Detection and Genome Sequencing of SARS-CoV-2 in a Domestic Cat with Respiratory Signs in Switzerland
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Blood Sample Collection
2.3. Swab Sample Collection
2.4. Nucleic Acid Extraction and Molecular Analysis
2.5. Confirmatiory Tests for Positive RT-qPCR Results
2.6. Next Generation Sequencing
2.7. Antibody Detection by Enzyme-Linked Immunosorbent Assay (ELISA)
2.8. Surrogate Virus Neutralization Test (sVNT)
2.9. Confirmatory Serological Tests
2.10. Testing for Further Viral Infections
2.11. Submission to GenBank, ProMed, and OIE
2.12. Statistical Analysis
3. Results
3.1. History and Clinical Signs
3.2. RT-qPCR
3.3. Next Generation Sequencing
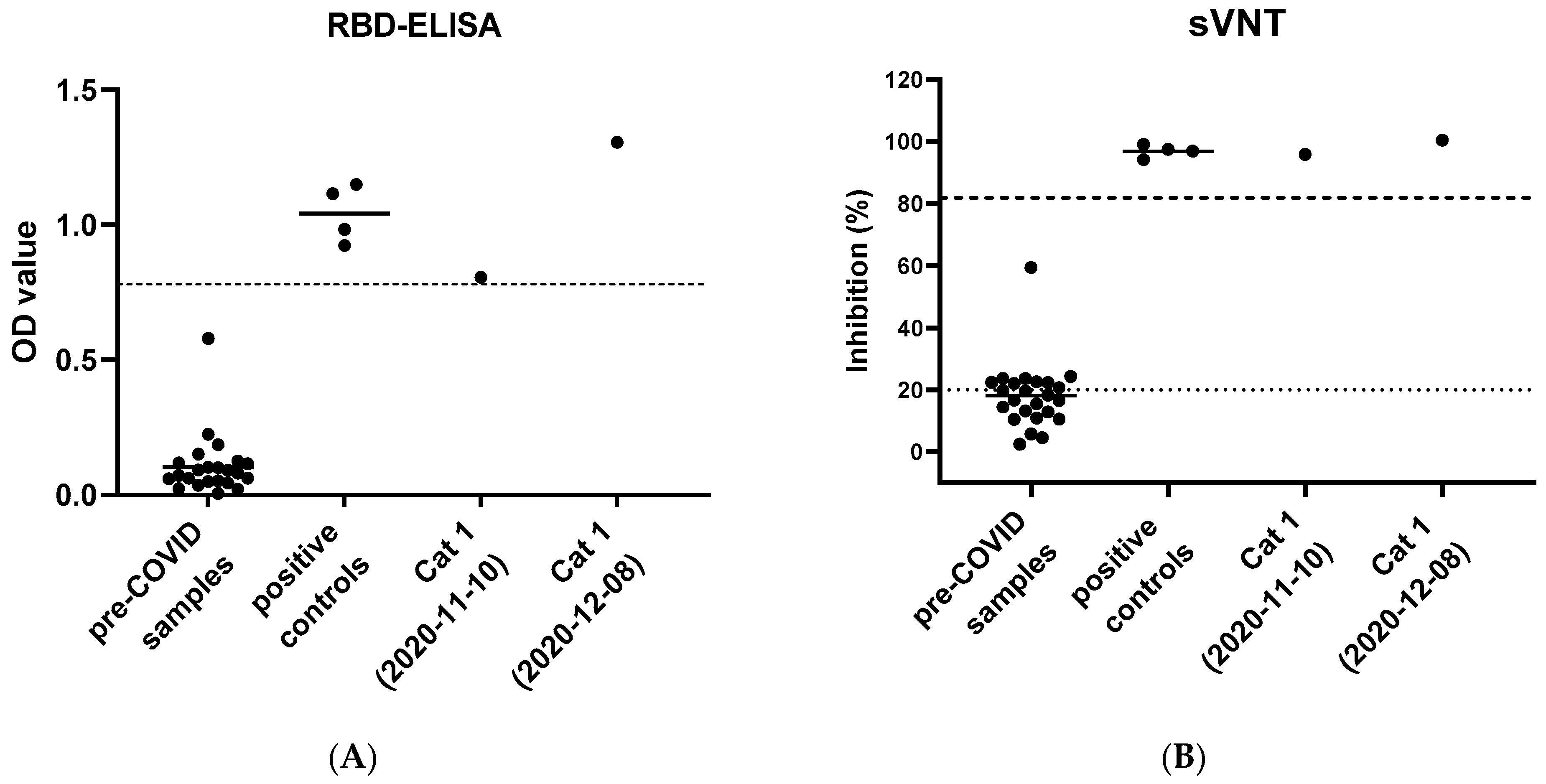
3.4. Serological Tests
3.5. Exclusion of Further Common Feline Viral Infections
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Sequence Identifier on GISAID | Submitting Laboratory | Originating Laboratory | Analysis |
|---|---|---|---|
| EPI_ISL_536400 | MRC-University of Glasgow Centre for Virus Research | Fareham Creek Veterinary Surgery | isolate from cat |
| EPI_ISL_487275 | Department of Veterinary Pathology, University of Liege-FARAH | Department of Veterinary Pathology, University of Liege-FARAH | isolate from cat |
| EPI_ISL_717979 | Laboratory of Biology, Department of Medicine, Democritus University of Thrace, Alexandroupolis, Greece | Laboratory of Microbiology and Infectious Diseases, Faculty of Veterinary Medicine, Aristotle University of Thessaloniki, University Campus, 541 24, Thessaloniki, Greece | isolate from cat |
| EPI_ISL_722300 | Erasmus Medical Center | Dutch COVID-19 response team | isolate from cat |
| EPI_ISL_683164 | Albertsen Lab, Department of Chemistry and Bioscience, Aalborg University, Denmark | Department of Virus and Microbiological Special Diagnostics, Statens Serum Institut, Copenhagen, Denmark | isolate from cat |
| EPI_ISL_683165 | Albertsen Lab, Department of Chemistry and Bioscience, Aalborg University, Denmark | Department of Virus and Microbiological Special Diagnostics, Statens Serum Institut, Copenhagen, Denmark | isolate from cat |
| EPI_ISL_683166 | Albertsen Lab, Department of Chemistry and Bioscience, Aalborg University, Denmark | Department of Virus and Microbiological Special Diagnostics, Statens Serum Institut, Copenhagen, Denmark | isolate from cat |
| EPI_ISL_759858 | School of Public Health, The University of Hong Kong | School of Public Health, The University of Hong Kong | isolate from cat |
| EPI_ISL_873040 | WHO National Influenza Centre Russian Federation | WHO National Influenza Centre Russian Federation | isolate from cat |
| EPI_ISL_873210 | Virology, Ecole Nationale Veterinaire de Toulouse | Virology, Ecole Nationale Veterinaire de Toulouse | isolate from cat |
| EPI_ISL_811147 | WHO National Influenza Centre Russian Federation | WHO National Influenza Centre Russian Federation | isolate from cat |
| EPI_ISL_437349 | Institut Pasteur CIBU-ERI | Ecole nationale vétérinaire d’Alfort-laboratoire de santé animale Anses UMR 1161 de virologie ENVA-Anses-INRAE | isolate from cat |
| EPI_ISL_699509 | Diagnostic Virology Laboratory, USDA National Veterinary Services Laboratories | Diagnostic Virology Laboratory, USDA National Veterinary Services Laboratories | isolate from cat |
| EPI_ISL_699507 | Diagnostic Virology Laboratory, USDA National Veterinary Services Laboratories | Diagnostic Virology Laboratory, USDA National Veterinary Services Laboratories | isolate from cat |
| EPI_ISL_699506 | Diagnostic Virology Laboratory, USDA National Veterinary Services Laboratories | Diagnostic Virology Laboratory, USDA National Veterinary Services Laboratories | isolate from cat |
| EPI_ISL_482820 | IrsiCaixa AIDS Research Lab | Centre de Recerca en Sanitat Animal (IRTA-CReSA) | isolate from cat |
| EPI_ISL_483063 | Virology, Ecole Nationale Veterinaire de Toulouse | unknown | isolate from cat |
| EPI_ISL_483064 | Virology, Ecole Nationale Veterinaire de Toulouse | unknown | isolate from cat |
| EPI_ISL_854043 | Bergthaler laboratory, CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences | Austrian Agency for Health and Food Safety (AGES) | phylogeny |
| EPI_ISL_854044 | Bergthaler laboratory, CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences | Austrian Agency for Health and Food Safety (AGES) | phylogeny |
| EPI_ISL_854047 | Bergthaler laboratory, CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences | Austrian Agency for Health and Food Safety (AGES) | phylogeny |
| EPI_ISL_854048 | Bergthaler laboratory, CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences | Austrian Agency for Health and Food Safety (AGES) | phylogeny |
| EPI_ISL_854056 | Bergthaler laboratory, CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences | Austrian Agency for Health and Food Safety (AGES) | phylogeny |
| EPI_ISL_854062 | Bergthaler laboratory, CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences | Austrian Agency for Health and Food Safety (AGES) | phylogeny |
| EPI_ISL_854064 | Bergthaler laboratory, CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences | Austrian Agency for Health and Food Safety (AGES) | phylogeny |
| EPI_ISL_854081 | Bergthaler laboratory, CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences | Austrian Agency for Health and Food Safety (AGES) | phylogeny |
| EPI_ISL_854082 | Bergthaler laboratory, CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences | Austrian Agency for Health and Food Safety (AGES) | phylogeny |
