Abstract
Retroviruses have a very complex and tightly controlled life cycle which has been studied intensely for decades. After a virus enters the cell, it reverse-transcribes its genome, which is then integrated into the host genome, and subsequently all structural and regulatory proteins are transcribed and translated. The proteins, along with the viral genome, assemble into a new virion, which buds off the host cell and matures into a newly infectious virion. If any one of these steps are faulty, the virus cannot produce infectious viral progeny. Recent advances in structural and molecular techniques have made it possible to better understand this class of viruses, including details about how they regulate and coordinate the different steps of the virus life cycle. In this review we summarize the molecular analysis of the assembly and maturation steps of the life cycle by providing an overview on structural and biochemical studies to understand these processes. We also outline the differences between various retrovirus families with regards to these processes.
1. Introduction
Retroviruses are positive RNA viruses; many of those cause diseases of major importance to humans and domestic animals. Their name comes from their ability to reverse transcribe their genome and insert it into the host cell. The retrovirus family can be divided into several subfamilies based on their nucleic acid sequences and their life cycles. Alpha (e.g., Rous Sarcoma Virus—RSV), Beta (e.g., Human Endogenous Retrovirus K—HERV-K), Gamma (e.g., Murine Leukaemia Virus—MLV), Delta (e.g., Human T-cell Leukaemia Virus—HTLV), Epsilon-retroviruses (e.g., Walleye Dermal Sarcoma Virus—WDSV), Lentiviruses (e.g., Human Immunodeficiency Virus—HIV), and Spumaretroviruses (e.g., Human Foamy Virus—HFV). All subfamilies encode the open reading frames gag, pol and env, but differences in their genomic organization and additional regulatory proteins make each family unique.
3. Immature Gag Lattice Structure
Apart from Spumaviruses, all retroviruses share a common morphology of the immature particle, with a dense, roughly spherical protein shell which stretches across approximately two-thirds of the internal surface of the virion (Figure 1a) [15]. The immature virus is formed exclusively by Gag hexamers that form a lattice through interactions of the CA domains as well as between SP1 or equivalent domains. An 8 nm spacing between hexamers is also a conserved feature of immature retroviral lattices [16,17,18,19]. However, a lattice consisting of only hexamers is mostly flat. To allow for curvature, it must incorporate holes or defects to fit onto a spherical surface [20]. The CA protein itself is divided into two subdomains: N-terminal (NTD) and C-terminal domains (CTD) separated by a flexible linker region. Despite low sequence conservation, the CA tertiary structure is highly conserved among retroviruses. The NTD generally consists of seven alpha helices, whereas the CTD is made up of four alpha-helices. These two domains are then joined by a flexible linker and oriented in such a way that they can form interactions to stabilize the hexamer [21]. The hexamer is the main building-block of the immature virus lattice. Intra-hexamer interactions are performed by a six-helix bundle (6HB) at the end of CTD and a spacer peptide downstream of capsid. Inter-hexamer interactions are performed by dimerization and trimerization of CA to form a lattice.
As the immature lattice is quite heterogenous in size and morphology, resolving its complete structure is difficult. Some domains of HIV-1 Gag were solved individually in earlier studies by X-ray crystallography and NMR [22,23,24]. Recently, the structure of HIV-1 CA CTD and the spacer peptide that recapitulates the immature lattice has been solved by X-ray crystallography [25]. For the structures of native Gag immature lattice assemblies, cryoET and subtomogram averaging (STA) provides a very powerful method [19,26,27,28,29], allowing for in situ structures to reach a resolution of ~3 Å [26]. These studies have shown that the hexameric assembly unit of immature CA lattice takes the shape of a wine glass, with the cup walls formed by the NTD, the cup bottom by the CTD, the stem by the 6HB formed by the last residues of CA and SP1 and the base by the amorphous NC/vRNA layer (Figure 1b) [30]. The neighbouring Gag hexamers are inter-connected to form a hexagonal lattice through trimeric interactions in the corners (blue circle). Three hexamers join, causing dimeric interactions (red circle) on the sides of the hexamer (cartoon overview Figure 1c). Helix 2 is responsible for the trimeric interactions between hexamers and the top of helix 4 in the NTD interacts with helices 5 and 6 in neighbouring hexamers (Figure 1c). In the CTD, the major homology region (MHR) is used for interactions within hexamers and the helix 9 contributes to the dimer interface between hexamers [18,31]. In contrast to the mature core, the Gag domains are organized in a linear fashion, meaning there are no contacts between NTDs and CTDs in immature lattices. Most viruses solved to date, such as HIV, RSV, EIAV and MLV, have the dimerization domain located in the CA CTD helix 9, while for HTLV-1 it seems to be in the NTD domain [17,18,19,32,33]. This unique feature may explain why HTLV-1 is the only retrovirus to display an immature lattice with straight facets [34].
