Abstract
Sequences derived from a novel toursvirus were identified from pooled genomic short read data from U.S. populations of southern corn rootworm (SCR, Diabrotica undecimpunctata howardi Barber) and northern corn rootworm (NCR, Diabrotica barberi Smith & Lawrence). Most viral sequences were identified from the SCR genomic dataset. As proteins encoded by toursvirus sequences from SCR and NCR were almost identical, the contig sets from SCR and NCR were combined to generate 26 contigs. A total of 108,176 bp were assembled from these contigs, with 120 putative toursviral ORFs identified indicating that most of the viral genome had been recovered. These ORFs included all 40 genes that are common to members of the Ascoviridae. Two genes typically present in Ascoviridae (ATP binding cassette transport system permeases and Baculovirus repeated open reading frame), were not detected. There was evidence for transposon insertion in viral sequences at different sites in the two host species. Phylogenetic analyses based on a concatenated set of 45 translated protein sequences clustered toursviruses into a distinct clade. Based on the combined evidence, we propose taxonomic separation of toursviruses from Ascoviridae.
1. Introduction
A complex of four corn rootworm species and subspecies are native to North America; western corn rootworm (WCR), Diabrotica virgifera virgifera (LeConte), southern corn rootworm (SCR), Diabrotica undecimpunctata howardi (Barber), Mexican corn rootworm, Diabrotica virgifera zeae (Krysan & Smith), and northern corn rootworm (NCR), Diabrotica barberi (Smith & Lawrence) [1]. These species pose significant threats to maize production in the United States, and cause estimated annual losses of $2 billion in cumulative yield and control costs [2]. Efforts to manage damage to maize crops have been hampered by adaptations to crop rotation in WCR [3] and NCR [4] populations. Furthermore, WCR has developed resistance to multiple insecticide chemistries [5]. More recently resistance to transgenic maize hybrids that express Bacillus thuringiensis (Bt) pesticidal proteins has been documented in WCR [6,7,8] and NCR [9]. Consequently, a more integrated approach to corn rootworm management has been proposed [10].
The use of viruses for suppression of corn rootworm populations provides an alternative strategy within an integrated approach for crop protection. The practical utility of viral-based biological control is exemplified by use of a nudivirus for control of the Asiatic rhinoceros beetle, Oryctes rhinoceros (L.) [11,12]. While microscopic observations have suggested the presence of DNA viruses in Diabrotica spp. including in SCR [13,14], none were defined as ascovirus-like nor were any associated pathologies observed [15,16].
Ascoviridae is a family comprised of viruses with circular, double-stranded DNA genomes that fall into one of two genera, Ascovirus and Toursvirus [17]. The genomes of Toursvirus are shorter (120–143 kbp) than those of Ascovirus (157–200 kbp). Eleven full length genome sequences of viruses belonging to five ascovirus species have been reported, with 119–194 predicted ORFs of which 40 are shared among them (Table 1). Phylogenetic analyses indicate that ascoviruses evolved from an ancestral iridovirus [18].

Table 1.
Characteristics of ascovirus genomes.
Ascoviruses in the genus Ascovirus primarily infect lepidopteran larvae in the family Noctuidae and are vectored during oviposition by parasitoid wasps. Importantly for their potential use as biocontrol agents, ascoviruses cause chronic and fatal disease in their larval lepidopteran hosts [28]. An ascovirus identified from a parasitoid wasp of lepidopterans, Diadromus pulchellus, originally named Diadromus pulchellus ascovirus 4a (DpAV) [29] was renamed Diadromus pulchellus toursvirus 1a (DpTV1a) on establishment of the Toursvirus genus [17]. This sole member of the genus Toursvirus replicates extensively in lepidopteran hosts and in the primary parasitoid vector D. pulchellus to a limited extent, with the viral genome existing in the wasp as unintegrated DNA. The first non-lepidopteran ascovirus, Dasineura jujubifolia toursvirus 2a (DjTV2a) was isolated from a dipteran, and is closely related to DpAV1a. However, no obvious disease symptoms were observed in DjTV2-infected insects [26].
To assess the potential for virus-based suppression of Diabrotica spp., we examined the associated virome drawing on both genomic and transcriptomic sequence data [30]. From this we found evidence for a diverse set of corn rootworm viruses. Findings include sequences derived from three novel small RNA viruses from WCR transcripts [31,32,33] and two from SCR transcripts; and DNA sequences of two novel nudiviruses derived from the SCR and WCR genomes [34]. Here we report a novel toursvirus, with the genome sequence assembled from short sequence reads derived from both SCR and NCR DNA extracts. This is the first putative member of Ascoviridae isolated from Coleoptera.
2. Materials and Methods
2.1. SCR and NCR Sample Collection and DNA Isolation
Adult SCR (n = 50) were collected from Ames, Iowa. Methods for SCR total genomic DNA isolation followed those previously described [34]. NCR samples were collected from a grower’s field near Monmouth, IL in late July of 2012. All samples were flash frozen in liquid nitrogen and stored at −80 °C. The NCR sample was comprised of 35 males and 36 females (n = 71) that were pooled. The sample was ground to a powder in liquid nitrogen. DNA was extracted from 3.0 mg of ground NCR material using the Qiagen DNeasy Blood and Tissue Extraction kit (Qiagen, Germantown, MD, USA), with modifications as described [35].