| EPI_ISL_854087 | Bergthaler laboratory, CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences | Austrian Agency for Health and Food Safety (AGES) | phylogeny |
| EPI_ISL_854108 | Bergthaler laboratory, CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences | Austrian Agency for Health and Food Safety (AGES) | phylogeny |
| EPI_ISL_603551 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_603729 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_603513 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_737750 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_857526 | Swiss National Reference Centre for Influenza | Swiss National Reference Centre for Influenza | phylogeny |
| EPI_ISL_830757 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830752 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830762 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_603459 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_603460 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_830872 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830879 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830881 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830882 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830884 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830888 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830891 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830901 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830903 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830906 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830907 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830914 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830992 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830993 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830925 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830928 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830929 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830998 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830959 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830965 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_830972 | University Hospital Basel, Clinical Bacteriology | University Hospital Basel, Clinical Virology | phylogeny |
| EPI_ISL_614899 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_614901 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_614902 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_614904 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_614906 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_721846 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_721853 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_768144 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_693893 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_693894 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_693936 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_796492 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_737782 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_899802 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_721921 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_603423 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_603490 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_603499 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_603509 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_603514 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_614967 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_614971 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_614974 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_693792 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_693838 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_729237 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_729214 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_693948 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_721950 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_729029 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_728979 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_729071 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_729074 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_729078 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_729084 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_737401 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_737919 | Department of Biosystems Science and Engineering, ETH Zurich | Viollier AG | phylogeny |
| EPI_ISL_402125 | National Institute for Communicable Disease Control and Prevention (ICDC) Chinese Center for Disease Control and Prevention (China CDC) | National Institute for Communicable Disease Control and Prevention (ICDC) Chinese Center for Disease Control and Prevention (China CDC) | phylogeny |
References
- World Health Organization. Novel Coronavirus—China. Available online: https://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/ (accessed on 18 December 2020).
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—From 11 March 2020. Available online: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (accessed on 18 December 2020).
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef]
- John Hopkins University. COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). Available online: https://coronavirus.jhu.edu/map.html (accessed on 18 December 2020).
- Bundesamt für Gesundheit BAG. COVID-19 Statistik. Available online: https://www.covid19.admin.ch/de/overview?ovTime=total (accessed on 30 November 2020).