A very important region within the immature lattice is the 6HB at the end of CA CTD and the beginning of SP1 (or a corresponding spacer peptide). This is the region where maturation inhibitors, such as Bevirimat (BVM) and PF46396 (PF96), bind [35,36]. In addition, a small molecule from the host cell, inositol hexaphosphate (IP6), was recently discovered to bind to the highly positive charged region at the top of 6HB and acts as a conserved assembly co-factor for different retroviruses, critical for the immature lattice assembly and viral infectivity [27,37]. The MHR, the loop between helix 9 and 10, and the beta-turn at the end of helix 11 further stabilize the 6HB. The 6HB could also be found in different retroviruses, such as MLV, EIAV and RSV [32,38]. NMR and molecular dynamic simulations have shown that this helix bundle is under a dynamic helix-coil equilibrium and that this is an important feature for optimal maturation and infectivity [39]. A second site compensatory mutation in the space peptide (T8I in HIV-1) in the BVM resistant mutant was shown to alter the dynamic property of 6HB and enable the formation of a fully extended stable 6HB connecting to the NC/vRNA layer [27]. Although all retroviral immature lattices solved to date share a hexameric immature lattice arrangement, the arrangement of CA NTD and its protein contacts differ between them, while the CA CTD interactions are highly conserved [16,17,18,19].
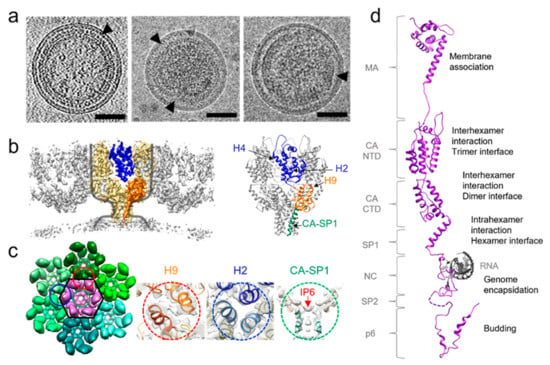
Figure 1.
Retroviral immature lattice organization. (a) Tomographic slice of an HIV-1 (left), and cryoEM images of RSV (center) [15] and HTLV-1 (right) [15] virus-like particles, black arrowheads point to Gag lattice. Scale bars, 50 nm. (b) HIV-1 immature Gag structure, shown as cryoET subtomoram averaging map (left) and atomic model of the hexamer (right). One hexamer is highlighted in yellow in cryoET STA map within which one monomer is colored in blue (NTD) and orange (CTD and SP1). Grey outline shows the wine glass profile of the Gag hexamer [27]. (c) HIV-1 immature lattice organization in top view. Details of the dimeric, trimeric and hexameric interfaces are shown in gold, blue and green dashes, respectively. (d) HIV-1 Gag polyprotein domain organization. Original PDB accession codes: 1UPH (MA) [5], 7ASH (CA and SP1) [27], 1F6U (NC and SL2 RNA loop) [40], 2C55 (p6) [41].
5. Mature Core Lattice Structure
Upon maturation, a subset of the CA protein assembles to form the mature core (Figure 2a). The mature core is a metastable structure responsible for protecting the viral genome from detection by cytoplasmic host innate immunity sensors, as well as to provide a compartment for reverse transcription initiation. The capsid core must also be able to uncoat to release the reverse transcribed genome for integration in the host genome. The capsid further acts as a docking platform for host proteins to facilitate the transport of virus core through the cytoplasm into the nucleus [64,65,66]. Many host restriction factors also recognize the mature capsid surface lattice and evoke inhibitory actions [67,68,69,70].
The mature retrovirus capsid core follows the fullerene geometry model. It is predominantly made up of hexamers (Figure 2b) and incorporates a few pentamers. These pentamers are necessary, as they allow for high curvature in the lattice. For lentiviruses, this core has a conical shape. For alpha, beta, gamma, and deltaretroviruses, this core is polyhedral or cylindrical in shape. The overall core shape is determined by the placement of the pentamers in the core. In a conical core, this is done by a partition of seven pentamers in the broad cone end, and five at the narrow end. A 6-6 partition leads to a cylindrical core, while a randomly distributed partition creates a polyhedron. There are also viruses which incorporate more than 12 pentamers, such as MLV. MLV has multi-layered or multiple cores as it incorporates almost all cleaved CA molecules, and the core is less tightly packed. The various MLV core morphologies require up to 24 pentamers but they are remarkably similar in structure to their neighbouring hexamers [17]. HTLV-1 mature cores are very often incomplete, which may explain the remarkably low infectivity of this retrovirus [71].
In HIV-1, the mature lattice has a thinner appearance (4 nm thick) in the cross section in comparison with the immature one (14 nm thick from CA NTD to NC/vRNA). The hexamer–hexamer spacing is larger than in the immature lattice, 10 nm instead of 8 nm. The key differences between immature and mature hexamers are the orientations of NTD and CTD and the intermolecular NTD–CTD contacts that stabilize the mature hexamer (Figure 2b,c). For the mature lattice assembly, the CTD is the main stabilizing domain between hexamers, specifically the 2-fold interface formed by helix 9 and the 3-fold interface formed by helices 10 and 11 (Figure 2d) [72,73]. The dimer interface appears stable and less flexible than the trimer interface, as suggested by the lower B-factor (Figure 2d). An important interface is formed by conserved positively charged residues at the center of the NTD hexamer. This position is reported to bind IP6 in an analogous manner as the positively charged ring found in the CTD at the top of 6HB in the immature lattice [37,74]. In HIV-1, above the mature hexamer center lies a beta-hairpin described to adopt different conformations depending on pH [75]. These conformations translate into different accessibilities to the central CA hexamer channel and have been hypothesized to be an important regulator of capsid core permeability to nucleotides necessary for reverse transcription of the viral genome [75].
