Abstract
Next-generation sequencing (NGS) is increasingly used for HIV-1 drug resistance genotyping. NGS methods have the potential for a more sensitive detection of low-abundance variants (LAV) compared to standard Sanger sequencing (SS) methods. A standardized threshold for reporting LAV that generates data comparable to those derived from SS is needed to allow for the comparability of data from laboratories using NGS and SS. Ten HIV-1 specimens were tested in ten laboratories using Illumina MiSeq-based methods. The consensus sequences for each specimen using LAV thresholds of 5%, 10%, 15%, and 20% were compared to each other and to the consensus of the SS sequences (protease 4–99; reverse transcriptase 38–247). The concordance among laboratories’ sequences at different thresholds was evaluated by pairwise sequence comparisons. NGS sequences generated using the 20% threshold were the most similar to the SS consensus (average 99.6% identity, range 96.1–100%), compared to 15% (99.4%, 88.5–100%), 10% (99.2%, 87.4–100%), or 5% (98.5%, 86.4–100%). The average sequence identity between laboratories using thresholds of 20%, 15%, 10%, and 5% was 99.1%, 98.7%, 98.3%, and 97.3%, respectively. Using the 20% threshold, we observed an excellent agreement between NGS and SS, but significant differences at lower thresholds. Understanding how variation in NGS methods influences sequence quality is essential for NGS-based HIV-1 drug resistance genotyping.
1. Introduction
Next-generation sequencing (NGS) is increasingly used in molecular diagnostic laboratories, including for HIV-1 drug resistance (HIVDR) genotyping [1,2,3,4]. NGS methods have several potential advantages over standard Sanger sequencing (SS) methods, including more a sensitive detection of low-abundance variants (LAV, here defined as variants detectable by NGS but not SS), potentially less subjective and more quantitative and automatable data processing steps, and a reduction in cost. Since virus populations within individuals include multiple variants (possibly including variants with mutations conferring drug resistance) that are present at frequencies below the minimum required for detection by SS, NGS has the potential to improve the utility of HIVDR genotyping [5,6]. However, NGS involves complex laboratory and analytic methods that are not yet well-standardized between laboratories, although recommendations for bioinformatic analysis pipelines have been proposed [7,8,9]. HIVDR genotyping tests based on different NGS platforms are also commercially available [10,11,12,13]. The clinical significance of LAV is largely unknown, although there is a general agreement that LAV detected by more sensitive methods such as NGS may increase the predictive value of HIVDR genotyping for clinical outcomes as compared to SS [14,15,16,17]. There is ongoing debate, along with a general lack of certainty, regarding the optimal LAV threshold for clinical applications [17,18,19].
The World Health Organization (WHO) HIVDR Laboratory Network supports the national surveillance of HIVDR in low- and middle-income countries (LMIC) [20,21,22]. Network laboratories currently employ a variety of SS-based methods including commercial kits and in-house developed procedures, but several laboratories are planning to adopt NGS methods. Since resistance prevalence trends over time and between countries and geographic regions are an important part of the survey results, the standardization of genotyping assay performance characteristics is crucial. Consistency is ensured by the implementation of a rigorous validation, quality assurance, and quality control system [23,24,25]. New technologies, such as NGS, must be introduced carefully, with consideration given to comparability to results from other laboratories in the network and to historical data. Currently, most laboratories located in LMIC do not have access to NGS platforms. Until all WHO Network laboratories have the capability to implement NGS, individual laboratories that are doing so are required to report consensus sequences that mimic those generated by SS as closely as possible. To support this approach during this transitional period, a standardized threshold for reporting LAV that generates data comparable to those derived from SS is needed.
The National Institute of Allergy and Infectious Diseases (NIAID) Virology Quality Assurance (VQA) program provides a comprehensive quality assessment program for virologic assays for HIV, including drug resistance genotyping [26]. A crucial function of the VQA program is to ensure the validity and inter- and intra-laboratory comparability of virologic laboratory data generated for NIAID-supported clinical trials and research by the provision and analysis of proficiency testing panels. The VQA program also implements standards of performance for existing and state-of-the-art new virologic assays, develops and evaluates biostatistical methods relating to the assays, and acquires, tests, stores, and dispenses quality control materials and reagents. Since 2007, the VQA has provided proficiency testing specimens to the WHO HIVDR Laboratory Network [24]. This resource is well-suited to the investigation of NGS LAV thresholds that maximize the comparability of sequences from SS assays.
In this paper, we report for the first time an inter-laboratory comparison of HIV protease and reverse transcriptase sequences from an external quality assurance panel, comparing NGS sequences to Sanger sequences and NGS sequences between laboratories.