- Fehr, A.R.; Perlman, S. Coronaviruses: An Overview of Their Replication and Pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar] [CrossRef]
- Herrewegh, A.A.; Smeenk, I.; Horzinek, M.C.; Rottier, P.J.; de Groot, R.J. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 1998, 72, 4508–4514. [Google Scholar] [CrossRef]
- Kipar, A.; Meli, M.L. Feline infectious peritonitis: Still an enigma? Vet. Pathol. 2014, 51, 505–526. [Google Scholar] [CrossRef]
- Pedersen, N.C.; Allen, C.E.; Lyons, L.A. Pathogenesis of feline enteric coronavirus infection. J. Feline Med. Surg. 2008, 10, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Mohd, H.A.; Al-Tawfiq, J.A.; Memish, Z.A. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) origin and animal reservoir. Virol. J. 2016, 13, 87. [Google Scholar] [CrossRef]
- Ye, Z.-W.; Yuan, S.; Yuen, K.-S.; Fung, S.-Y.; Chan, C.-P.; Jin, D.-Y. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020, 16, 1686–1697. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Zhang, T.; Wu, Q.; Zhang, Z. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr. Biol. 2020, 30, 1346–1351. [Google Scholar] [CrossRef]
- Qiu, Y.; Zhao, Y.B.; Wang, Q.; Li, J.Y.; Zhou, Z.J.; Liao, C.H.; Ge, X.Y. Predicting the angiotensin converting enzyme 2 (ACE2) utilizing capability as the receptor of SARS-CoV-2. Microbes Infect. 2020, 22, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Gu, L.; Ma, L.; Duan, Y. Atlas of ACE2 gene expression in mammals reveals novel insights in transmisson of SARS-Cov-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Stout, A.E.; André, N.M.; Jaimes, J.A.; Millet, J.K.; Whittaker, G.R. Coronaviruses in cats and other companion animals: Where does SARS-CoV-2/COVID-19 fit? Vet. Microbiol. 2020, 247, 108777. [Google Scholar] [CrossRef]
- OIE—World Organization for Animal Health. Events in Animals. Available online: https://www.oie.int/en/scientific-expertise/specific-information-and-recommendations/questions-and-answers-on-2019novel-coronavirus/events-in-animals/ (accessed on 4 January 2021).
- Sit, T.H.C.; Brackman, C.J.; Ip, S.M.; Tam, K.W.S.; Law, P.Y.T.; To, E.M.W.; Yu, V.Y.T.; Sims, L.D.; Tsang, D.N.C.; Chu, D.K.W.; et al. Infection of dogs with SARS-CoV-2. Nature 2020, 586, 776–778. [Google Scholar] [CrossRef]
- Newman, A.; Smith, D.; Ghai, R.R.; Wallace, R.M.; Torchetti, M.K.; Loiacono, C.; Murrell, L.S.; Carpenter, A.; Moroff, S.; Rooney, J.A.; et al. First reported cases of SARS-CoV-2 infection in companion animals–New York, March–April 2020. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 710–713. [Google Scholar] [CrossRef]
- World Health Organization. Transmission of SARS-CoV-2: Implications for Infection Prevention Precautions. Available online: https://www.who.int/news-room/commentaries/detail/transmission-of-sars-cov-2-implications-for-infection-prevention-precautions (accessed on 18 December 2020).
- Barrs, V.R.; Peiris, M.; Tam, K.W.S.; Law, P.Y.T.; Brackman, C.J.; To, E.M.W.; Yu, V.Y.T.; Chu, D.K.W.; Perera, R.; Sit, T.H.C. SARS-CoV-2 in Quarantined Domestic Cats from COVID-19 Households or Close Contacts, Hong Kong, China. Emerg. Infect. Dis. 2020, 26. [Google Scholar] [CrossRef]
- Oreshkova, N.; Molenaar, R.J.; Vreman, S.; Harders, F.; Oude Munnink, B.B.; Hakze-van der Honing, R.W.; Gerhards, N.; Tolsma, P.; Bouwstra, R.; Sikkema, R.S.; et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eur. Surveill. 2020, 25. [Google Scholar] [CrossRef]
- Kim, Y.I.; Kim, S.G.; Kim, S.M.; Kim, E.H.; Park, S.J.; Yu, K.M.; Chang, J.H.; Kim, E.J.; Lee, S.; Casel, M.A.B.; et al. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe 2020, 27, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef] [PubMed]
- Le Bras, A. SARS-CoV-2 causes COVID-19-like disease in cynomolgus macaques. Lab. Anim. 2020, 49, 174. [Google Scholar] [CrossRef] [PubMed]
- Schaecher, S.R.; Stabenow, J.; Oberle, C.; Schriewer, J.; Buller, R.M.; Sagartz, J.E.; Pekosz, A. An immunosuppressed Syrian golden hamster model for SARS-CoV infection. Virology 2008, 380, 312–321. [Google Scholar] [CrossRef]
- Sia, S.F.; Yan, L.M.; Chin, A.W.H.; Fung, K.; Choy, K.T.; Wong, A.Y.L.; Kaewpreedee, P.; Perera, R.; Poon, L.L.M.; Nicholls, J.M.; et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020, 583, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, P.J.; Hatta, M.; Chiba, S.; Maemura, T.; Fan, S.; Takeda, M.; Kinoshita, N.; Hattori, S.I.; Sakai-Tagawa, Y.; Iwatsuki-Horimoto, K.; et al. Transmission of SARS-CoV-2 in Domestic Cats. N. Engl. J. Med. 2020, 383, 592–594. [Google Scholar] [CrossRef] [PubMed]