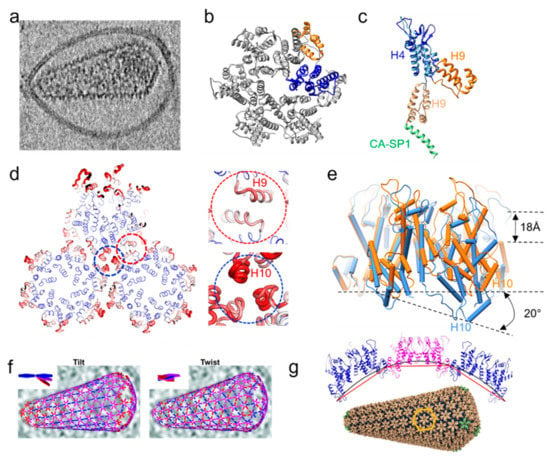
Figure 2.
HIV-1 mature capsid organization. (a) Tomographic slice of a mature HIV-1 particle with a capsid cone [76]. (b) Atomic model of an HIV-1 mature CA hexamer derived from tubular assemblies (PDB 6SKK [77]). One CA monomer is coloured in blue (NTD) and orange (CTD). (c) Comparison of immature CA-SP1 (lighter shade) and mature CA structure, aligned at the NTD (blue). (d) HIV-1 mature lattice organization in top view. The width and color of the sausage are directly proportional to the B-factor, from blue (−30) to red (−100). Details of the dimeric and trimeric interfaces are shown in red and blue dashes, respectively. (e) Intrinsic curvature of the mature CA hexamer by superposition of a planar CA hexamer (orange, PDB 4XFX [78]) and a highly curved hexamer (blue, PDB 6SKK [77]). (f) Tilt (left) and twist (right) angles between hexamers within a single core. Insets show a schematic illustration of tilt and twist angles [76]. The tilt/twist angle is indicated by the colour of the connecting lines between hexamer positions, from blue (less tilt along the long axis of the core) to red (more tilt along the circumference). (g) All-atom atomic model of theHIV-1 conical core (bottom) and a cross-section of three hexamers along the curved direction (top). The black line illustrates continuous curvature of the lattice given by both intra and inter-hexamer curvature, while the red line illustrates discrete curvature given by inter-hexamer curvature alone [77]. The lattice unit is marked with orange hexagon; pentamers shown in green [72].
The pentameric structure recently determined from native capsid by cryoET and subtomogram averaging is different from the previous crystal structure of the cross-linked pentamer [72,76]. Its NTDs are rotated by approximately 19 degrees compared to its hexameric counterpart. In doing so, it excludes helix 3 from the interface and forms a 10-helix bundle with its neighbouring hexamer instead. Moreover, the binding site for host factors and small molecules such as PF74 is more open at the pentamer NTD-CTD interface in comparison to the hexameric one [76]. The five arginine residues (R18) at the center of the pentamer in HIV CA (or the corresponding residue K17 in RSV CA) have been proposed to regulate the transition between hexamer and pentamer by balancing its electrostatic destabilization with stabilizing the lattice [38,79].
The CA domain of different viruses has very low sequence conservation; nonetheless, there is strong structural conservation between the CA proteins of different retroviruses [21]. This structurally conserved protein has a remarkable ability to accommodate different lattice curvatures. Recent cryoEM studies have shown that this ability is enabled by two features: the different tilt and twist orientations between CA hexamers [76], and more prominently by intrinsic curvatures of the hexamer itself (Figure 2e), enabled by the flexible linker between CA NTD and CTD [77] (Figure 2f,g). This curvature was also observed in the mature RSV capsid-like particles, as RSV has a highly variable core. This flexibility can adapt to the different requirements of the hexamers at various curvatures and a more random distribution of pentamers [38,80]. The curvature of the RSV capsid was thought to be derived from variable inter-capsomer interfaces; however, more recent structure data suggest the capsomers are intrinsically curved [38,77].
6. Future Perspective
Even with all the recent structural findings on the replication cycle of retroviruses, there are still many poorly understood processes. It is still unclear where Gag initially dimerizes, how it is transported to the assembly sites and how exactly maturation is triggered. The pathway of how the virus transitions from the immature to the mature capsid is also not completely understood. In addition, HIV-1 has served as the model organism to understand these concepts. Our understanding of other types of retroviruses has been catching up in recent years in terms of their mature and immature capsid architecture. Nonetheless, there is still much to be done to understand exactly how each virus regulates its assembly and maturation. Novel technical advances, such as correlative light and electron microscopy (CLEM) [81,82,83,84,85], cryoFIB lamella [86,87] cellular tomography [83,86], integrative imaging [83,88,89], and computational advances on subtomogram averaging [26,90] will play an important role filling these knowledge gaps in the near future.
Funding
This research was funded by the Wellcome Trust Investigator Award (206422/Z/17/Z), the BBSRC grant (BB/S003339/1), the ERC AdG grant (101021133) and the NIH grants (AI150481, AI129862). The APC was funded by the Wellcome Trust Investigator Award (206422/Z/17/Z).