2.2. Sequencing Library Preparation and Illumina Sequencing
Purified DNA was submitted to the Iowa State University DNA Facility (Ames, IA, USA). Genomic DNA was size selected and used to generate ~500 bp insert libraries using Illumina TruSeq v2 Library Construction Kits (Illumina, San Diego, CA, USA). Single-end 100-bp Illumina HiSeq2500 reads were generated, with SCR and NCR libraries run in separate lanes. Data were received in raw fastq format and were submitted to the National Center for Biotechnology Information (NCBI) Short Read Archive (SRR13364002 for SCR, SRR13363759 for NCR). Raw reads were trimmed to remove low-quality nucleotides as previously described [35] prior to further use in this study.
2.3. Sequence Assembly and Annotation
SCR and NCR sequence data were assembled as described previously [34,36]. Briefly, the trimmed DNA sequence reads from SCR and NCR were assembled using Trinity (v2.6.6) [37], followed by reduction in sequence redundancy using CAP3 [38]. Contigs over 450 bp were selected and used for viral sequence annotation. The selected contigs were first used as queries against a local insect DNA viral protein sequence database with the BLASTx algorithm [39] embedded in Bioedit v.7.2 [40] (https://bioedit.software.informer.com/; accessed on 15 November 2020). BLASTx results were filtered for E values ≤ 0.0001. These contigs were further used as queries against the NCBI nr database with the BLASTx algorithm. Contigs with “hits” to viral sequences were sorted based on putative virus species of the hit. DNA fragments with alignments to ascovirus and iridovirus sequences were selected for further analyses. As the putative toursvirus sequences derived from NCR and SCR encoded proteins had 100% identity, the two sets of contigs were merged. Potential coding sequences (CDS) (≥50 aa) were translated using SnapGene Viewer (SnapGene software—Insightful Science; available at snapgene.com; accessed 2-1-21). Individual protein translations were then aligned to those in the NCBI nr protein database using BLASTp as described previously [34].
2.4. Viral Sequence Analysis
Further sequence alignments and other manipulations were performed using Bioedit [40]. Details of SCR and NCR viral protein translation, protein molecular mass, location of an ORF within the genome fragments and related information were generated by SnapGene Viewer (GSL BioTech LLC, San Diego, CA, USA). The ORF and other features in SCR and NCR viral DNA fragments were visualized using maps generated by SnapGene Viewer. Methods for viral sequence mapping with the sequencing reads have been previously described [34,36]. The protein sequences encoded by 45 genes derived from a total of 26 viral genomes were selected on the basis of the BLAST analysis, for phylogenetic analysis (Table S1). Phylogenetic tree construction was performed with PyloSuite (v1.2.2) [41]. IQ-TREE methods were used to build the phylogenetic tree [42] including Edge-lined partition models for 5000 ultrafast bootstraps [43,44] and the Shimodaira–Hasegawa–like approximate likelihood-ratio test [45]. The resulting phylogeny was viewed using FigTree (v1.4.4) (http://tree.bio.ed.ac.uk/software/figtree/; accessed on 3 June 2021).
2.5. Analysis of Similairy between Toursvirus, Ascovirus and Other Invertebrate DNA Viruses
To infer phylogenetic relationships between toursviruses and related DNA viruses, the putative protein sequences of DpTV1a (119 protein sequences) and DjTV3a (141 protein sequences) were aligned to the NCBI nr database with the BLASTp algorithm. The species associated with the 10 most similar proteins were extracted and ranked from most- (1) to least- (10) similar.
3. Results
3.1. Novel Toursvirus-like Sequences Identified from SCR and NCR
Processing of short read genome sequence data from the SCR and NCR samples generated 118.5 and 9.7 million trimmed reads, respectively. Subsequent assemblies for SCR and NCR read data yielded 1,604,773 and 26,984 contigs (≥200 nt), respectively. Results from BLASTx searches against our local insect DNA viral protein sequence database showed “hits” among contigs for SCR samples with a previously identified novel nudivirus (Diabrotica undecimpunctata howardi nudivirus) [34]. Additionally, 28 unique DNA fragments, ranging from 469–19,547 bp from the SCR assembly showed significant identity with toursviruses. These putative novel toursvirus-like DNA fragments predicted a cumulative total of 115 protein coding sequences (CDS ≥ 50 aa). Similarly, BLASTx analysis of assembled NCR contigs revealed 42 unique DNA fragments ranging from 516–8299 bp, that showed “hits” to toursviral DNA accessions. Putative CDS translations for these NCR contigs predicted 117 putative viral protein coding genes.