- Gaudreault, N.N.; Trujillo, J.D.; Carossino, M.; Meekins, D.A.; Morozov, I.; Madden, D.W.; Indran, S.V.; Bold, D.; Balaraman, V.; Kwon, T.; et al. SARS-CoV-2 infection, disease and transmission in domestic cats. Emerg. Microbes Infect. 2020, 9, 2322–2332. [Google Scholar] [CrossRef] [PubMed]
- Munster, V.J.; Feldmann, F.; Williamson, B.N.; van Doremalen, N.; Pérez-Pérez, L.; Schulz, J.; Meade-White, K.; Okumura, A.; Callison, J.; Brumbaugh, B.; et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 2020, 585, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Bosco-Lauth, A.M.; Hartwig, A.E.; Porter, S.M.; Gordy, P.W.; Nehring, M.; Byas, A.D.; VandeWoude, S.; Ragan, I.K.; Maison, R.M.; Bowen, R.A. Experimental infection of domestic dogs and cats with SARS-CoV-2: Pathogenesis, transmission, and response to reexposure in cats. Proc. Natl. Acad. Sci. USA 2020, 117, 26382–26388. [Google Scholar] [CrossRef]
- Hosie, M.J.; Epifano, I.; Herder, V.; Orton, R.J.; Stevenson, A.; Johnson, N.; MacDonald, E.; Dunbar, D.; McDonald, M.; Howie, F.; et al. Respiratory disease in cats associated with human-to-cat transmission of SARS-CoV-2 in the UK. bioRxiv 2020. [Google Scholar] [CrossRef]
- Garigliany, M.; Van Laere, A.-S.; Clercx, C.; Giet, D.; Escriou, N.; Huon, C.; van der Werf, S.; Eloit, M.; Desmecht, D. SARS-CoV-2 Natural Transmission from Human to Cat, Belgium, March 2020. Emerg. Infect. Dis. J. 2020, 26, 3069. [Google Scholar] [CrossRef] [PubMed]
- Sailleau, C.; Dumarest, M.; Vanhomwegen, J.; Delaplace, M.; Caro, V.; Kwasiborski, A.; Hourdel, V.; Chevaillier, P.; Barbarino, A.; Comtet, L.; et al. First detection and genome sequencing of SARS-CoV-2 in an infected cat in France. Transbound. Emerg. Dis. 2020, 67, 2324–2328. [Google Scholar] [CrossRef]
- Segalés, J.; Puig, M.; Rodon, J.; Avila-Nieto, C.; Carrillo, J.; Cantero, G.; Terrón, M.T.; Cruz, S.; Parera, M.; Noguera-Julián, M.; et al. Detection of SARS-CoV-2 in a cat owned by a COVID-19-affected patient in Spain. Proc. Natl. Acad. Sci. USA 2020, 117, 24790–24793. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, H.; Gao, J.; Huang, K.; Yang, Y.; Hui, X.; He, X.; Li, C.; Gong, W.; Zhang, Y.; et al. A serological survey of SARS-CoV-2 in cat in Wuhan. Emerg. Microbes Infect. 2020, 9, 2013–2019. [Google Scholar] [CrossRef] [PubMed]
- Patterson, E.I.; Elia, G.; Grassi, A.; Giordano, A.; Desario, C.; Medardo, M.; Smith, S.L.; Anderson, E.R.; Prince, T.; Patterson, G.T.; et al. Evidence of exposure to SARS-CoV-2 in cats and dogs from households in Italy. Nat. Commun. 2020, 11, 6231. [Google Scholar] [CrossRef]
- Deng, J.; Liu, Y.; Sun, C.; Bai, J.; Sun, J.; Hao, L.; Li, X.; Tian, K. SARS-CoV-2 Serological Survey of Cats in China before and after the Pandemic. Virol. Sinica 2020, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Stevanovic, V.; Vilibic-Cavlek, T.; Tabain, I.; Benvin, I.; Kovac, S.; Hruskar, Z.; Mauric, M.; Milasincic, L.; Antolasic, L.; Skrinjaric, A.; et al. Seroprevalence of SARS-CoV-2 infection among pet animals in Croatia and potential public health impact. Transbound. Emerg. Dis. 2020. [Google Scholar] [CrossRef]
- Fritz, M.; Rosolen, B.; Krafft, E.; Becquart, P.; Elguero, E.; Vratskikh, O.; Denolly, S.; Boson, B.; Vanhomwegen, J.; Gouilh, M.A.; et al. High prevalence of SARS-CoV-2 antibodies in pets from COVID-19+ households. One Health 2021, 11, 100192. [Google Scholar] [CrossRef]
- Michelitsch, A.; Hoffmann, D.; Wernike, K.; Beer, M. Occurrence of Antibodies against SARS-CoV-2 in the Domestic Cat Population of Germany. Vaccines 2020, 8, 772. [Google Scholar] [CrossRef]
- Hamer, S.A.; Pauvolid-Corrêa, A.; Zecca, I.B.; Davila, E.; Auckland, L.D.; Roundy, C.M.; Tang, W.; Torchetti, M.; Killian, M.L.; Jenkins-Moore, M.; et al. Natural SARS-CoV-2 infections, including virus isolation, among serially tested cats and dogs in households with confirmed human COVID-19 cases in Texas, USA. bioRxiv 2020. [Google Scholar] [CrossRef]
- Verband für Heimtiernahrung. Statistik Heimtierpopulation Schweiz. Available online: https://www.vhn.ch/statistiken/heimtiere-schweiz/ (accessed on 18 December 2020).
