrossRef]
- Vivet-Boudou, V.; Isel, C.; El Safadi, Y.; Smyth, R.P.; Laumond, G.; Moog, C.; Paillart, J.-C.; Marquet, R. Evaluation of anti-HIV-1 mutagenic nucleoside analogues. J. Biol. Chem. 2015, 290, 371–383. [Google Scholar] [CrossRef]
- Rawson, J.M.O.; Landman, S.R.; Reilly, C.S.; Bonnac, L.; Patterson, S.E.; Mansky, L.M. Lack of mutational hot spots during decitabine-mediated HIV-1 mutagenesis. Antimicrob. Agents Chemother. 2015, 59, 6834–6843. [Google Scholar] [CrossRef][Green Version]
- Menéndez-Arias, L. Targeting HIV: Antiretroviral therapy and development of drug resistance. Trends Pharmacol. Sci. 2002, 23, 381–388. [Google Scholar] [CrossRef]
- Menéndez-Arias, L.; Delgado, R. Update and latest advances in antiretroviral therapy. Trends Pharmacol. Sci. 2022, 43, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Tapia, N.; Fernàndez, G.; Parera, M.; Gómez-Mariano, G.; Clotet, B.; Quiñones-Mateu, M.; Domingo, E.; Martínez, M.A. Combination of a mutagenic agent with a reverse transcriptase inhibitor results in systematic inhibition of HIV-1 infection. Virology 2005, 338, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Seier, T.; Zilberberg, G.; Zeiger, D.M.; Lovett, S.T. Azidothymidine and other chain terminators are mutagenic for template-switch-generated genetic mutations. Proc. Natl. Acad. Sci. USA 2012, 109, 6171–6174. [Google Scholar] [CrossRef] [PubMed]
- Harris, K.S.; Brabant, W.; Styrchak, S.; Gall, A.; Daifuku, R. KP-1212/1461, a nucleoside designed for the treatment of HIV by viral mutagenesis. Antivir. Res. 2005, 67, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Fedeles, B.I.; Singh, V.; Peng, C.S.; Silvestre, K.J.; Simi, A.K.; Simpson, J.H.; Tokmakoff, A.; Essigmann, J.M. Tautomerism provides a molecular explanation for the mutagenic properties of the anti-HIV nucleoside 5-aza-5,6-dihydro-2′-deoxycytidine. Proc. Natl. Acad. Sci. USA 2014, 111, E3252–E3259. [Google Scholar] [CrossRef]
- Mullins, J.I.; Heath, L.; Hughes, J.P.; Kicha, J.; Styrchak, S.; Wong, K.G.; Rao, U.; Hansen, A.; Harris, K.S.; Laurent, J.-P.; et al. Mutation of HIV-1 genomes in a clinical population treated with the mutagenic nucleoside KP1461. PLoS ONE 2011, 6, e15135. [Google Scholar] [CrossRef]
- Fontecave, M.; Lepoivre, M.; Elleingand, E.; Gerez, C.; Guittet, O. Resveratrol, a remarkable inhibitor of ribonucleotide reductase. FEBS Lett. 1998, 421, 277–279. [Google Scholar] [CrossRef]
- Rawson, J.M.; Heineman, R.H.; Beach, L.B.; Martin, J.L.; Schnettler, E.K.; Dapp, M.J.; Patterson, S.E.; Mansky, L.M. 5,6-Dihydro-5-aza-2′-deoxycytidine potentiates the anti-HIV-1 activity of ribonucleotide reductase inhibitors. Bioorg. Med. Chem. 2013, 21, 7222–7228. [Google Scholar] [CrossRef]
- Guarino, E.; Salguero, I.; Kearsey, S.E. Cellular regulation of ribonucleotide reductase in eukaryotes. Semin. Cell Dev. Biol. 2014, 30, 97–103. [Google Scholar] [CrossRef]
- Musiałek, M.W.; Rybaczek, D. Hydroxyurea-The good, the bad and the ugly. Genes 2021, 12, 1096. [Google Scholar] [CrossRef]
- Clouser, C.L.; Patterson, S.E.; Mansky, L.M. Exploiting drug repositioning for discovery of a novel HIV combination therapy. J. Virol. 2010, 84, 9301–9309. [Google Scholar] [CrossRef] [PubMed]