3.2. Toursvirus Sequences Identified from SCR and NCR Derived from the Same Virus
Annotation of CDS translations from the SCR and NCR contigs with initial putative BLASTx “hits” to toursvirus-like accessions by a secondary BLASTp query against the NCBI nr database further indicated similarities to toursviruses. Specifically, toursvirus protein accessions were the top BLASTp matches for our CDS translations from putative virus-derived contigs of SCR and NCR (Table S2). Due to the 100% amino acid identity between putative CDS translations from SCR and NCR identified by interspecific BLASTp alignment, these annotations were merged across SCR and NCR contigs (results not shown). This showed that, although the toursviral-like contigs varied in size, the order and orientation of the 120 CDS were conserved between the 28 and 42 genomic fragments from SCR (Figure S1) and NCR (Figure S2). As these data indicated that the toursvirus isolates from SCR and NCR derive from the same or similar viruses, the toursviral sequence fragments from SCR and NCR were merged to generate 26 unique toursviral consensus sequence fragments (F1-F26) for further analysis (Figure 1; Table 1 and Table S2). As the virus was identified from two different Diabrotica species, this novel toursvirus is named Diabrotica toursvirus 3a (DiTV3a).
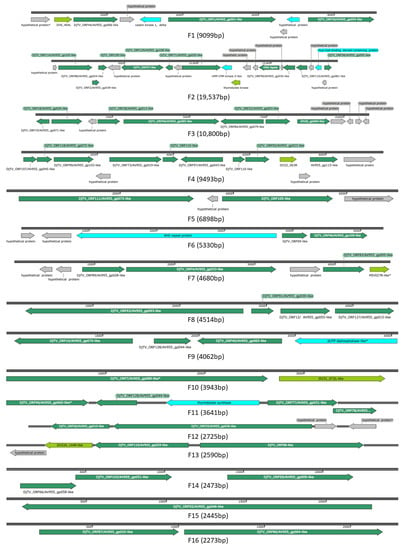

Figure 1.
Map of the 26 DiTV3a genomic fragments. Fragments derived from both SCR and NCR were combined. Arrows indicate ORFs and ORF orientation. Dark green, ORFs that hit toursvirus genes (DjTV2a/DpTV1a); light green, similar to other viral genes; blue, ORFs that hit non-viral genes; grey, unknown ORFs. *, partial sequence. The fragments identified from SCR and NCR isolates are provided in Figures S1 and S2.
3.3. Annotation of the Novel Toursvirus Sequences
The 26 toursvirus genome sequence fragments totaled 108,176 bp, nearly 10 kbp less than that of the DpTV1a genome, the shorter genome of the two toursviruses characterized to date (Table 1). This suggests that some of the genome sequence of the new toursvirus may not have been recovered from the SCR and NCR samples. The C+G content of the virus genome is 30% (Table 2), which is less than that of the two known toursviruses (Table 1). Mapping DNA sequence reads to the 26 DiTV3a genome fragments showed nucleotide coverages of ~52-fold and ~16-fold for SCR and NCR, respectively. The lower coverage from NCR may partially account for the shorter contig lengths in the assembly identified as toursvirus-like when compared to those in the SCR sample (Figures S1 and S2). One hundred and seven of the 120 putative ORFs (≥50 aa) identified in the 26 fragments of DiTV3a (Table 3 and Table S2) were full length based on comparisons to those from other toursviruses (Table S2), with 13 ORFs encoding putative partial protein sequences. Fifty nine percent of the 120 putative ORFS (71 ORFs) have similarity to proteins encoded by known toursviruses (DpTV1a and DjTV2a), while 25% (30 ORFs) lacked similarity to any protein sequences in GenBank (Table 3). The organization of ORFs in the toursviral DNA fragments is shown in Figure 1, and the corresponding ORFs found in the SCR and NCR are presented in Figures S1 and S2, respectively.

Table 2.
Sequence coverage and nucleotide content of DiTV3a genome fragments (F1 to F26).

Table 3.
Putative genes of DiTV3a per genome fragment.
3.4. Analysis of the Putative DiTV3a Genes
Annotations assigned to the accession of top BLASTp “hit” were used to attribute potential function of putative DiTV3a ORFs (Table S2). The ORFs with similarity to known toursviruses are indicated in Figure 1. About 70% of the putative DiTV3a ORFs returned significant hits (E values < 0.001) to proteins in the NCBI nr database, with 51 and 20 of the top “hits” from DjAV2a (and DpTV1a, respectively (Table S2). Best matches were also predicted to viral genes from iridoviruses (5 hits), a mimivirus (Acanthamoeba polyphaga mimivirus), and a poxvirus (Fowlpox virus). Seven BLASTp “hits” were from non-viral proteins (bacterial, insect, a protozoan, and a nematode protein), and the remaining 30 putative DiTV3a proteins returned no hits (listed as “hypothetical proteins” in Table S2).
The sequences of these dsDNA viruses were similar to toursviral, ascoviral and iridoviral genes [25,26]. These genes were grouped into 7 functional categories based on their putative biological functions derived from gene annotations from related viruses (Table 3). Fifty nine percent of the 120 putative ORFS (71 ORFs) have similarity to proteins encoded by known toursviruses (DpTV1a and DjTV2a), while 25% (30 ORFs) lacked similarity to any protein sequences in GenBank (Table 3). From the 120 putative ORFs of DiTV3a, 40 are shared among known members of the family Ascoviridae (indicated in bold in Table 3 and Table S2). The presence of all 40 of these shared genes along with the number of putative ORFs identified relative to those of other ascoviruses (119 in DpTV1a and 141 in DjTV2a; Table 1), suggests that the vast majority of DiTV3a genes were recovered from the assembly of DNA sequencing reads. Notably, two types of genes commonly found in toursviruses and ascoviruses, ATP binding cassette (ABC) transport system permeases and Baculovirus repeated open reading frame (bro) [46] were not present in the recovered DiTV3a genomes.