- Studer, N.; Lutz, H.; Saegerman, C.; Gonczi, E.; Meli, M.L.; Boo, G.; Hartmann, K.; Hosie, M.J.; Moestl, K.; Tasker, S.; et al. Pan-European Study on the Prevalence of the Feline Leukaemia Virus Infection—Reported by the European Advisory Board on Cat Diseases (ABCD Europe). Viruses 2019, 11, 993. [Google Scholar] [CrossRef]
- Gomes-Keller, M.A.; Tandon, R.; Gonczi, E.; Meli, M.L.; Hofmann-Lehmann, R.; Lutz, H. Shedding of feline leukemia virus RNA in saliva is a consistent feature in viremic cats. Vet. Microbiol. 2006, 112, 11–21. [Google Scholar] [CrossRef]
- Berger, A.; Willi, B.; Meli, M.L.; Boretti, F.S.; Hartnack, S.; Dreyfus, A.; Lutz, H.; Hofmann-Lehmann, R. Feline calicivirus and other respiratory pathogens in cats with Feline calicivirus-related symptoms and in clinically healthy cats in Switzerland. BMC Vet. Res. 2015, 11, 282. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef]
- Gultom, M.; Licheri, M.; Laloli, L.; Wider, M.; Strässle, M.; Steiner, S.; Kratzel, A.; Thao, T.T.N.; Stalder, H.; Portmann, J.; et al. Susceptibility of well-differentiated airway epithelial cell cultures from domestic and wildlife animals to SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- World Health Organization. Protocol: Real-Time RT-PCR Assays for the Detection of SARS-CoV-2; Institut Pasteur, Paris. Available online: https://www.who.int/docs/default-source/coronaviruse/real-time-rt-pcr-assays-for-the-detection-of-sars-cov-2-institut-pasteur-paris.pdf?sfvrsn=3662fcb6_2 (accessed on 17 December 2020).
- Nadeau, S.; Beckmann, C.; Topolsky, I.; Vaughan, T.; Hodcroft, E.; Schär, T.; Nissen, I.; Santacroce, N.; Burcklen, E.; Ferreira, P.; et al. Quantifying SARS-CoV-2 spread in Switzerland based on genomic sequencing data. medRxiv 2020. [Google Scholar] [CrossRef]
- Quick, J.; Grubaugh, N.D.; Pullan, S.T.; Claro, I.M.; Smith, A.D.; Gangavarapu, K.; Oliveira, G.; Robles-Sikisaka, R.; Rogers, T.F.; Beutler, N.A.; et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 2017, 12, 1261–1276. [Google Scholar] [CrossRef]
- Posada-Céspedes, S.; Seifert, D.; Topolsky, I.; Jablonski, K.P.; Metzner, K.J.; Beerenwinkel, N. V-pipe: A computational pipeline for assessing viral genetic diversity from high-throughput data. Bioinformatics 2021. [Google Scholar] [CrossRef]
- Hitslib. SAMTOOLS Merge—Merges Multiple Sorted Input Files Into a Single Output. Available online: http://www.htslib.org/doc/samtools-merge.html (accessed on 1 February 2021).
- Github. Bcftools—Utilities for Variant Calling and Manipulating VCFs and BCFs. Available online: Samtools.github.io/bcftools/bcftools.html (accessed on 1 February 2021).
- Github. VCF Consensus Builder. Available online: https://github.com/peterk87/vcf_consensus_builder (accessed on 1 February 2021).
- Pangolin. Phylogenetic Assignment of Named Global Outbreak Lineages. Available online: https://github.com/hCoV-2019/pangolin (accessed on 26 January 2021).
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.i.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Boenzli, E.; Hadorn, M.; Hartnack, S.; Huder, J.; Hofmann-Lehmann, R.; Lutz, H. Detection of antibodies to the feline leukemia Virus (FeLV) transmembrane protein p15E: An alternative approach for serological FeLV detection based on antibodies to p15E. J. Clin. Microbiol. 2014, 52, 2046–2052. [Google Scholar] [CrossRef] [PubMed]
- Engvall, E.; Perlmann, P. Enzyme-linked immunosorbent assay, Elisa. Quantitation of specific antibodies by enzyme-labeled anti-immunoglobulin in antigen-coated tubes. J. Immunol. 1972, 109, 129–135. [Google Scholar] [PubMed]
- Voller, A.; Bidwell, D.E.; Bartlett, A. Enzyme immunoassays in diagnostic medicine. Theory and practice. Bull. World Health Org. 1976, 53, 55–65. [Google Scholar] [PubMed]
- Spiri, A.M.; Meli, M.L.; Riond, B.; Herbert, I.; Hosie, M.J.; Hofmann-Lehmann, R. Environmental Contamination and Hygienic Measures After Feline Calicivirus Field Strain Infections of Cats in a Research Facility. Viruses 2019, 11, 958. [Google Scholar] [CrossRef]
- Okba, N.M.A.; Müller, M.; Li, W.; Wang, C.; GeurtsvanKessel, C.; Corman, V.; Lamers, M.; Sikkema, R.; de Bruin, E.; Chandler, F.; et al. Severe Acute Respiratory Syndrome Coronavirus 2−Specific Antibody Responses in Coronavirus Disease Patients. Emerg. Infect. Dis. J. 2020, 26, 1478. [Google Scholar] [CrossRef]
- Tan, C.W.; Chia, W.N.; Qin, X.; Liu, P.; Chen, M.I.; Tiu, C.; Hu, Z.; Chen, V.C.; Young, B.E.; Sia, W.R.; et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020, 38, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, W.; Drabek, D.; Okba, N.M.A.; van Haperen, R.; Osterhaus, A.D.M.E.; van Kuppeveld, F.J.M.; Haagmans, B.L.; Grosveld, F.; Bosch, B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020, 11, 2251. [Google Scholar] [CrossRef]
- Zhao, S.; Schuurman, N.; Li, W.; Wang, C.; Smit, L.A.; Broens, E.M.; Wagenaar, J.A.; van Kuppeveld, F.J.; Bosch, B.J.; Egberink, H. Serologic screening of severe acute respiratory syndrome coronavirus 2 infection in cats and dogs during first coronavirus disease wave, the Netherlands. Emerg. Infect. Dis 2021. [Google Scholar] [CrossRef]
- Wernike, K.; Aebischer, A.; Michelitsch, A.; Hoffmann, D.; Freuling, C.; Balkema-Buschmann, A.; Graaf, A.; Müller, T.; Osterrieder, N.; Rissmann, M.; et al. Multi-species ELISA for the detection of antibodies against SARS-CoV-2 in animals. Transbound. Emerg. Dis. 2020. [Google Scholar] [CrossRef]
- Vögtlin, A.; Fraefel, C.; Albini, S.; Leutenegger, C.M.; Schraner, E.; Spiess, B.; Lutz, H.; Ackermann, M. Quantification of Feline Herpesvirus 1 DNA in Ocular Fluid Samples of Clinically Diseased Cats by Real-Time TaqMan PCR. J. Clin. Microbiol. 2002, 40, 519–523. [Google Scholar] [CrossRef]