- Rawson, J.M.O.; Roth, M.E.; Xie, J.; Daly, M.B.; Clouser, C.L.; Landman, S.R.; Reilly, C.S.; Bonnac, L.; Kim, B.; Patterson, S.E.; et al. Synergistic reduction of HIV-1 infectivity by 5-azacytidine and inhibitors of ribonucleotide reductase. Bioorg. Med. Chem. 2016, 24, 2410–2422. [Google Scholar] [CrossRef] [PubMed]
- Beach, L.B.; Rawson, J.M.; Kim, B.; Patterson, S.E.; Mansky, L.M. Novel inhibitors of human immunodeficiency virus type 2 infectivity. J. Gen. Virol. 2014, 95 Pt 12, 2778–2783. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, M.; Sebastián-Martín, A.; García-Marquina, G.; Menéndez-Arias, L. Fidelity of classwide-resistant HIV-2 reverse transcriptase and differential contribution of K65R to the accuracy of HIV-1 and HIV-2 reverse transcriptases. Sci. Rep. 2017, 7, 44834. [Google Scholar] [CrossRef]
- McDaniel, Y.Z.; Patterson, S.E.; Mansky, L.M. Distinct dual antiviral mechanism that enhances hepatitis B virus mutagenesis and reduces viral DNA synthesis. Antiviral. Res. 2019, 170, 104540. [Google Scholar] [CrossRef]
- Holland, J.; Spindler, K.; Horodyski, F.; Grabau, E.; Nichol, S.; VandePol, S. Rapid evolution of RNA genomes. Science 1982, 215, 1577–1585. [Google Scholar] [CrossRef]
- Zhou, S.; Hill, C.S.; Sarkar, S.; Tse, L.V.; Woodburn, B.M.D.; Schinazi, R.F.; Sheahan, T.P.; Baric, R.S.; Heise, M.T.; Swanstrom, R. β-D-N4-hydroxycytidine inhibits SARS-CoV-2 through lethal mutagenesis but is also mutagenic to mammalian cells. J. Infect. Dis. 2021, 224, 415–419. [Google Scholar] [CrossRef]
- Crotty, S.; Maag, D.; Arnold, J.J.; Zhong, W.; Lau, J.Y.; Hong, Z.; Andino, R.; Cameron, C.E. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 2000, 6, 1375–1379. [Google Scholar] [CrossRef]
- Harki, D.A.; Graci, J.D.; Galarraga, J.E.; Chain, W.J.; Cameron, C.E.; Peterson, B.R. Synthesis and antiviral activity of 5-substituted cytidine analogues: Identification of a potent inhibitor of viral RNA-dependent RNA polymerases. J. Med. Chem. 2006, 49, 6166–6169. [Google Scholar] [CrossRef]
- Graci, J.D.; Harki, D.A.; Korneeva, V.S.; Edathil, J.P.; Too, K.; Franco, D.; Smidansky, E.D.; Paul, A.V.; Peterson, B.R.; Brown, D.M.; et al. Lethal mutagenesis of poliovirus mediated by a mutagenic pyrimidine analogue. J. Virol. 2007, 81, 11256–11266. [Google Scholar] [CrossRef]
- Graci, J.D.; Too, K.; Smidansky, E.D.; Edathil, J.P.; Barr, E.W.; Harki, D.A.; Galarraga, J.E.; Bollinger, J.M., Jr.; Peterson, B.R.; Loakes, D.; et al. Lethal mutagenesis of picornaviruses with N-6-modified purine nucleoside analogues. Antimicrob. Agents Chemother. 2008, 52, 971–979. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Graci, J.D.; Gnädig, N.F.; Galarraga, J.E.; Castro, C.; Vignuzzi, M.; Cameron, C.E. Mutational robustness of an RNA virus influences sensitivity to lethal mutagenesis. J. Virol. 2012, 86, 2869–2873. [Google Scholar] [CrossRef] [PubMed]
- Moreno, H.; Tejero, H.; de la Torre, J.C.; Domingo, E.; Martín, V. Mutagenesis-mediated virus extinction: Virus-dependent effect of viral load on sensitivity to lethal defection. PLoS ONE 2012, 7, e32550. [Google Scholar] [CrossRef] [PubMed]