Acknowledgments
The author thanks Anka Lucic for critical reading of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sundquist, W.I.; Kräusslich, H.-G. HIV-1 Assembly, Budding, and Maturation. Cold Spring Harb. Perspect. Med. 2012, 2, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Flügel, R.M.; Pfrepper, K.I. Proteolytic processing of foamy virus Gag and Pol proteins. Curr. Top. Microbiol. Immunol. 2003, 277, 63–88. [Google Scholar]
- Garcia-Montojo, M.; Doucet-O’Hare, T.; Henderson, L.; Nath, A. Human endogenous retrovirus-K (HML-2): A comprehensive review. Crit. Rev. Microbiol. 2018, 44, 715–738. [Google Scholar] [CrossRef]
- Bussienne, C.; Marquet, R.; Paillart, J.C.; Bernacchi, S. Post-Translational Modifications of Retroviral HIV-1 Gag Precursors: An Overview of Their Biological Role. Int. J. Mol. Sci. 2021, 22, 2871. [Google Scholar] [CrossRef]
- Tang, C.; Loeliger, E.; Luncsford, P.; Kinde, I.; Beckett, D.; Summers, M.F. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. USA 2004, 101, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Inlora, J.; Collins, D.R.; Trubin, M.E.; Chung, J.Y.J.; Ono, A. Membrane Binding and Subcellular Localization of Retroviral Gag Proteins Are Differentially Regulated by MA Interactions with Phosphatidylinositol-(4,5)-Bisphosphate and RNA. mBio 2014, 5. [Google Scholar] [CrossRef]
- Martin, J.L.; Mendonça, L.M.; Angert, I.; Mueller, J.D.; Zhang, W.; Mansky, L.M. Disparate Contributions of Human Retrovirus Capsid Subdomains to Gag-Gag Oligomerization, Virus Morphology, and Particle Biogenesis. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Woodward, C.L.; Cheng, S.N.; Jensen, G.J. Electron Cryotomography Studies of Maturing HIV-1 Particles Reveal the Assembly Pathway of the Viral Core. J. Virol. 2015, 89, 1267–1277. [Google Scholar] [CrossRef]
- Mailler, E.; Bernacchi, S.; Marquet, R.; Paillart, J.C.; Vivet-Boudou, V.; Smyth, R.P. The life-cycle of the HIV-1 gag–RNA complex. Viruses 2016, 8, 248. [Google Scholar] [CrossRef]
- Chen, J.; Liu, Y.; Wu, B.; Nikolaitchik, O.A.; Mohan, P.R.; Chen, J.; Pathak, V.K.; Hu, W.-S. Visualizing the translation and packaging of HIV-1 full-length RNA. Proc. Natl. Acad. Sci. USA 2020, 117, 6145–6155. [Google Scholar] [CrossRef]
- Chen, J.; Rahman, S.A.; Nikolaitchik, O.A.; Grunwald, D.; Sardo, L.; Burdick, R.C.; Plisov, S.; Liang, E.; Tai, S.; Pathak, V.K.; et al. HIV-1 RNA genome dimerizes on the plasma membrane in the presence of Gag protein. Proc. Natl. Acad. Sci. USA 2016, 113, E201–E208. [Google Scholar] [CrossRef]
- Kutluay, S.B.; Zang, T.; Blanco-Melo, D.; Powell, C.; Jannain, D.; Errando, M.; Bieniasz, P.D. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell 2014, 159, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Todd, G.C.; Duchon, A.; Inlora, J.; Olson, E.D.; Musier-Forsyth, K.; Ono, A. Inhibition of HIV-1 Gag-membrane interactions by specific RNAs. RNA 2017, 23, 395–405. [Google Scholar] [CrossRef]
- Carlson, L.A.; de Marco, A.; Oberwinkler, H.; Habermann, A.; Briggs, J.A.G.; Kräusslich, H.G.; Grünewald, K. Cryo electron tomography of native HIV-1 budding sites. PLoS Pathog. 2010, 6, e1001173. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.L.; Cao, S.; Maldonado, J.O.; Zhang, W.; Mansky, L.M. Distinct Particle Morphologies Revealed through Comparative Parallel Analyses of Retrovirus-Like Particles. J. Virol. 2016, 90, 8074–8084. [Google Scholar] [CrossRef]
- Bharat, T.A.M.; Castillo Menendez, L.R.; Hagen, W.J.H.; Lux, V.; Igonet, S.; Schorb, M.; Schur, F.K.M.; Kräusslich, H.-G.; Briggs, J.A.G. Cryo-electron microscopy of tubular arrays of HIV-1 Gag resolves structures essential for immature virus assembly. Proc. Natl. Acad. Sci. USA 2014, 111, 8233–8238. [Google Scholar] [CrossRef] [PubMed]
- Qu, K.; Glass, B.; Dolezal, M.; Schur, F.K.M.; Murciano, B.; Rein, A.; Rumlová, M.; Ruml, T.; Kräusslich, H.G.; Briggs, J.A.G. Structure and architecture of immature and mature murine leukemia virus capsids. Proc. Natl. Acad. Sci. USA 2018, 115, E11751–E11760. [Google Scholar] [CrossRef]
- Schur, F.K.M.; Hagen, W.J.H.; Rumlová, M.; Ruml, T.; Müller, B.; Kraüsslich, H.G.; Briggs, J.A.G. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 Å resolution. Nature 2015, 517, 505–508. [Google Scholar] [CrossRef]