3.5. Putative DiTV3a Genes Associated with Retrotransposon Elements
Some DiTV3a ORFs were associated with putative retrotransposon elements. Genomic fragments comprised of DiTV3a genes and retrotransposon-related genes were observed in both SCR and NCR samples (Figure 2), but integrations varied between contigs derived from SCR and NCR. For instance, DiTV3a_F14_ORF2 and ORF3 were assembled with a DNA fragment of 7219 bp, wherein an ORF encoding an endonuclease-reverse transcriptase was predicted in the SCR contig. The other putative ORFs in DiTV3a_F14 “hit” three uncharacterized protein coding loci in the WCR genome assembly, LOC114344791, LOC114341432 and LOC114348326 (Figure 2). Similarly, an NCR-derived 8295 bp contig was assembled containing DiTV3a_F4_ORF2 and ORF3 genes together with genes encoding a retrovirus-related activating signal cointegrator 1 complex subunit (Pol polyprotein family) from transposon 412-like protein and a GATA zinc finger domain-containing protein 14-like protein (Figure 2).

Figure 2.
DiTV3a fragments fused to retrotransposon elements. Green, DiTV3a ORFs; blue, host genes with retrotransposon-related genes labeled (endonuclease-reverse transcriptase, activating signal cointegrator 1 complex subunit, GATA zinc finger domain-containing protein 14-like); gray, hypothetical proteins.
3.6. Toursvirus Proteins Are More Similar to Those of Iridoviruses Than Ascoviruses
Our BLASTp searches resulted in identification of 90 putative DiTV3a gene translations that matched known viral proteins (Table 3). Seventy-one DiTV3a genes were similar to known toursviral genes. There were also 19 ORFs that hit the genes of other viruses (mainly iridoviruses) or proteins of non-viral origin. Surprisingly, none of the top hits were from ascoviral genes. Assessment of similarity among the 119 DpTV1a proteins, 141 DjTV2a proteins, 120 DiTV3a proteins and those of related DNA viruses showed that the majority (~70%) of the BLASTp top hits to DpTV1a proteins were to proteins of DjTV2a, and vice versa (Figure 3). The majority of the top 1 and top 2 hits from queried DiTV3a proteins were to either DpTV1a or DjTV2a, and the top 3 to top 10 hit viral species were iridoviruses (Table S3). Less than 10% of the top 1 to top 10 hits for DiTV3a were to ascoviruses. While entomopoxviruses were frequent among the “hits”, almost all of these were to bro genes in DpTV1a or DjTV2a, which were not identified in DiTV3a. Only one entomopoxvirus hit was observed in the top 10 hit species of DiTV3a. A few DpTV1a proteins hit ichnovirus proteins. Hits to marseilleviruses were frequently observed, and protein sequences of Pithovirus, a group of giant DNA viruses, were frequently hit by toursviruses in the BLASTp search. Interestingly, at least 25% of the top 2–10 hits of the toursviral proteins were from bacteria and other non-viral organisms, demonstrating the diversity in the composition of toursviral genomes. Taken together, our analysis of similarity among all putative proteins encoded by the three toursviruses showed greatest DiTV3 protein similarity to iridovirus proteins, with relatively little similarity to Ascovirus proteins.
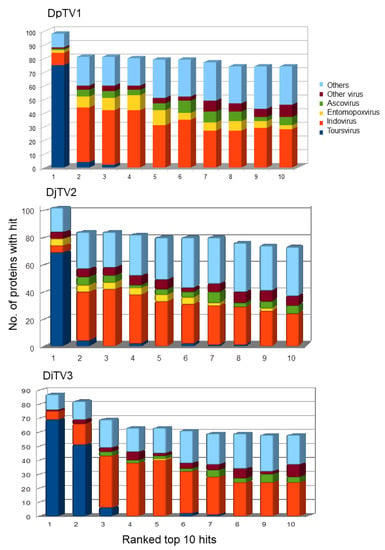
Figure 3.
Similarity of toursvirus proteins to proteins from other virus species. The virus group associated with the top 10 non-redundant hits from 1 to 10 from BLASTp analysis against the NCBI nr database are shown for putative ORFs of DpTV1, DjTV2 and DiTV3a. The numbers of proteins with similarity to the different virus groups or to other proteins are shown.
3.7. Phylogenetic Analyses Indicate That Toursviruses form a Distinct Clade
To assess the evolutionary relationships among ascoviruses, toursviruses, and iridoviruses, a phylogenetic tree was generated based on the concatenated protein sequences in silico translated from 45 genes encoded by 26 viruses. The sequences used were derived from four genera of Iridoviridae, specifically Lymphocystivirus, Ranavirus (Alphairidovirinae), Chloriridovirus, and Iridovirus (Betairidovirinae). Sequences derived from Meglocytivirus (Alphairidovirnae) and Decapodiridovirus (Betairidovirinae) were not included in the phylogenetic analysis due to low sequence similarity to those of toursviruses. The tree predicts clustering of toursviruses into a distinct clade (Figure 4). The tree supports the premise that toursviruses are phylogenetically closer to members of Iridovirus than to those of Ascovirus (Figure 4).