- Frankenfeld, J.; Meili, T.; Meli, M.L.; Riond, B.; Helfer-Hungerbuehler, A.K.; Bönzli, E.; Pineroli, B.; Hofmann-Lehmann, R. Decreased Sensitivity of the Serological Detection of Feline Immunodeficiency Virus Infection Potentially Due to Imported Genetic Variants. Viruses 2019, 11, 697. [Google Scholar] [CrossRef]
- Calzolari, M.; Young, E.; Cox, D.; Davis, D.; Lutz, H. Serological diagnosis of feline immunodeficiency virus infection using recombinant transmembrane glycoprotein. Vet. Immunol. Immunopathol. 1995, 46, 83–92. [Google Scholar] [CrossRef]
- Egberink, H.F.; Lutz, H.; Horzinek, M.C. Use of western blot and radioimmunoprecipitation for diagnosis of feline leukemia and feline immunodeficiency virus infections. J. Am. Vet. Med. Assoc. 1991, 199, 1339–1342. [Google Scholar]
- Lutz, H.; Pedersen, N.C.; Durbin, R.; Theilen, G.H. Monoclonal antibodies to three epitopic regions of feline leukemia virus p27 and their use in enzyme-linked immunosorbent assay of p27. J. Immunol. Methods 1983, 56, 209–220. [Google Scholar] [CrossRef]
- Tandon, R.; Cattori, V.; Gomes-Keller, M.A.; Meli, M.L.; Golder, M.C.; Lutz, H.; Hofmann-Lehmann, R. Quantitation of feline leukaemia virus viral and proviral loads by TaqMan real-time polymerase chain reaction. J. Virol. Methods 2005, 130, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Federal Food Safety and Veterinary Office FSVO. Research on SARS-CoV-2—Confirmation in a Cat in the Canton of Zurich. Available online: https://www.oie.int/fileadmin/Home/MM/Switzerland_03.12.2020_Research_on_SARS-CoV-2_-_confirmation_in_a_cat_in_the_Canton_of_Zurich.pdf (accessed on 28 December 2020).
- ProMed. Coronavirus Disease 2019 Update (519): Switzerland (Zurich) Animal, Cat, Oie. Available online: https://promedmail.org/promed-post/?id=20201204.7993204 (accessed on 28 December 2020).
- Elbe, S.; Buckland-Merrett, G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017, 1, 33–46. [Google Scholar] [CrossRef]
- Ruiz-Arrondo, I.; Portillo, A.; Palomar, A.M.; Santibáñez, S.; Santibáñez, P.; Cervera, C.; Oteo, J.A. Detection of SARS-CoV-2 in pets living with COVID-19 owners diagnosed during the COVID-19 lockdown in Spain: A case of an asymptomatic cat with SARS-CoV-2 in Europe. Transbound. Emerg. Dis. 2020. [Google Scholar] [CrossRef]
- ProMed. Coronavirus Disease 2019 Update (181): Germany (Bavaria), France (Nouvelle-Aquitaine), Cat, Oie Animal Case Definition. Available online: https://promedmail.org/promed-post/?id=20200513.7332909 (accessed on 28 December 2020).
- Musso, N.; Costantino, A.; La Spina, S.; Finocchiaro, A.; Andronico, F.; Stracquadanio, S.; Liotta, L.; Visalli, R.; Emmanuele, G. New SARS-CoV-2 Infection Detected in an Italian Pet Cat by RT-qPCR from Deep Pharyngeal Swab. Pathogens 2020, 9, 746. [Google Scholar] [CrossRef]
- United States Department of Agriculture. SARS-CoV-2 Case Definition. Available online: https://www.aphis.usda.gov/animal_health/one_health/downloads/SARS-CoV-2-case-definition.pdf (accessed on 18 December 2020).
- Jefferson, T.; Spencer, E.; Brassey, J.; Heneghan, C. Viral cultures for COVID-19 infectivity assessment. Systematic review. medRxiv 2020. [Google Scholar] [CrossRef]
- Kim, M.C.; Cui, C.; Shin, K.R.; Bae, J.Y.; Kweon, O.J.; Lee, M.K.; Choi, S.H.; Jung, S.Y.; Park, M.S.; Chung, J.W. Duration of Culturable SARS-CoV-2 in Hospitalized Patients with Covid-19. N. Engl. J. Med. 2021. [Google Scholar] [CrossRef] [PubMed]
- McAloose, D.; Laverack, M.; Wang, L.; Killian, M.L.; Caserta, L.C.; Yuan, F.; Mitchell, P.K.; Queen, K.; Mauldin, M.R.; Cronk, B.D.; et al. From People to Panthera: Natural SARS-CoV-2 Infection in Tigers and Lions at the Bronx Zoo. mBio 2020, 11, e02220. [Google Scholar] [CrossRef] [PubMed]
- Chiocchetti, R.; Galiazzo, G.; Fracassi, F.; Giancola, F.; Pietra, M. ACE2 Expression in the Cat and the Tiger Gastrointestinal Tracts. Front. Vet. Sci. 2020, 7, 514. [Google Scholar] [CrossRef]
- Bulfamante, G.P.; Perrucci, G.L.; Falleni, M.; Sommariva, E.; Tosi, D.; Martinelli, C.; Songia, P.; Poggio, P.; Carugo, S.; Pompilio, G. Evidence of SARS-CoV-2 Transcriptional Activity in Cardiomyocytes of COVID-19 Patients without Clinical Signs of Cardiac Involvement. Biomedicines 2020, 8, 626. [Google Scholar] [CrossRef]
- Andersson, M.I.; Arancibia-Carcamo, C.V.; Auckland, K.; Baillie, J.K.; Barnes, E.; Beneke, T.; Bibi, S.; Brooks, T.; Carroll, M.; Crook, D.; et al. SARS-CoV-2 RNA detected in blood products from patients with COVID-19 is not associated with infectious virus. Wellcome Open Res. 2020, 5, 181. [Google Scholar] [CrossRef] [PubMed]
- Perera, R.; Ko, R.; Tsang, O.T.Y.; Hui, D.S.C.; Kwan, M.Y.M.; Brackman, C.J.; To, E.M.W.; Yen, H.L.; Leung, K.; Cheng, S.M.S.; et al. Evaluation of a SARS-CoV-2 surrogate virus neutralization test for detection of antibody in human, canine, cat and hamster sera. J. Clin. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Felten, S.; Klein-Richers, U.; Hofmann-Lehmann, R.; Bergmann, M.; Unterer, S.; Leutenegger, C.M.; Hartmann, K. Correlation of Feline Coronavirus Shedding in Feces with Coronavirus Antibody Titer. Pathogens 2020, 9, 598. [Google Scholar] [CrossRef]
- Tehrani, Z.R.; Saadat, S.; Saleh, E.; Ouyang, X.; Constantine, N.; DeVico, A.L.; Harris, A.D.; Lewis, G.K.; Kottilil, S.; Sajadi, M.M. Specificity and Performance of Nucleocapsid and Spike-based SARS-CoV-2 Serologic Assays. medRxiv Preprint Server Health Sci. 2020. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef] [PubMed]
- Kampf, G.; Lemmen, S.; Suchomel, M. Ct values and infectivity of SARS-CoV-2 on surfaces. Lancet Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention CDC. SARS-CoV-2 Variants. Available online: https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html (accessed on 3 February 2021).