- Airaksinen, A.; Pariente, N.; Menéndez-Arias, L.; Domingo, E. Curing of foot-and-mouth disease virus from persistently infected cells by ribavirin involves enhanced mutagenesis. Virology 2003, 311, 339–349. [Google Scholar] [CrossRef]
- Pariente, N.; Airaksinen, A.; Domingo, E. Mutagenesis versus inhibition in the efficiency of extinction of foot-and-mouth disease virus. J. Virol. 2003, 77, 7131–7138. [Google Scholar] [CrossRef][Green Version]
- Sierra, M.; Airaksinen, A.; González-López, C.; Agudo, R.; Arias, A.; Domingo, E. Foot-and-mouth disease virus mutant with decreased sensitivity to ribavirin: Implications for error catastrophe. J. Virol. 2007, 81, 2012–2024. [Google Scholar] [CrossRef]
- Agudo, R.; Ferrer-Orta, C.; Arias, A.; de la Higuera, I.; Perales, C.; Pérez-Luque, R.; Verdaguer, N.; Domingo, E. A multi-step process of viral adaptation to a mutagenic nucleoside analogue by modulation of transition types leads to extinction-escape. PLoS Pathog. 2010, 6, e1001072. [Google Scholar] [CrossRef]
- de Avila, A.I.; Moreno, E.; Perales, C.; Domingo, E. Favipiravir can evoke lethal mutagenesis and extinction of foot-and-mouth disease virus. Virus Res. 2017, 233, 105–112. [Google Scholar] [CrossRef]
- Arias, A.; Thorne, L.; Goodfellow, I. Favipiravir elicits antiviral mutagenesis during virus replication in vivo. Elife 2014, 3, e03679. [Google Scholar] [CrossRef]
- Qiu, L.; Patterson, S.E.; Bonnac, L.F.; Geraghty, R.J. Nucleobases and corresponding nucleosides display potent antiviral activities against dengue virus possibly through viral lethal mutagenesis. PLoS Negl. Trop. Dis. 2018, 12, e0006421. [Google Scholar] [CrossRef]
- Bassi, M.R.; Sempere, R.N.; Meyn, P.; Polacek, C.; Arias, A. Extinction of Zika virus and Usutu virus by lethal mutagenesis reveals different patterns of sensitivity to three mutagenic drugs. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [PubMed]
- Day, C.W.; Smee, D.F.; Julander, J.G.; Yamshchikov, V.F.; Sidwell, R.W.; Morrey, J.D. Error-prone replication of West Nile virus caused by ribavirin. Antiviral. Res. 2005, 67, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Escribano-Romero, E.; Jiménez de Oya, N.; Domingo, E.; Saiz, J.C. Extinction of West Nile virus by favipiravir through lethal mutagenesis. Antimicrob. Agents Chemother. 2017, 61, e01400-17. [Google Scholar] [CrossRef] [PubMed]
- Lanford, R.E.; Chavez, D.; Guerra, B.; Lau, J.Y.; Hong, Z.; Brasky, K.M.; Beames, B. Ribavirin induces error-prone replication of GB virus B in primary tamarin hepatocytes. J. Virol. 2001, 75, 8074–8081. [Google Scholar] [CrossRef] [PubMed]
- Contreras, A.M.; Hiasa, Y.; He, W.; Terella, A.; Schmidt, E.V.; Chung, R.T. Viral RNA mutations are region specific and increased by ribavirin in a full-length hepatitis C virus replication system. J. Virol. 2002, 76, 8505–8517. [Google Scholar] [CrossRef]
- Zhou, S.; Liu, R.; Baroudy, B.M.; Malcolm, B.A.; Reyes, G.R. The effect of ribavirin and IMPDH inhibitors on hepatitis C virus subgenomic replicon RNA. Virology 2003, 310, 333–342. [Google Scholar] [CrossRef]
- Cuevas, J.M.; González-Candelas, F.; Moya, A.; Sanjuán, R. Effect of ribavirin on the mutation rate and spectrum of hepatitis C virus in vivo. J. Virol. 2009, 83, 5760–5764. [Google Scholar] [CrossRef]