Figure 4.
Phylogenetic tree based on the concatenated sequences of 45 proteins encoded by 26 toursviruses, iridoviruses and ascoviruses. Protein sequences were selected based on BLASTp results (E value ≤ 0.001) and downloaded from the NCBI protein database. Methods for phylogenetic tree construction are as described in materials and methods. Bootstrap values (percent) are indicated. Virus groups are shown at right. Corresponding protein accession numbers for each virus are provided in Table S1.
4. Discussion
We previously reported two novel DNA viruses (nudiviruses) identified from genome sequence data of SCR and WCR [34]. Here we identify the third DNA virus sequence from Diabrotica spp., which is from a novel toursvirus in SCR and NCR. An estimated 90% of the DiTV3a genomic DNA was recovered following assembly of short read sequencing data from the host genomes. However, relatively short DNA fragments were assembled with many gaps, and further work will be required to generate the complete genome sequence. DiTV3a sequences isolated from SCR and NCR were almost identical, indicating these two isolates may be derived from closely related lineages of the same virus. One hundred and twenty putative ORFs were predicted from the 26 DiTV3a genomic fragments. Sequences of DiTV3a were found in SCR and NCR, but not from the previously analyzed WCR genomic sequences [34]. DiTV3 is the first toursvirus identified in Coleoptera.
4.1. Genome Assembly
The DiTV3a sequences were assembled into twenty-six fragments, with the longest less than 20 kbp. It is not clear why longer fragments of DiTV3a were not assembled. Technical parameters that could account for this include 100 bp single end reads being less tractable for assembly than paired end reads, repetitive regions within the genome hindering assembly, and assembly parameters. Some genes commonly found in other ascoviruses (e.g., ABC transport system permease and bro) were not identified from the assembled DiTV3a fragments, suggesting either that some sequence regions of the DiTV3 genome were not assembled, or that these genes were absent from this virus. One possible explanation is that viral sequence coverage was insufficient for recovery of sequence reads in all regions of the DiTV3 genome. We previously discovered a near full length nudivirus genome sequence (DuhNV) from the same SCR DNA sample. The average base coverage of DuhNV was less than 19-fold [34], ~3-fold less than that of DiTV3a, suggesting that the number of reads derived from DiTV3a should be sufficient to generate longer fragments. Therefore, additional factors likely account for the poor DiTV3a genome assembly. In contrast to DiTV3a sequences which were associated with retrotransposon elements in both SCR and NCR, no retroviral elements were associated with the DuhNV sequences. It is conceivable that the DiTV3a genome sequence has been disrupted by retrotransposon activity, potentially resulting in our inability to identify genes such as ABC transport system permease and bro using the BLAST parameters employed.
Both ABC transport system permease and bro are multi-gene families, members of which are commonly found in toursviruses and ascoviruses. Based on sequences deposited in NCBI, 2 to 6 ABC transport system permease genes are found in toursviruses and ascoviruses, except for the Spodoptera frugiperda ascovirus 1a, SfAV 1a. Three to 25 bro genes are found in toursviruses and ascoviruses. DpTV1 and DjTV2 each encode two ABC-type transport system permease genes, and 9 and 5 copies of bro, respectively. It is unknown why these two genes were not identified from the recovered DiTV3a genome sequence. To address whether partial sequences of these genes are present in the short contigs, we translated the SCR and NCR contigs with the six-frame translation option. The resulting protein sequences were aligned by BLASTp using protein sequences encoded by DpTV1 and DjTV2 as reference. No sequences encoding potential ABC-type transport system permease or Bro proteins were detected. Therefore, it is unlikely that these sequences were missed due to poor DiTV3a genome assembly. It is possible that DiTV3a either does not encode ABC-type transport system permease related proteins or Bro proteins, or that these genes have been disrupted by transposon activity. It is notable in this context that Spodoptera frugiperda ascovirus 1a lacks the permease gene [26].
4.2. Sequence Integration
Differential association of transposon-related sequences with DiTV3a sequences recovered from SCR and NCR indicates structural variation between the two isolates, even though the virus-derived sequences are highly similar. As the three putative WCR genes in Figure 2 are not on the same scaffold in the WCR genome assembly, these host genes could have been acquired and integrated into the viral genome. Such integration of host sequences into viral genomes has been described previously in large DNA viruses of insects (Baculoviridae) [47,48].
The integration of viral genomes into host genomes is not uncommon for DNA viruses, including a member of Iridoviridae, Frog virus 3 [49]. However, the underlying mechanisms of integration events are poorly understood [49,50]. It is unclear whether the genome sequences of DiTV3a were integrated into the genomes of SCR or NCR in the samples used for this work. However, previous analysis of SCR transcriptomes [33] did not reveal toursvirus RNAs, which would be expected for intact DiTV3a ORFs if the virus was integrated into the host genome.