- Schur, F.K.M.; Obr, M.; Hagen, W.J.H.; Wan, W.; Jakobi, A.J.; Kirkpatrick, J.M.; Sachse, C.; Kräusslich, H.-G.; Briggs, J.A.G. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science 2016, 353, 506–508. [Google Scholar] [CrossRef]
- Briggs, J.A.G.; Kräusslich, H.G. The molecular architecture of HIV. J. Mol. Biol. 2011, 410, 491–500. [Google Scholar] [CrossRef]
- Bharat, T.A.M.; Davey, N.E.; Ulbrich, P.; Riches, J.D.; De Marco, A.; Rumlova, M.; Sachse, C.; Ruml, T.; Briggs, J.A.G. Structure of the immature retroviral capsid at 8Å resolution by cryo-electron microscopy. Nature 2012, 487, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Kelly, B.N.; Howard, B.R.; Wang, H.; Robinson, H.; Sundquist, W.I.; Hill, C.P. Implications for viral capsid assembly from crystal structures of HIV-1 Gag(1-278) and CA(N)(133-278). Biochemistry 2006, 45, 11257–11266. [Google Scholar] [CrossRef] [PubMed]
- Morellet, N.; Druillennec, S.; Lenoir, C.; Bouaziz, S.; Roques, B.P. Helical structure determined by NMR of the HIV-1 (345-392)Gag sequence, surrounding p2: Implications for particle assembly and RNA packaging. Protein Sci. 2005, 14, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Ndassa, Y.; Summers, M.F. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 2002, 9, 537–543. [Google Scholar] [CrossRef]
- Wagner, J.M.; Zadrozny, K.K.; Chrustowicz, J.; Purdy, M.D.; Yeager, M.; Ganser-Pornillos, B.K.; Pornillos, O. Crystal structure of an HIV assembly and maturation switch. Elife 2016, 5, e17063. [Google Scholar] [CrossRef] [PubMed]
- Himes, B.A.; Zhang, P. emClarity: Software for high-resolution cryo-electron tomography and subtomogram averaging. Nat. Methods 2018, 15, 955–961. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, L.; Sun, D.; Ning, J.; Liu, J.; Kotecha, A.; Olek, M.; Frosio, T.; Fu, X.; Himes, B.A.; Kleinpeter, A.B.; et al. CryoET structures of immature HIV Gag reveal six-helix bundle. Commun. Biol. 2021, 4, 481. [Google Scholar] [CrossRef]
- Turoňová, B.; Schur, F.K.M.; Wan, W.; Briggs, J.A.G. Efficient 3D-CTF correction for cryo-electron tomography using NovaCTF improves subtomogram averaging resolution to 3.4Å. J. Struct. Biol. 2017, 199, 187–195. [Google Scholar] [CrossRef]
- Wright, E.R.; Schooler, J.B.; Ding, H.J.; Kieffer, C.; Fillmore, C.; Sundquist, W.I.; Jensen, G.J. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 2007, 26, 2218–2226. [Google Scholar] [CrossRef]
- Campbell, S.; Fisher, R.J.; Towler, E.M.; Fox, S.; Issaq, H.J.; Wolfe, T.; Phillips, L.R.; Rein, A. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc. Natl. Acad. Sci. USA 2001, 98, 10875–10879. [Google Scholar] [CrossRef] [PubMed]
- Novikova, M.; Zhang, Y.; Freed, E.O.; Peng, K. Multiple Roles of HIV-1 Capsid during the Virus Replication Cycle. Virol. Sin. 2019, 34, 119–134. [Google Scholar] [CrossRef]
- Dick, R.A.; Xu, C.; Morado, D.R.; Kravchuk, V.; Ricana, C.L.; Lyddon, T.D.; Broad, A.M.; Feathers, J.R.; Johnson, M.C.; Vogt, V.M.; et al. Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly. PLoS Pathog. 2020, 16, e1008277. [Google Scholar] [CrossRef]
- Martin, J.L.; Mendonça, L.M.; Marusinec, R.; Zuczek, J.; Angert, I.; Blower, R.J.; Mueller, J.D.; Perilla, J.R.; Zhang, W.; Mansky, L.M. Critical Role of the Human T-Cell Leukemia Virus Type 1 Capsid N-Terminal Domain for Gag-Gag Interactions and Virus Particle Assembly. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, J.O.; Cao, S.; Zhang, W.; Mansky, L.M. Distinct Morphology of Human T-Cell Leukemia Virus Type 1-Like Particles. Viruses 2016, 8, 132. [Google Scholar] [CrossRef] [PubMed]
- Keller, P.W.; Adamson, C.S.; Heymann, J.B.; Freed, E.O.; Steven, A.C. HIV-1 maturation inhibitor bevirimat stabilizes the immature Gag lattice. J. Virol. 2011, 85, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Spearman, P. HIV-1 Gag as an Antiviral Target: Development of Assembly and Maturation Inhibitors. Curr. Top. Med. Chem. 2016, 16, 1154–1166. [Google Scholar] [CrossRef] [PubMed]
- Dick, R.A.; Zadrozny, K.K.; Xu, C.; Schur, F.K.M.; Lyddon, T.D.; Ricana, C.L.; Wagner, J.M.; Perilla, J.R.; Ganser-Pornillos, B.K.; Johnson, M.C.; et al. Inositol phosphates are assembly co-factors for HIV-1. Nature 2018, 560, 509–512. [Google Scholar] [CrossRef] [PubMed]