- Ortega-Prieto, A.M.; Sheldon, J.; Grande-Pérez, A.; Tejero, H.; Gregori, J.; Quer, J.; Esteban, J.I.; Domingo, E.; Perales, C. Extinction of hepatitis C virus by ribavirin in hepatoma cells involves lethal mutagenesis. PLoS ONE 2013, 8, e71039. [Google Scholar] [CrossRef]
- Dietz, J.; Schelhorn, S.-E.; Fitting, D.; Mihm, U.; Susser, S.; Welker, M.-W.; Füller, C.; Däumer, M.; Teuber, G.; Wedemeyer, H.; et al. Deep sequencing reveals mutagenic effects of ribavirin during monotherapy of hepatitis C virus genotype 1-infected patients. J. Virol. 2013, 87, 6172–6181. [Google Scholar] [CrossRef]
- Gallego, I.; Soria, M.E.; Gregori, J.; de Ávila, A.I.; García-Crespo, C.; Moreno, E.; Gadea, I.; Esteban, J.; Fernández-Roblas, R.; Esteban, J.I.; et al. Synergistic lethal mutagenesis of hepatitis C virus. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef]
- de Ávila, A.I.; Gallego, I.; Soria, M.E.; Gregori, J.; Quer, J.; Esteban, J.I.; Rice, C.M.; Domingo, E.; Perales, C. Lethal mutagenesis of hepatitis C virus induced by favipiravir. PLoS ONE 2016, 11, e0164691. [Google Scholar] [CrossRef] [PubMed]
- Todt, D.; Walter, S.; Brown, R.; Steinmann, E. Mutagenic effects of ribavirin on hepatitis E virus—Viral extinction versus selection of fitness-enhancing mutations. Viruses 2016, 8, 283. [Google Scholar] [CrossRef]
- Urakova, N.; Kuznetsova, V.; Crossman, D.K.; Sokratian, A.; Guthrie, D.B.; Kolykhalov, A.A.; Lockwood, M.A.; Natchus, M.G.; Crowley, M.R.; Painter, G.R.; et al. β-D-N4-hydroxycytidine is a potent anti-alphavirus compound that induces a high level of mutations in the viral genome. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Shannon, A.; Selisko, B.; Le, N.-T.-T.; Huchting, J.; Touret, F.; Piorkowski, G.; Fattorini, V.; Ferron, F.; Decroly, E.; Meier, C.; et al. Rapid incorporation of favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat. Commun. 2020, 11, 4682. [Google Scholar] [CrossRef]
- Agostini, M.L.; Pruijssers, A.J.; Chappell, J.D.; Gribble, J.; Lu, X.; Andres, E.L.; Bluemling, G.R.; Lockwood, M.A.; Sheahan, T.P.; Sims, A.C.; et al. Small-molecule antiviral β-D-N4-hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J. Virol. 2019, 93, e01348-19. [Google Scholar] [CrossRef]
- Díaz-Martínez, L.; Brichette-Mieg, I.; Pineño-Ramos, A.; Domínguez-Huerta, G.; Grande-Pérez, A. Lethal mutagenesis of an RNA plant virus via lethal defection. Sci. Rep. 2018, 8, 1444. [Google Scholar] [CrossRef]
- Pauly, M.D.; Lauring, A.S. Effective lethal mutagenesis of influenza virus by three nucleoside analogs. J. Virol. 2015, 89, 3584–3597. [Google Scholar] [CrossRef]
- Toots, M.; Yoon, J.-J.; Cox, R.M.; Hart, M.; Sticher, Z.M.; Makhsous, N.; Plesker, R.; Barrena, A.H.; Reddy, P.G.; Mitchell, D.G.; et al. Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci. Transl. Med. 2019, 11, eaax5866. [Google Scholar] [CrossRef]
- Grande-Pérez, A.; Lázaro, E.; Lowenstein, P.; Domingo, E.; Manrubia, S.C. Suppression of viral infectivity through lethal defection. Proc. Natl. Acad. Sci. USA 2005, 102, 4448–4452. [Google Scholar] [CrossRef]
- Severson, W.E.; Schmaljohn, C.S.; Javadian, A.; Jonsson, C.B. Ribavirin causes error catastrophe during Hantaan virus replication. J. Virol. 2003, 77, 481–488. [Google Scholar] [CrossRef]