4.3. Evolutionary Relationships
A close relationship between Ascoviridae and Iridoviridae was previously observed by phylogenetic analysis of their DNA polymerases [51]. DpTV1 was also shown at the evolutionary intersection of iridoviruses, ascoviruses, and ichnoviruses [25]. Currently, Ascoviridae, Iridoviridae, and Marseilleviridae are assigned to the Order Pimascovirales (in Realm: Varidanviria, Kingdom: Bamfordvirae, and Phylum: Nucleocytoviricoda) (https://talk.ictvonline.org/taxonomy; accessed on 27 January 2021). At present, DpTV1a is the only toursvirus recognized by the International Committee on Taxonomy of Viruses (ICTV). The recently reported DjTV2a [26] and DiTV3a presented in this manuscript are two new members of the Toursvirus genus.
The comprehensive phylogenetic analysis based on 45 viral protein sequences showed greater similarity of toursvirus proteins to those of iridoviruses, than to those of ascoviruses (Figure 4). This result is consistent with the high numbers of iridovirus hits on BLASTp analysis of toursvirus proteins, with relatively few from ascovirus proteins (Figure 3). Indeed, phylogenetic analyses for each of 28 core genes for the first identified toursvirus (DpTV1) showed that 17 core genes supported the hypothesis that DpTV1 is more closely related to iridoviruses and belongs to a clade distinct from Ascovirus [25].
Based on this analysis, Toursvirus and Ascovirus should not be taxonomically grouped together in Ascoviridae. The extensive sequence divergence of the Ascovirus genus from the Toursvirus genus since their evolution from a common ancestor forms the basis for this recommendation. We propose that the genus Toursvirus be separated from Ascoviridae, and a new family Toursviridae be created within the order Pimascovirales. Pimascovirales would then contain four families: Ascoviridae, Iridoviridae, Marseilleviridaes, and Toursviridae.
Supplementary Materials
The following Supplementary Materials are available online at https://www.mdpi.com/article/10.3390/v14020397/s1. Table S1. Accession numbers for viral proteins used for construction of the phylogenetic tree. Table S2. Summary of DiTV3a sequence analysis. Table S3. Species associated with the 10 most similar proteins to putative proteins encoded by DpTV1, DjPV2 and DiTV3a as depicted in Figure 3. DiTV3a sequence, text file containing sequences of DiTV3 identified in this study. Figure S1. DiTV3a genomic fragments (28) isolated from the southern corn rootworm. Figure S2. DiTV3a genomic fragments (42) isolated from the northern corn rootworm. File S1. DiTV3a sequence.
Author Contributions
Conceptualization, B.C.B. and S.L.; methodology, S.L. and B.S.C.; formal analysis, S.L.; investigation, S.L., T.W.S., B.S.C. and B.C.B.; resources, T.W.S. and B.S.C.; writing—original draft preparation, S.L. and B.C.B.; writing—review and editing, T.W.S., B.S.C. and B.C.B.; visualization, S.L. and B.C.B.; project administration, B.C.B.; funding acquisition, B.C.B. and T.W.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Monsanto Company’s Corn Rootworm Knowledge Research Program, USDA National Institute of Food and Agriculture, Hatch project IOW05450, USDA Agricultural Research Service project 5030-22000-019-00D, by State of Iowa funds, and by the University of Florida Institute of Food and Agricultural Sciences. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply a recommendation or endorsement by USDA. USDA ARS is an equal opportunity provider and employer.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Raw sequence data used for this analysis are available as indicated in Section 2.2 of the manuscript. DiTV3a sequence is provided as a text file in Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Krysan, J.L.; Smith, R.F. Systematics of the virgifera species group of Diabrotica (Coleoptera: Chrysomelidae: Galerucinae). Entomography 1987, 5, 375–484. [Google Scholar]
- Wechsler, S.; Smith, D. Has resistance taken root in U.S. corn fields? Demand for insect control. Am. J. Agric. Econ. 2018, 100, 1136–1150. [Google Scholar] [CrossRef]
- Sammons, A.E.; Edwards, C.R.; Bledsoe, L.W.; Boeve, P.J.; Stuart, J.J. Behavioral and feeding assays reveal a western corn rootworm (Coleoptera: Chyromelidae) variant that is attracted to soybean. Environ. Entomol. 1997, 26, 1336–1342. [Google Scholar] [CrossRef]
- Levine, E.; Oloumi-Sadeghi, H.; Fisher, J.R. Discovery of multiyear diapause in Illinois and South Dakota northern corn rootworm (Coleoptera: Chrysomelidae) eggs and incidence of the prolonged diapause trait in Illinois. J. Econ. Entomol. 1992, 85, 262–267. [Google Scholar] [CrossRef]