- Obr, M.; Ricana, C.L.; Nikulin, N.; Feathers, J.-P.R.; Klanschnig, M.; Thader, A.; Johnson, M.C.; Vogt, V.M.; Schur, F.K.M.; Dick, R.A. Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer. Nat. Commun. 2021, 12, 3226. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Quinn, C.M.; Perilla, J.R.; Zhang, H.; Shirra, R., Jr.; Hou, G.; Byeon, I.-J.; Suiter, C.L.; Ablan, S.; Urano, E.; et al. Quenching protein dynamics interferes with HIV capsid maturation. Nat. Commun. 2017, 8, 1779. [Google Scholar] [CrossRef]
- Amarasinghe, G.K.; De Guzman, R.N.; Turner, R.B.; Chancellor, K.J.; Wu, Z.R.; Summers, M.F. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2000, 301, 491–511. [Google Scholar] [CrossRef]
- Fossen, T.; Wray, V.; Bruns, K.; Rachmat, J.; Henklein, P.; Tessmer, U.; Maczurek, A.; Klinger, P.; Schubert, U. Solution structure of the human immunodeficiency virus type 1 p6 protein. J. Biol. Chem. 2005, 280, 42515–42527. [Google Scholar] [CrossRef]
- Mattei, S.; Anders, M.; Konvalinka, J.; Kräusslich, H.-G.; Briggs, J.A.G.; Müller, B.; Krausslich, H.-G.; Briggs, J.A.G.; Muller, B. Induced Maturation of Human Immunodeficiency Virus. J. Virol. 2014, 88, 13722–13731. [Google Scholar] [CrossRef]
- Mattei, S.; Tan, A.; Glass, B.; Müller, B.; Kräusslich, H.-G.; Briggs, J.A.G. High-resolution structures of HIV-1 Gag cleavage mutants determine structural switch for virus maturation. Proc. Natl. Acad. Sci. USA 2018, 115, E9401–E9410. [Google Scholar] [CrossRef] [PubMed]
- Hoyte, A.C.; Jamin, A.V.; Koneru, P.C.; Kobe, M.J.; Larue, R.C.; Fuchs, J.R.; Engelman, A.N.; Kvaratskhelia, M. Resistance to pyridine-based inhibitor KF116 reveals an unexpected role of integrase in HIV-1 Gag-Pol polyprotein proteolytic processing. J. Biol. Chem. 2017, 292, 19814–19825. [Google Scholar] [CrossRef]
- Sadiq, S.K.; Mirambeau, G.; Meyerhans, A. Equilibrium Model of Drug-Modulated GagPol-Embedded HIV-1 Reverse Transcriptase Dimerization to Enhance Premature Protease Activation. AIDS Res. Hum. Retrovir. 2018, 34, 804–807. [Google Scholar] [CrossRef] [PubMed]
- Sudo, S.; Haraguchi, H.; Hirai, Y.; Gatanaga, H.; Sakuragi, J.-i.; Momose, F.; Morikawa, Y. Efavirenz enhances HIV-1 gag processing at the plasma membrane through Gag-Pol dimerization. J. Virol. 2013, 87, 3348–3360. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Louis, J.M.; Aniana, A.; Suh, J.Y.; Clore, G.M. Visualizing transient events in amino-terminal autoprocessing of HIV-1 protease. Nature 2008, 455, 693–696. [Google Scholar] [CrossRef]
- Pettit, S.C.; Everitt, L.E.; Choudhury, S.; Dunn, B.M.; Kaplan, A.H. Initial cleavage of the human immunodeficiency virus type 1 GagPol precursor by its activated protease occurs by an intramolecular mechanism. J. Virol. 2004, 78, 8477–8485. [Google Scholar] [CrossRef]
- Ludwig, C.; Leiherer, A.; Wagner, R. Importance of Protease Cleavage Sites within and Flanking Human Immunodeficiency Virus Type 1 Transframe Protein p6* for Spatiotemporal Regulation of Protease Activation. J. Virol. 2008, 82, 4573–4584. [Google Scholar] [CrossRef] [PubMed]
- Bardy, M.; Gay, B.; Pébernard, S.; Chazal, N.; Courcoul, M.; Vigne, R.; Decroly, E.; Boulanger, P. Interaction of human immunodeficiency virus type 1 Vif with Gag and Gag-Pol precursors: Co-encapsidation and interference with viral protease-mediated Gag processing. J. Gen. Virol. 2001, 82, 2719–2733. [Google Scholar] [CrossRef][Green Version]
- Mendonça, L.M.; Poeys, S.C.; Abreu, C.M.; Tanuri, A.; Costa, L.J. HIV-1 Nef inhibits Protease activity and its absence alters protein content of mature viral particles. PLoS ONE 2014, 9, e95352. [Google Scholar] [CrossRef]
- Louis, J.M.; Clore, G.M.; Gronenborn, A.M. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nat. Struct. Biol. 1999, 6, 868–875. [Google Scholar]
- Qu, K.; Ke, Z.; Zila, V.; Anders-Össwein, M.; Glass, B.; Mücksch, F.; Müller, R.; Schultz, C.; Müller, B.; Kräusslich, H.G.; et al. Maturation of the matrix and viral membrane of HIV-1. Science 2021, 373, 700–704. [Google Scholar] [CrossRef] [PubMed]
- Kessl, J.; Kutluay, S.; Townsend, D.; Rebensburg, S.; Slaughter, A.; Larue, R.; Shkriabai, N.; Bakouche, N.; Fuchs, J.; Bieniasz, P.; et al. HIV-1 Integrase Binds the Viral RNA Genome and Is Essential during Virion Morphogenesis. Cell 2016, 166, 1257–1268. [Google Scholar] [CrossRef] [PubMed]