2. Materials and Methods
2.1. Specimens
Ten VQA HIVDR genotyping proficiency testing panel specimens (five from each of the two panels) were used. The specimens were prepared from patient plasma or cell culture virus stocks, and belonged to HIV-1 subtypes B, C, D, or F, at viral load loads ranging from 3656 to 29,139 copies/mL. Several specimens contained multiple drug resistance-associated mutations (DRMs), some of which were present as mixtures (Table 1).

Table 1.
Characteristics of Virology Quality Assurance (VQA) specimens used in this study.
2.2. Sequencing Methods
Ten laboratories participated in this evaluation study. The laboratories are numbered from 1 to 10. Six of the laboratories were from the WHO HIVDR Laboratory Network, and four were extra-network laboratories with extensive HIVDR testing and NGS experience. Each laboratory used its own RNA extraction, RT-PCR amplification, raw sequencing data analysis, and post-testing QA procedures (Table A1), but all used the Illumina MiSeq platform (Foster City, CA, USA). One laboratory (#1) used a unique molecular identifier approach to more accurately quantitate the number of amplified templates in each reaction [27,28].
The laboratories submitted consensus sequences for each specimen using LAV thresholds of 5%, 10%, 15%, and 20% (i.e., minor nucleic acid variations with frequencies below these thresholds were ignored, and all variations with frequencies above the threshold are included in the base call at that position). Lower thresholds were not evaluated because of the lack of data demonstrating the clinical relevance of LAV at less than 5%. The software used to generate the consensus sequences was not able to do so using the 20% threshold in laboratory 4, so there are no data for this laboratory in the 20% group. The consensus sequences spanned the protease (PR)-reverse transcriptase (RT) regions that encompass all DRM sites that contribute to the resistance to PR and RT inhibitors of interest to the WHO HIVDR surveillance program (PR 10–93 and RT 41–238), except those from laboratory 1 which did not cover RT amino acids 123 to 151.
2.3. Sequence Comparison
The SS consensus sequences for each specimen were generated by VQA based on over 30 results from independent laboratories that used an SS-based, FDA-approved commercial genotyping kit (ViroSeq or TruGene), using an 80% identity threshold. Where an 80% absolute agreement was not reached, an “N” was inserted at that position, and these positions were excluded from identity percentage calculations. The VQA SS consensus sequence covers protease codons 4–99 and reverse transcriptase 38–247; portions of the NGS sequences outside this region were excluded from the analysis of identity to the VQA consensus. A secondary analysis evaluating only the sequence at DRM codons (any position with a potential impact on the penalty score in the Stanford HIVdb algorithm, version 8.5) was also performed.
The sequences were aligned using Geneious software (version 11.1; San Diego, CA, USA) and analyzed in Microsoft Excel. To assess the extent to which the sequences between labs agreed with each other, without comparison to SS, the sequence identity at all positions was determined between all possible pairs of sequences for each specimen and threshold. Missing data (gaps) were ignored.
Sequence quality evaluation (i.e., assessing the presence of anomalies such as frameshifts, stop codons, APOBEC mutations, and unusual mutations) was performed with Stanford HIVdb (https://hivdb.stanford.edu/). The anomalies reported in the region not covered by laboratory 1 (RT 123–151) were ignored for this laboratory only.
Comparisons of percent identity between thresholds were performed using the Wilcoxon matched-pairs signed rank test and paired t-test (Prism 7, GraphPad, San Diego, CA, USA), and a random effects model with laboratory and specimen as random effects, cut-off values as fixed effects, and pairwise adjusted estimates of differences between cut-off values using SAS PROC MIXED.
3. Results
3.1. Comparison of NGS Sequences to VQA Sanger Consensus
Six of the ten laboratories generated results from all ten specimens. Three laboratories (4, 7, and 9) did not report results for one specimen each, and laboratory 8 did not report data for three specimens. There was no obvious association between assay failure and virus subtype or viral load.
The NGS sequences generated using the four different thresholds were compared to the VQA SS consensus sequence. Figure 1 displays the percentage nucleotide identity for each specimen, and for each laboratory. There was a strong tendency for the percent identity to increase as the threshold increased, especially in laboratories 1, 2, 3, 4, 9, and 10. The identity appeared to increase substantially from 5% to 15%, then increase only slightly at 20%. In laboratories 5, 6, 7, and 8, there was relatively less impact of threshold on agreement with the SS consensus for most specimens, and the identity at the lower thresholds was higher than in the other laboratories. For example, the mean percent identity at the 5% threshold was 99.5% in laboratory 8 and 99.1% in laboratory 7, compared to 98.2% in laboratories 1 and 9. The overall percent identity was slightly lower for specimen 24.1, which had 2.3% mixtures and a viral load of 7815 copies/mL.