- Souza, D.; Peterson, J.A.; Wright, R.J.; Meinke, L.J. Field efficacy of soil insecticides on pyrethroid-resistant western corn rootworms (Diabrotica virgifera virgifera LeConte). Pest Manag. Sci. 2020, 76, 827–833. [Google Scholar] [CrossRef] [PubMed]
- Gassmann, A.J.; Petzold-Maxwell, J.L.; Keweshan, R.S.; Dunbar, M.W. Field-evolved resistance to Bt maize by western corn rootworm. PLoS ONE 2011, 6, e22629. [Google Scholar] [CrossRef]
- Gassmann, A.J. Resistance to Bt maize by western corn rootworm: Insights from the laboratory and the field. Curr. Opin. Insect Sci. 2016, 15, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Gassmann, A.J.; Shrestha, R.B.; Kropf, A.L.; St Clair, C.R.; Brenizer, B.D. Field-evolved resistance by western corn rootworm to Cry34/35Ab1 and other Bacillus thuringiensis traits in transgenic maize. Pest Manag. Sci. 2020, 76, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Calles-Torrez, V.; Knodel, J.J.; Boetel, M.A.; French, B.W.; Fuller, B.W.; Ransom, J.K. Field-evolved resistance of northern and western corn rootworm (Coleoptera: Chrysomelidae) populations to corn hybrids expressing single and pyramided Cry3Bb1 and Cry34/35Ab1 Bt proteins in North Dakota. J. Econ. Entomol. 2019, 112, 1875–1886. [Google Scholar] [CrossRef] [PubMed]
- Gassmann, A.J. Resistance to Bt Maize by western corn rootworm: Effects of pest biology, the pest-crop interaction and the agricultural landscape on resistance. Insects 2021, 12, 136. [Google Scholar] [CrossRef]
- Bedford, G.O. Biology and management of palm dynastid beetles: Recent advances. Annu. Rev. Entomol. 2013, 58, 353–372. [Google Scholar] [CrossRef]
- Jackson, T.A.; Crawford, A.M.; Glare, T.R. Oryctes virus—Time for a new look at a useful biocontrol agent. J. Invertebr. Pathol. 2005, 89, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Kitajima, E.W. Nonoccluded baculovirus-like and filamentous virus-like particles in the spotted cucumber beetle, Diabrotica-undecimpunctata (Coleoptera, Chrysomelid). J. Invertebr. Pathol. 1984, 43, 234–241. [Google Scholar] [CrossRef]
- Degrugillier, M.E.; Degrugillier, S.S.; Jackson, J.J. Nonoccluded, cytoplasmic virus-particles and Rickettsia-like organisms in testes and spermathecae of Diabrotica-virgifera. J. Invertebr. Pathol. 1991, 57, 50–58. [Google Scholar] [CrossRef]
- Kim, K.S. Cytopathology of spotted cucumber beetle hemocytes containing virus-like particles. J. Invertebr. Pathol. 1980, 36, 292–301. [Google Scholar] [CrossRef]
- Kim, K.S.; Scott, H.A.; Fulton, J.P. Viruses discovered in beetle pests. Ark. Farm Res. 1981, 30, 19. [Google Scholar]
- Asgari, S.; Bideshi, D.K.; Bigot, Y.; Federici, B.A.; Cheng, X.W.; ICTV Report, C. ICTV Virus Taxonomy Profile: Ascoviridae. J. Gen. Virol. 2017, 98, 4–5. [Google Scholar] [CrossRef]
- Piegu, B.; Asgari, S.; Bideshi, D.; Federici, B.A.; Bigot, Y. Evolutionary relationships of iridoviruses and divergence of ascoviruses from invertebrate iridoviruses in the superfamily Megavirales. Mol. Phylogenet. Evol. 2015, 84, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Bideshi, D.K.; Demattei, M.V.; Rouleux-Bonnin, F.; Stasiak, K.; Tan, Y.; Bigot, S.; Bigot, Y.; Federici, B.A. Genomic sequence of Spodoptera frugiperda Ascovirus 1a, an enveloped, double-stranded DNA insect virus that manipulates apoptosis for viral reproduction. J. Virol. 2006, 80, 11791–11805. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xue, J.; Seaborn, C.P.; Arif, B.M.; Cheng, X.W. Sequence and organization of the Trichoplusia ni ascovirus 2c (Ascoviridae) genome. Virology 2006, 354, 167–177. [Google Scholar] [CrossRef]
- Asgari, S.; Davis, J.; Wood, D.; Wilson, P.; McGrath, A. Sequence and organization of the Heliothis virescens ascovirus genome. J. Gen. Virol. 2007, 88, 1120–1132. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.H.; Wang, Y.S.; Wang, X.; Garretson, T.A.; Dai, L.Y.; Zhang, C.X.; Cheng, X.W. Genomic sequence of Heliothis virescens ascovirus 3g isolated from Spodoptera exigua. J. Virol. 2012, 86, 12467–12468. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, G.H.; Garretson, T.A.; Cheng, X.H.; Holztrager, M.S.; Li, S.J.; Wang, X.; Cheng, X.W. Phylogenetic position and replication kinetics of Heliothis virescens ascovirus 3h (HvAV-3h) isolated from Spodoptera exigua. PLoS ONE 2012, 7, e40225. [Google Scholar] [CrossRef]