- Jurado, K.; Wang, H.; Slaughter, A.; Feng, L.; Kessl, J.; Koh, Y.; Wang, W.; Ballandras-Colas, A.; Patel, P.; Fuchs, J.; et al. Allosteric integrase inhibitor potency is determined through the inhibition of HIV-1 particle maturation. Proc. Natl. Acad. Sci. USA 2013, 110, 8690–8695. [Google Scholar] [CrossRef] [PubMed]
- Chojnacki, J.; Staudt, T.; Glass, B.; Bingen, P.; Engelhardt, J.; Anders, M.; Schneider, J.; Müller, B.; Hell, S.W.; Kräusslich, H.G. Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science 2012, 338, 524–528. [Google Scholar] [CrossRef]
- Frank, G.A.; Narayan, K.; Bess, J.W.; Del Prete, G.Q.; Wu, X.; Moran, A.; Hartnell, L.M.; Earl, L.A.; Lifson, J.D.; Subramaniam, S. Maturation of the HIV-1 core by a non-diffusional phase transition. Nat. Commun. 2015, 6, 5854. [Google Scholar] [CrossRef]
- Meng, X.; Zhao, G.; Yufenyuy, E.; Ke, D.; Ning, J.; DeLucia, M.; Ahn, J.; Gronenborn, A.M.; Aiken, C.; Zhang, P. Protease Cleavage Leads to Formation of Mature Trimer Interface in HIV-1 Capsid. PLoS Pathog. 2012, 8, 1002886. [Google Scholar] [CrossRef]
- Ning, J.; Erdemci-Tandogan, G.; Yufenyuy, E.L.; Wagner, J.; Himes, B.A.; Zhao, G.; Aiken, C.; Zandi, R.; Zhang, P. In vitro protease cleavage and computer simulations reveal the HIV-1 capsid maturation pathway. Nat. Commun. 2016, 7, 13689. [Google Scholar] [CrossRef]
- Benjamin, J.; Ganser-Pornillos, B.K.; Tivol, W.F.; Sundquist, W.I.; Jensen, G.J. Three-dimensional Structure of HIV-1 Virus-like Particles by Electron Cryotomography. J. Mol. Biol. 2005, 346, 577–588. [Google Scholar] [CrossRef]
- Briggs, J.A.G.; Grünewald, K.; Glass, B.; Förster, F.; Kräusslich, H.G.; Fuller, S.D. The Mechanism of HIV-1 Core Assembly: Insights from Three-Dimensional Reconstructions of Authentic Virions. Structure 2006, 14, 15–20. [Google Scholar] [CrossRef]
- Levandovsky, A.; Zandi, R. Nonequilibirum assembly, retroviruses, and conical structures. Phys. Rev. Lett. 2009, 102, 198102. [Google Scholar] [CrossRef]
- Yu, Z.; Dobro, M.J.; Woodward, C.L.; Levandovsky, A.; Danielson, C.M.; Sandrin, V.; Shi, J.; Aiken, C.; Zandi, R.; Hope, T.J.; et al. Unclosed HIV-1 Capsids Suggest a Curled Sheet Model of Assembly. J. Mol. Biol. 2013, 425, 112–123. [Google Scholar] [CrossRef]
- Liu, C.; Perilla, J.R.; Ning, J.; Lu, M.; Hou, G.; Ramalho, R.; Himes, B.A.; Zhao, G.; Bedwell, G.J.; Byeon, I.-J.; et al. Cyclophilin A stabilizes the HIV-1 capsid through a novel non-canonical binding site. Nat. Commun. 2016, 7, 10714. [Google Scholar] [CrossRef]
- Ning, J.; Zhong, Z.; Fischer, D.K.; Harris, G.; Watkins, S.C.; Ambrose, Z.; Zhang, P. Truncated CPSF6 Forms Higher-Order Complexes That Bind and Disrupt HIV-1 Capsid. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Zhong, Z.; Ning, J.; Boggs, E.A.; Jang, S.; Wallace, C.; Telmer, C.; Bruchez, M.P.; Ahn, J.; Engelman, A.N.; Zhang, P.; et al. Cytoplasmic CPSF6 Regulates HIV-1 Capsid Trafficking and Infection in a Cyclophilin A-Dependent Manner. mBio 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, F.J.D.; He, S.; Perilla, J.R.; Jang, S.; Schulten, K.; Engelman, A.N.; Scheres, S.H.W.; Zhang, P. CryoEM structure of MxB reveals a novel oligomerization interface critical for HIV restriction. Sci. Adv. 2017, 3. [Google Scholar] [CrossRef] [PubMed]
- Wilbourne, M.; Zhang, P. Visualizing HIV-1 Capsid and Its Interactions with Antivirals and Host Factors. Viruses 2021, 13, 246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Mendonça, L.M.; Mansky, L.M. The Retrovirus Capsid Core. Subcell. Biochem. 2018, 88, 169–187. [Google Scholar]
- Zhao, G.; Ke, D.; Vu, T.; Ahn, J.; Shah, V.B.; Yang, R.; Aiken, C.; Charlton, L.M.; Gronenborn, A.M.; Zhang, P. Rhesus TRIM5α Disrupts the HIV-1 Capsid at the Inter-Hexamer Interfaces. PLoS Pathog. 2011, 7, e1002009. [Google Scholar] [CrossRef]
- Meissner, M.E.; Mendonça, L.M.; Zhang, W.; Mansky, L.M. Polymorphic Nature of Human T-Cell Leukemia Virus Type 1 Particle Cores as Revealed through Characterization of a Chronically Infected Cell Line. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Ni, T.; Zhu, Y.; Yang, Z.; Xu, C.; Chaban, Y.; Nesterova, T.; Ning, J.; Böcking, T.; Parker, M.; Monnie, C.; et al. Structure of Native HIV-1 Cores and Their Interactions with IP6 and CypA. Sci. Adv. 2021, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Perilla, J.R.; Yufenyuy, E.L.; Meng, X.; Chen, B.; Ning, J.; Ahn, J.; Gronenborn, A.M.; Schulten, K.; Aiken, C.; et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 2013, 497, 643–646. [Google Scholar] [CrossRef] [PubMed]