- ProMed. SARS-CoV-2 Infection in Mink, Netherlands. Available online: https://promedmail.org/promed-post/?id=7272289 (accessed on 17 December 2020).
- World Health Organization. SARS-CoV-2 Mink-Associated Variant Strain—Denmark. Available online: https://www.who.int/csr/don/06-november-2020-mink-associated-sars-cov2-denmark/en/ (accessed on 1 December 2020).
- Oude Munnink, B.B.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; van der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2021, 371, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Totton, S.C.; Sargeant, J.M.; O’Connor, A.M. How could we conclude cat-to-human transmission of SARS-CoV-2? Zoonoses Public Health 2020. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention CDC. COVID-19, If You Have Pets. Available online: https://www.cdc.gov/coronavirus/2019-ncov/daily-life-coping/pets.html (accessed on 2 December 2020).
- Hosie, M.J.; Hofmann-Lehmann, R.; Hartmann, K.; Egberink, H.; Truyen, U.; Addie, D.D.; Belák, S.; Boucraut-Baralon, C.; Frymus, T.; Lloret, A.; et al. Anthropogenic Infection of Cats during the 2020 COVID-19 Pandemic. Viruses 2021, 13, 185. [Google Scholar] [CrossRef] [PubMed]
- European Advisory Board on Cat Diseases (ABCD). ABCD Guidelines SARS-CoV-2 in Cats. Available online: http://www.abcdcatsvets.org/ (accessed on 18 December 2020).
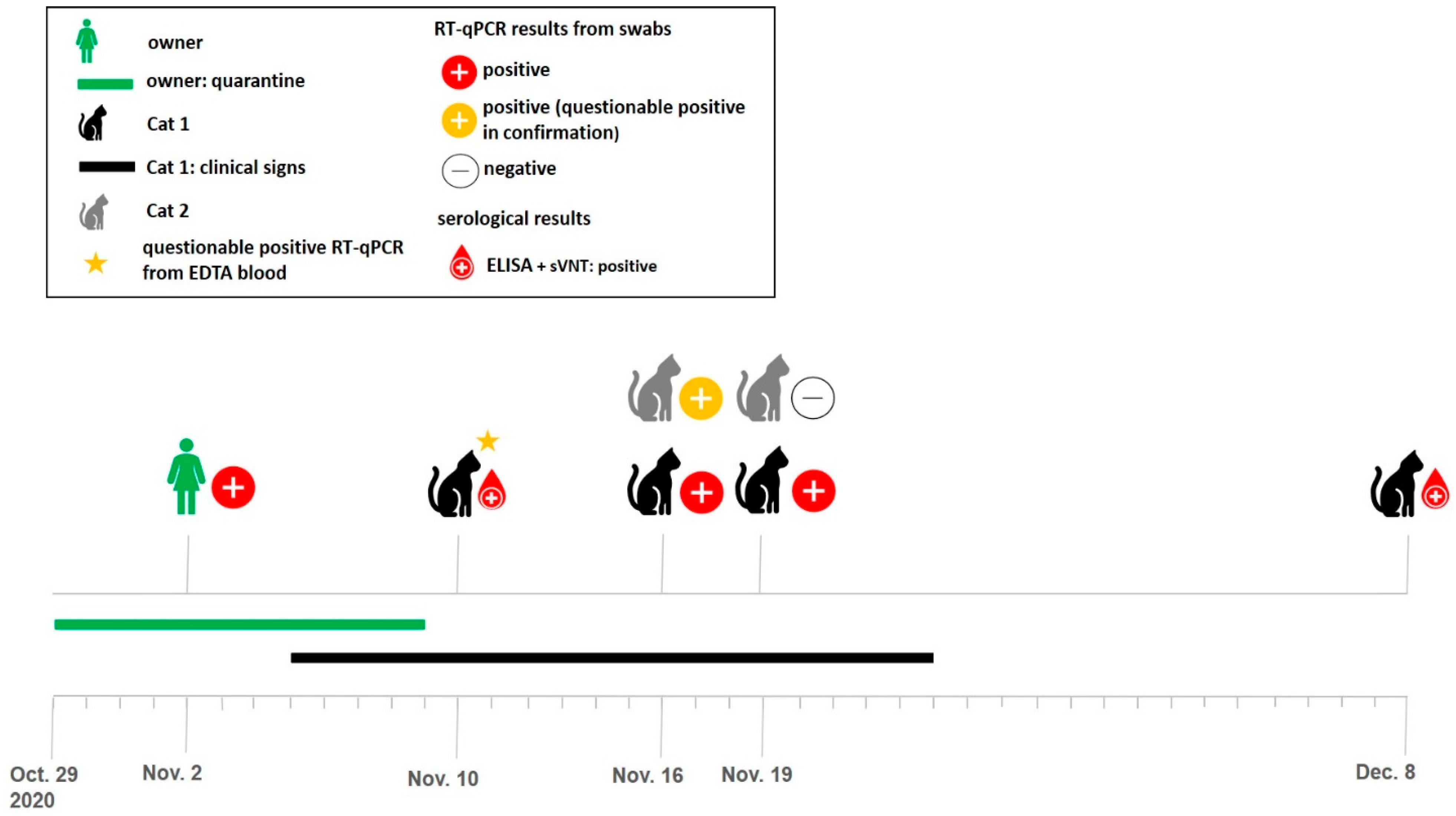


| Cat ID | Collection Date | Material | E-Assay (CT-Value) 1 | RdRp-Assay (CT-Value) 1 | Interpretation of the Results 2 |
|---|---|---|---|---|---|
| Cat 1 | 10 November 2020 | EDTA blood | 43.2 | 42.2 | questionable positive |
| 16 November 2020 | oral swab | negative | 36.6 | negative | |
| nasal swab | 32.6 | 33.2 | Positive | ||
| fecal swab | negative | 44 | negative | ||
| fur swab | 34.4 | 34.1 | positive | ||
| bedding swab | 31.8 | 31.7 | positive | ||
| 19 November 2020 | oral swab | negative | negative | negative | |
| nasalswab | 32.5 | 32.4 | positive | ||
| fecal swab | negative | negative | negative | ||
| fur swab | 36.9 | 35.7 | positive | ||
| Cat 2 | 16 November 2020 | oral swab | 37.5 | 36.2 | positive |
| nasal swab | negative | negative | negative | ||
| fecal swab | negative | negative | negative | ||
| fur swab | 44.9 | 35.8 | questionable positive | ||
| bedding swab | 32.9 | 33.1 | positive | ||
| 19 November 2020 | oral swab | negative | negative | negative | |
| nasal swab | negative | negative | negative | ||
| fecal swab | negative | negative | negative | ||
| fur swab | 33.8 | 35.9 | positive |