- Chung, D.-H.; Sun, Y.; Parker, W.B.; Arterburn, J.B.; Bartolucci, A.; Jonsson, C.B. Ribavirin reveals a lethal threshold of allowable mutation frequency for Hantaan virus. J. Virol. 2007, 81, 11722–11729. [Google Scholar] [CrossRef] [PubMed]
- Borrego, B.; de Ávila, A.I.; Domingo, E.; Brun, A. Lethal mutagenesis of Rift Valley fever virus induced by favipiravir. Antimicrob. Agents Chemother. 2019, 63, e00669-19. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Jarabo, C.M.; Ly, C.; Domingo, E.; de la Torre, J.C. Lethal mutagenesis of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV). Virology 2003, 308, 37–47. [Google Scholar] [CrossRef]
- Espy, N.; Nagle, E.; Pfeffer, B.; Garcia, K.; Chitty, A.J.; Wiley, M.; Sanchez-Lockhart, M.; Bavari, S.; Warren, T.; Palacios, G. T-705 induces lethal mutagenesis in Ebola and Marburg populations in macaques. Antiviral. Res. 2019, 170, 104529. [Google Scholar] [CrossRef] [PubMed]
- Vignuzzi, M.; Stone, J.K.; Andino, R. Ribavirin and lethal mutagenesis of poliovirus: Molecular mechanisms, resistance and biological implications. Virus Res. 2005, 107, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Olivencia, G.; Estébanez, M.; Membrillo, F.J.; Ybarra, M.D.C. Uso de ribavirina en virus distintos de la hepatitis C. una revisión de la evidencia. Enferm. Infecc. Microbiol. Clin. (Engl.) 2019, 37, 602–608. [Google Scholar] [CrossRef]
- Graci, J.D.; Cameron, C.E. Mechanisms of action of ribavirin against distinct viruses. Rev. Med. Virol. 2006, 16, 37–48. [Google Scholar] [CrossRef]
- Paeshuyse, J.; Dallmeier, K.; Neyts, J. Ribavirin for the treatment of chronic hepatitis C virus infection: A review of the proposed mechanisms of action. Curr. Opin. Virol. 2011, 1, 590–598. [Google Scholar] [CrossRef]
- Kentsis, A.; Topisirovic, I.; Culjkovic, B.; Shao, L.; Borden, K.L.B. Ribavirin suppresses EIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc. Natl. Acad. Sci. USA 2004, 101, 18105–18110. [Google Scholar] [CrossRef]
- Sidwell, R.W.; Huffman, J.H.; Khare Lois, G.P.; Allen, B.; Witkowski Roland, J.T.; Robins, K. Broad-spectrum antiviral activity of virazole: 1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science 1972, 177, 705–706. [Google Scholar] [CrossRef]
- Geraghty, R.J.; Aliota, M.T.; Bonnac, L.F. Broad-spectrum antiviral strategies and nucleoside analogues. Viruses 2021, 13, 667. [Google Scholar] [CrossRef]
- Arnold, J.J.; Cameron, C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol) is sufficient for template switching in vitro. J. Biol. Chem. 1999, 274, 2706–2716. [Google Scholar] [CrossRef] [PubMed]
- Gohara, D.W.; Crotty, S.; Arnold, J.J.; Yoder, J.D.; Andino, R.; Cameron, C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): Structural, biochemical, and biological analysis of conserved structural motifs a and b. J. Biol. Chem. 2000, 275, 25523–25532. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, J.K.; Kirkegaard, K. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl. Acad. Sci. USA 2003, 100, 7289–7294. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Orta, C.; Sierra, M.; Agudo, R.; de la Higuera, I.; Arias, A.; Pérez-Luque, R.; Escarmís, C.; Domingo, E.; Verdaguer, N. Structure of foot-and-mouth disease virus mutant polymerases with reduced sensitivity to ribavirin. J. Virol. 2010, 84, 6188–6199. [Google Scholar] [CrossRef] [PubMed]