- Mallery, D.L.; Marquez, C.L.; McEwan, W.A.; Dickson, C.F.; Jacques, D.A.; Anandapadamanaban, M.; Bichel, K.; Towers, G.J.; Saiardi, A.; Bocking, T.; et al. IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis. eLife 2018, 7, e35335. [Google Scholar] [CrossRef] [PubMed]
- Jacques, D.A.; McEwan, W.A.; Hilditch, L.; Price, A.J.; Towers, G.J.; James, L.C. HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis. Nature 2016, 536, 349–353. [Google Scholar] [CrossRef]
- Mattei, S.; Glass, B.; Hagen, W.J.H.; Kräusslich, H.-G.; Briggs, J.A.G. The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 2016, 354, 1434–1437. [Google Scholar] [CrossRef]
- Ni, T.; Gerard, S.; Zhao, G.; Dent, K.; Ning, J.; Zhou, J.; Shi, J.; Anderson-Daniels, J.; Li, W.; Jang, S.; et al. Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A. Nat. Struct. Mol. Biol. 2020, 27, 855–862. [Google Scholar] [CrossRef]
- Gres, A.T.; Kirby, K.A.; Kewalramani, V.N.; Tanner, J.J.; Pornillos, O.; Sarafianos, S.G. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Science 2015, 349, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Pornillos, O.; Ganser-Pornillos, B.K.; Yeager, M. Atomic-level modelling of the HIV capsid. Nature 2011, 469, 424–427. [Google Scholar] [CrossRef]
- Acton, O.; Grant, T.; Nicastro, G.; Ball, N.J.; Goldstone, D.C.; Robertson, L.E.; Sader, K.; Nans, A.; Ramos, A.; Stoye, J.P.; et al. Structural basis for Fullerene geometry in a human endogenous retrovirus capsid. Nat. Commun. 2019, 10, 5822. [Google Scholar] [CrossRef]
- Fu, X.; Ning, J.; Zhong, Z.; Ambrose, Z.; Watkins, S.C.; Zhang, P. AutoCLEM: An Automated Workflow for Correlative Live-Cell Fluorescence Microscopy and Cryo-Electron Tomography. Sci. Rep. 2019, 9, 19207. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.; Ke, D.; Debiec, K.; Zhao, G.; Meng, X.; Ambrose, Z.; Gibson, G.A.; Watkins, S.C.; Zhang, P. Direct visualization of HIV-1 with correlative live-cell microscopy and cryo-electron tomography. Structure 2011, 19, 1573–1581. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, L.; Howe, A.; Gilchrist, J.B.; Sheng, Y.; Sun, D.; Knight, M.L.; Zanetti-Domingues, L.C.; Bateman, B.; Krebs, A.-S.; Chen, L.; et al. Correlative multi-scale cryo-imaging unveils SARS-CoV-2 assembly and egress. Nat. Commun. 2021, 12, 4629. [Google Scholar] [CrossRef] [PubMed]
- Tao, C.-L.; Liu, Y.-T.; Sun, R.; Zhang, B.; Qi, L.; Shivakoti, S.; Tian, C.-L.; Zhang, P.; Lau, P.-M.; Zhou, Z.H.; et al. Differentiation and Characterization of Excitatory and Inhibitory Synapses by Cryo-electron Tomography and Correlative Microscopy. J. Neurosci. 2018, 38, 1493–1510. [Google Scholar] [CrossRef]
- Zhang, P. Correlative cryo-electron tomography and optical microscopy of cells. Curr. Opin. Struct. Biol. 2013, 23, 763–770. [Google Scholar] [CrossRef]
- Sutton, G.; Sun, D.; Fu, X.; Kotecha, A.; Hecksel, C.W.; Clare, D.K.; Zhang, P.; Stuart, D.I.; Boyce, M. Assembly intermediates of orthoreovirus captured in the cell. Nat. Commun. 2020, 11, 4445. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Strunk, K.; Zhao, G.; Gray, J.L.; Zhang, P. 3D structure determination of native mammalian cells using cryo-FIB and cryo-electron tomography. J. Struct. Biol. 2012, 180, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Mendonça, L.; Allen, E.R.; Howe, A.; Lee, M.; Allen, J.D.; Chawla, H.; Pulido, D.; Donnellan, F.; Davies, H.; et al. Native-like SARS-CoV-2 Spike Glycoprotein Expressed by ChAdOx1 nCoV-19/AZD1222 Vaccine. ACS Cent. Sci. 2021, 7, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Sun, D.; Schertel, A.; Ning, J.; Fu, X.; Gwo, P.P.; Watson, A.M.; Zanetti-Domingues, L.C.; Martin-Fernandez, M.L.; Freyberg, Z.; et al. Serial cryoFIB/SEM Reveals Cytoarchitectural Disruptions in Leigh Syndrome Patient Cells. Structure 2021, 29, 82–87. [Google Scholar] [CrossRef]
- Zhang, P. Advances in cryo-electron tomography and subtomogram averaging and classification. Curr. Opin. Struct. Biol. 2019, 58, 249–258. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).