- Chen, Z.S.; Hou, D.H.; Cheng, X.W.; Wang, X.; Huang, G.H. Genomic analysis of a novel isolate Heliothis virescens ascovirus 3i (HvAV-3i) and identification of ascoviral repeat ORFs (aros). Arch. Virol. 2018, 163, 2849–2853. [Google Scholar] [CrossRef]
- Bigot, Y.; Renault, S.; Nicolas, J.; Moundras, C.; Demattei, M.V.; Samain, S.; Bideshi, D.K.; Federici, B.A. Symbiotic virus at the evolutionary intersection of three types of large DNA viruses; iridoviruses, ascoviruses, and ichnoviruses. PLoS ONE 2009, 4, e6397. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, M.; Xiao, H.; Huang, G.H.; Deng, F.; Hu, Z. Genome analysis of Dasineura jujubifolia toursvirus 2, a novel Ascovirus. Virol. Sin. 2020, 35, 134–142. [Google Scholar] [CrossRef]
- Bigot, Y.; Rabouille, A.; Periquet, G. Particle and genomic characteristics of a new member of the Ascoviridae: Diadromus pulchellus ascovirus. J. Gen. Virol. 1997, 78, 5. [Google Scholar] [CrossRef]
- Federici, B.A.; Bideshi, D.K.; Tan, Y.; Spears, T.; Bigot, Y. Ascoviruses: Superb manipulators of apoptosis for viral replication and transmission. Curr. Top. Microbiol. Immunol. 2009, 328, 171–196. [Google Scholar]
- Bigot, Y.; Rabouille, A.; Periquet, G. Biological and molecular features of the relationships between Diadromus pulchellus ascovirus, a parasitoid hymenopteran wasp (Diadromus pulchellus) and its lepidopteran host, Acrolepiopsis assectella. J. Gen. Virol. 1997, 78, 5. [Google Scholar] [CrossRef]
- Bonning, B.C. The insect virome: Opportunities and challenges. Curr. Issues Mol. Biol. 2020, 34, 1–12. [Google Scholar] [CrossRef]
- Liu, S.; Chen, Y.; Sappington, T.W.; Bonning, B.C. Genome sequence of a novel positive-sense, single-stranded RNA virus isolated from western corn rootworm, Diabrotica virgifera virgifera LeConte. Genome Announc. 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chen, Y.; Sappington, T.W.; Bonning, B.C. Genome sequence of Diabrotica virgifera virgifera virus2, a novel small RNA virus of the western corn rootworm, Diabrotica virgifera virgifera LeConte. Genome Announc. 2017, 5. [Google Scholar] [CrossRef]
- Liu, S.; Chen, Y.; Sappington, T.W.; Bonning, B.C. Genome sequence of the first coleopteran iflavirus isolated from western corn rootworm, Diabrotica virgifera virgifera LeConte. Genome Announc. 2017, 5. [Google Scholar] [CrossRef]
- Liu, S.; Sappington, T.W.; Coates, B.S.; Bonning, B.C. Nudivirus sequences identified from the southern and western corn rootworms (Coleoptera: Chrysomelidae). Viruses 2021, 13, 269. [Google Scholar] [CrossRef] [PubMed]
- Coates, B.S. Assembly and annotation of full mitochondrial genomes for the corn rootworm species, Diabrotica virgifera virgifera and Diabrotica barberi (Insecta: Coleoptera: Chrysomelidae), using Next Generation Sequence data. Gene 2014, 542, 190–197. [Google Scholar] [CrossRef]
- Liu, S.; Coates, B.S.; Bonning, B.C. Endogenous viral elements integrated into the genome of the soybean aphid, Aphis glycines. Insect Biochem. Mol. Biol. 2020, 123, 103405. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.M.; Eyers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Zhang, D.; Gao, F.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Bideshi, D.K.; Renault, S.; Stasiak, K.; Federici, B.A.; Bigot, Y. Phylogenetic analysis and possible function of bro-like genes, a multigene family widespread among large double-stranded DNA viruses of invertebrates and bacteria. J. Gen. Virol. 2003, 84, 2531–2544. [Google Scholar] [CrossRef] [PubMed]
- Rappoport, N.; Linial, M. Viral proteins acquired from a host converge to simplified domain architectures. PLoS Comput. Biol. 2012, 8, e1002364. [Google Scholar] [CrossRef]
- Bezier, A.; Annaheim, M.; Herbiniere, J.; Wetterwald, C.; Gyapay, G.; Bernard-Samain, S.; Wincker, P.; Roditi, I.; Heller, M.; Belghazi, M.; et al. Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science 2009, 323, 926–930. [Google Scholar] [CrossRef]
- Desfarges, S.; Ciuffi, A. Viral integration and consequences on host gene expression. In Viruses: Essential Agents of Life; Witzany, G., Ed.; Springer: Dordrecht, The Netherlands, 2012. [Google Scholar]
- Desfarges, S.; Ciuffi, A. Retroviral integration site selection. Viruses 2010, 2, 111–130. [Google Scholar] [CrossRef]
- Stasiak, K.; Demattei, M.V.; Federici, B.A.; Bigot, Y. Phylogenetic position of the Diadromus pulchellus ascovirus DNA polymerase among viruses with large double-stranded DNA genomes. J. Gen. Virol. 2000, 81, 3059–3072. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).