- Young, K.-C.; Lindsay, K.L.; Lee, K.-J.; Liu, W.-C.; He, J.-W.; Milstein, S.L.; Lai, M.M.C. Identification of a ribavirin-resistant NS5B mutation of hepatitis C virus during ribavirin monotherapy. Hepatology 2003, 38, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Lutchman, G.; Danehower, S.; Song, B.-C.; Liang, T.J.; Hoofnagle, J.H.; Thomson, M.; Ghany, M.G. Mutation rate of the hepatitis C virus NS5B in patients undergoing treatment with ribavirin monotherapy. Gastroenterology 2007, 132, 1757–1766. [Google Scholar] [CrossRef] [PubMed]
- Mejer, N.; Galli, A.; Ramirez, S.; Fahnøe, U.; Benfield, T.; Bukh, J. Ribavirin inhibition of cell-culture infectious hepatitis C genotype 1-3 viruses is strain-dependent. Virology 2020, 540, 132–140. [Google Scholar] [CrossRef]
- Mejer, N.; Fahnøe, U.; Galli, A.; Ramirez, S.; Weiland, O.; Benfield, T.; Bukh, J. Mutations identified in the hepatitis C virus (HCV) polymerase of patients with chronic HCV treated with ribavirin cause resistance and affect viral replication fidelity. Antimicrob. Agents Chemother. 2020, 64. [Google Scholar] [CrossRef]
- Gong, P.; Peersen, O.B. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 2010, 107, 22505–22510. [Google Scholar] [CrossRef]
- Gallego, I.; Gregori, J.; Soria, M.E.; García-Crespo, C.; García-Álvarez, M.; Gómez-González, A.; Valiergue, R.; Gómez, J.; Esteban, J.I.; Quer, J.; et al. Resistance of high fitness hepatitis C virus to lethal mutagenesis. Virology 2018, 523, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Delang, L.; Abdelnabi, R.; Neyts, J. Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antiviral. Res. 2018, 153, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Jordan, P.C.; Stevens, S.K.; Deval, J. Nucleosides for the treatment of respiratory RNA virus infections. Antivir. Chem. Chemother. 2018, 26, 2040206618764483. [Google Scholar] [CrossRef] [PubMed]
- Sangawa, H.; Komeno, T.; Nishikawa, H.; Yoshida, A.; Takahashi, K.; Nomura, N.; Furuta, Y. Mechanism of action of T-705 ribosyl triphosphate against influenza virus RNA polymerase. Antimicrob. Agents Chemother. 2013, 57, 5202–5208. [Google Scholar] [CrossRef]
- Jin, Z.; Smith, L.K.; Rajwanshi, V.K.; Kim, B.; Deval, J. The ambiguous base-pairing and high substrate efficiency of T-705 (favipiravir) ribofuranosyl 5’-triphosphate towards influenza A virus polymerase. PLoS ONE 2013, 8, e68347. [Google Scholar] [CrossRef]
- Baranovich, T.; Wong, S.-S.; Armstrong, J.; Marjuki, H.; Webby, R.J.; Webster, R.G.; Govorkova, E.A. T-705 (favipiravir) induces lethal mutagenesis in influenza A H1N1 viruses in vitro. J. Virol. 2013, 87, 3741–3751. [Google Scholar] [CrossRef]
- Goldhill, D.H.; Te Velthuis, A.J.W.; Fletcher, R.A.; Langat, P.; Zambon, M.; Lackenby, A.; Barclay, W.S. The mechanism of resistance to favipiravir in influenza. Proc. Natl. Acad. Sci. USA 2018, 115, 11613–11618. [Google Scholar] [CrossRef]