| Gene | SNP (nt) | aa Change | Cat 1 PCR 1 | Cat 1 PCR 2 | Cat 2 PCR 1 | Cat 2 PCR 2 |
|---|---|---|---|---|---|---|
| 5UTR | C241T * | noncoding | T | T | n | n |
| ORF1ab | A1236G * | ORF1a: D324G | G | G | n | n |
| ORF1ab | C3037T * | synonymous | T | T | n | n |
| ORF1ab | C3738T * | ORF1a: P1158L | T | T | n | n |
| ORF1ab | T7122C * | ORF1a: I2286T | Y (C or T) | C | n | n |
| ORF1ab | G9130T * | synonymous | T | T | n | n |
| ORF1ab | T14349C | synonymous | n | n | n | C |
| ORF1ab | C14408T * | ORF1b: P314L | T | T | n | T |
| ORF1ab | C16111T * | synonymous | T | T | Y (C or T) | n |
| ORF1ab | G18186T * | ORF1b: M1573I | n | T | n | n |
| ORF1ab | A19137G * | synonymous | G | G | n | n |
| S | G22801T * | synonymous | T | K (T or G) | T | n |
| S | A23403G * | S: D614G | n | G | G | n |
| S | G23580C * | S: S673T | C | C | n | n |
| ORF6 | T27319C * | synonymous | C | n | C | n |
| N | G28487A * | N: V72I | A | A | A | n |
| N | C28868T * | N: P199S | T | T | n | n |
| N | G28881A * | N: R203K | A | A | n | n |
| N | G28882A * | N: R203K | A | A | n | n |
| N | G28883C * | N: G204R | C | C | n | n |
| N | C29091T | synonymous | n | n | T | n |
| 3UTR | G29706T | noncoding | n | n | T | n |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klaus, J.; Meli, M.L.; Willi, B.; Nadeau, S.; Beisel, C.; Stadler, T.; ETH SARS-CoV-2 Sequencing Team; Egberink, H.; Zhao, S.; Lutz, H.; et al. Detection and Genome Sequencing of SARS-CoV-2 in a Domestic Cat with Respiratory Signs in Switzerland. Viruses 2021, 13, 496. https://doi.org/10.3390/v13030496
Klaus J, Meli ML, Willi B, Nadeau S, Beisel C, Stadler T, ETH SARS-CoV-2 Sequencing Team, Egberink H, Zhao S, Lutz H, et al. Detection and Genome Sequencing of SARS-CoV-2 in a Domestic Cat with Respiratory Signs in Switzerland. Viruses. 2021; 13(3):496. https://doi.org/10.3390/v13030496
Chicago/Turabian StyleKlaus, Julia, Marina L. Meli, Barbara Willi, Sarah Nadeau, Christian Beisel, Tanja Stadler, ETH SARS-CoV-2 Sequencing Team, Herman Egberink, Shan Zhao, Hans Lutz, and et al. 2021. "Detection and Genome Sequencing of SARS-CoV-2 in a Domestic Cat with Respiratory Signs in Switzerland" Viruses 13, no. 3: 496. https://doi.org/10.3390/v13030496
APA StyleKlaus, J., Meli, M. L., Willi, B., Nadeau, S., Beisel, C., Stadler, T., ETH SARS-CoV-2 Sequencing Team, Egberink, H., Zhao, S., Lutz, H., Riond, B., Rösinger, N., Stalder, H., Renzullo, S., & Hofmann-Lehmann, R. (2021). Detection and Genome Sequencing of SARS-CoV-2 in a Domestic Cat with Respiratory Signs in Switzerland. Viruses, 13(3), 496. https://doi.org/10.3390/v13030496