- Goldhill, D.H.; Yan, A.; Frise, R.; Zhou, J.; Shelley, J.; Gallego Cortés, A.; Miah, S.; Akinbami, O.; Galiano, M.; Zambon, M.; et al. Favipiravir-resistant influenza A virus shows potential for transmission. PLoS Pathog. 2021, 17, e1008937. [Google Scholar] [CrossRef]
- Delang, L.; Segura Guerrero, N.; Tas, A.; Quérat, G.; Pastorino, B.; Froeyen, M.; Dallmeier, K.; Jochmans, D.; Herdewijn, P.; Bello, F.; et al. Mutations in the Chikungunya virus non-structural proteins cause resistance to favipiravir (T-705), a broad-spectrum antiviral. J. Antimicrob. Chemother. 2014, 69, 2770–2784. [Google Scholar] [CrossRef]
- Wang, Y.; Li, G.; Yuan, S.; Gao, Q.; Lan, K.; Altmeyer, R.; Zou, G. In vitro assessment of combinations of enterovirus inhibitors against enterovirus 71. Antimicrob. Agents Chemother. 2016, 60, 5357–5367. [Google Scholar] [CrossRef]
- Naydenova, K.; Muir, K.W.; Wu, L.-F.; Zhang, Z.; Coscia, F.; Peet, M.J.; Castro-Hartmann, P.; Qian, P.; Sader, K.; Dent, K.; et al. Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP. Proc. Natl. Acad. Sci. USA 2021, 118, e2021946118. [Google Scholar] [CrossRef] [PubMed]
- Padhi, A.K.; Dandapat, J.; Saudagar, P.; Uversky, V.N.; Tripathi, T. Interface-based design of the favipiravir-binding site in SARS-CoV-2 RNA-dependent RNA polymerase reveals mutations conferring resistance to chain termination. FEBS Lett. 2021, 595, 2366–2382. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Guo, S.; Yi, D.; Li, Q.; Ma, L.; Zhang, Y.; Wang, J.; Li, X.; Guo, F.; Lin, R.; et al. A cell-based assay to discover inhibitors of SARS-CoV-2 RNA dependent RNA polymerase. Antiviral. Res. 2021, 190, 105078. [Google Scholar] [CrossRef] [PubMed]
- Sheahan, T.P.; Sims, A.C.; Zhou, S.; Graham, R.L.; Pruijssers, A.J.; Agostini, M.L.; Leist, S.R.; Schäfer, A.; Dinnon, K.H., 3rd; Stevens, L.J.; et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020, 12, eabb5883. [Google Scholar] [CrossRef] [PubMed]
- Gordon, C.J.; Tchesnokov, E.P.; Schinazi, R.F.; Götte, M. Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template. J. Biol. Chem. 2021, 297, 100770. [Google Scholar] [CrossRef] [PubMed]
- Kabinger, F.; Stiller, C.; Schmitzová, J.; Dienemann, C.; Kokic, G.; Hillen, H.S.; Höbartner, C.; Cramer, P. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 2021, 28, 740–746. [Google Scholar] [CrossRef]
- Jena, N.R. Role of different tautomers in the base-pairing abilities of some of the vital antiviral drugs used against COVID-19. Phys. Chem. Chem. Phys. 2020, 22, 28115–28122. [Google Scholar] [CrossRef]
- Rosenke, K.; Hansen, F.; Schwarz, B.; Feldmann, F.; Haddock, E.; Rosenke, R.; Barbian, K.; Meade-White, K.; Okumura, A.; Leventhal, S.; et al. Orally delivered MK-4482 inhibits SARS-CoV-2 replication in the Syrian hamster model. Nat. Commun. 2021, 12, 2295. [Google Scholar] [CrossRef]
- Cox, R.M.; Wolf, J.D.; Plemper, R.K. Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat. Microbiol. 2021, 6, 11–18. [Google Scholar] [CrossRef]
- Jayk Bernal, A.; Gomes da Silva, M.M.; Musungaie, D.B.; Kovalchuk, E.; Gonzalez, A.; De los Reyes, V.; Martín-Quirós, A.; Caraco, Y.; Williams-Diaz, A.; Brown, M.L.; et al. Molnupiravir for oral treatment of COVID-19 in nonhospitalized patients. N. Engl. J. Med. 2022, 386, 509–520. [Google Scholar] [CrossRef]
- Fischer, W.A., 2nd; Eron, J.J., Jr.; Holman, W.; Cohen, M.S.; Fang, L.; Szewczyk, L.J.; Sheahan, T.P.; Baric, R.; Mollan, K.R.; Wolfe, C.R.; et al. A phase 2a clinical trial of molnupiravir in patients with COVID-19 shows accelerated SARS-CoV-2 RNA clearance and elimination of infectious virus. Sci. Transl. Med. 2022, 14, eabl7430. [Google Scholar] [CrossRef] [PubMed]
- Abdelnabi, R.; Foo, C.S.; De Jonghe, S.; Maes, P.; Weynand, B.; Neyts, J. Molnupiravir inhibits replication of the emerging SARS-CoV-2 variants of concern in a hamster infection model. J. Infect. Dis. 2021, 224, 749–753. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wang, Y.; Lavrijsen, M.; Lamers, M.M.; de Vries, A.C.; Rottier, R.J.; Bruno, M.J.; Peppelenbosch, M.P.; Haagmans, B.L.; Pan, Q. SARS-CoV-2 omicron variant is highly sensitive to molnupiravir, nirmatrelvir, and the combination. Cell Res. 2022, 32, 322–324. [Google Scholar] [CrossRef] [PubMed]
- Vangeel, L.; Chiu, W.; De Jonghe, S.; Maes, P.; Slechten, B.; Raymenants, J.; André, E.; Leyssen, P.; Neyts, J.; Jochmans, D. Remdesivir, molnupiravir and nirmatrelvir remain active against SARS-CoV-2 omicron and other variants of concern. Antiviral. Res. 2022, 198, 105252. [Google Scholar] [CrossRef]
- Menéndez-Arias, L. Decoding molnupiravir-induced mutagenesis in SARS-CoV-2. J. Biol. Chem. 2021, 297, 100867. [Google Scholar] [CrossRef]
- Abdelnabi, R.; Foo, C.S.; Kaptein, S.J.F.; Zhang, X.; Do, T.N.D.; Langendries, L.; Vangeel, L.; Breuer, J.; Pang, J.; Williams, R.; et al. The combined treatment of molnupiravir and favipiravir results in a potentiation of antiviral efficacy in a SARS-CoV-2 hamster infection model. EBioMedicine 2021, 72, 103595. [Google Scholar] [CrossRef]
- Schultz, D.C.; Johnson, R.M.; Ayyanathan, K.; Miller, J.; Whig, K.; Kamalia, B.; Dittmar, M.; Weston, S.; Hammond, H.L.; Dillen, C.; et al. Pyrimidine inhibitors synergize with nucleoside analogues to block SARS-CoV-2. Nature 2022, 604, 134–140. [Google Scholar] [CrossRef]
- Painter, G.R.; Natchus, M.G.; Cohen, O.; Holman, W.; Painter, W.P. Developing a direct acting, orally available antiviral agent in a pandemic: The evolution of molnupiravir as a potential treatment for COVID-19. Curr. Opin. Virol. 2021, 50, 17–22. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, T.; Xi, J.; Zhang, G.; Wang, T.; Liu, W.; You, X.; Zhang, X.; Xia, Z.; Luan, Y. PIG-A gene mutation as a genotoxicity biomarker in human population studies: An investigation in lead-exposed workers. Environ. Mol. Mutagen. 2020, 61, 611–621. [Google Scholar] [CrossRef]
- Waters, M.D.; Warren, S.; Hughes, C.; Lewis, P.; Zhang, F. Human genetic risk of treatment with antiviral nucleoside analog drugs that induce lethal mutagenesis: The special case of molnupiravir. Environ. Mol. Mutagen. 2022, 63, 37–63. [Google Scholar] [CrossRef]
- Ma, Y.; Frutos-Beltrán, E.; Kang, D.; Pannecouque, C.; De Clercq, E.; Menéndez-Arias, L.; Liu, X.; Zhan, P. Medicinal chemistry strategies for discovering antivirals effective against drug-resistant viruses. Chem. Soc. Rev. 2021, 50, 4514–4540. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).