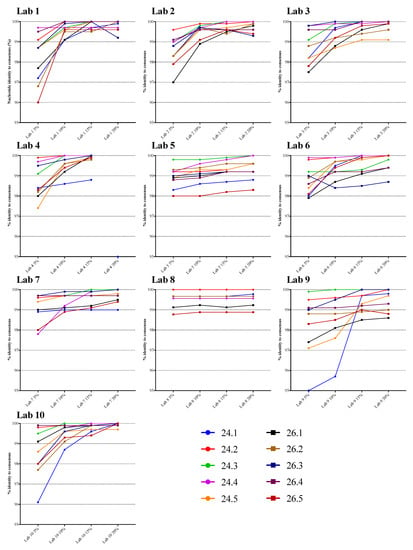
Figure 1.
Plots of next-generation sequencing (NGS)-derived PR-RT nucleotide sequence identity vs. VQA Sanger consensus at various thresholds. Each line represents one specimen from panel 24 (24.1 through 24.5) or 26 (26.1 through 26.5).
The mean percent nucleotide identity with the VQA SS consensus sequence across all results (specimens and laboratories) at the four different thresholds was highest at the 20% threshold (99.7%, the range in values was 98.3–100%), compared to 15% (99.6%, range of 98.2–100%), 10% (99.4%, range of 95.7–100%) or 5% (98.7%, range of 95.0–100%) (Table A2). In general, and as expected, the increased number of mismatches in the NGS sequences was a result of more mixed bases in the lower threshold NGS sequence but not in the VQA SS consensus. These mixed bases at lower thresholds may indicate LAV that SS is unable to detect, or they may reflect sequencing or analytic errors. All comparisons of percent identity between thresholds were statistically significant (p < 0.0001, Wilcoxon test or paired t-test; p < 0.01, random effects model; Table A3). The mean identity of each laboratory’s sequences was over 99% for all laboratories at the 15% and 20% thresholds, nine of ten at 10%, and two of ten at 5%.
The analysis of nucleotide sequence identity compared to SS was also performed considering only HIVDR-associated codons. The observed trends were similar, although the differences between the 15% and 20% thresholds were smaller.
3.2. Comparison of NGS Sequences Between Laboratories
While analyzing the level of identity between NGS and SS consensus sequences, we observed that the positions at which differences were located were not always the same between laboratories. Figure 2 shows a portion of the sequence alignment for specimen 24.1 in reverse transcriptase between amino acids 218 and 222. Mixed bases in the third position of codon 219 and 220 were more often detected at lower thresholds, but in different codons for different laboratories. This suggests that the reduced agreement at low thresholds described above is not simply a result of a consistent increase in sensitivity for the detection of LAV by NGS, but that variation in LAV detection between laboratories may be position dependent—especially at low thresholds.
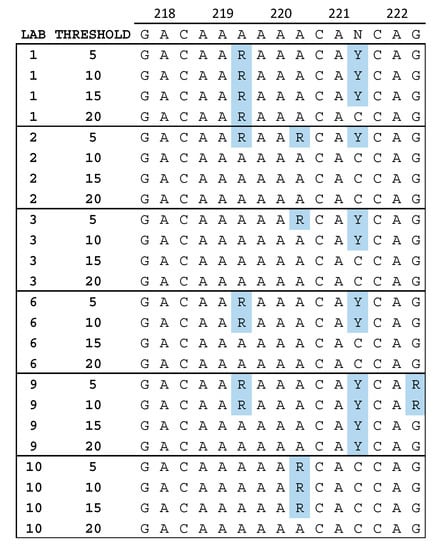
Figure 2.
Nucleotide sequence alignment for six laboratories. The VQA Sanger sequencing (SS) consensus is shown at the top. Mixtures of A and G (R) or C and T (Y) that were reported by some but not all laboratories are highlighted in blue. The sequences from laboratories 5, 7, and 8 did not contain any mixtures in this region, and those from laboratory 4 contained the Y in codon 221 at all thresholds reported (5%, 10%, and 15%).
To assess the degree of inter-laboratory sequence agreement, the individual sequences for each specimen were compared to the corresponding sequences from the other laboratories, for each specimen and threshold, and the pairwise sequence identity was calculated. The number of comparisons ranged from 28 (because of missing data from some laboratories) to 45. The mean percent identity between laboratories for each specimen and threshold is shown in Figure 3. There was a clear increase in sequence agreement as the threshold was raised for all specimens, except 26.5 for which the results for 10%, 15%, and 20% were similar to each other, but still higher than for 5%. The highest inter-laboratory agreement was observed for specimens 24.2 (99.9% at the 20% threshold), 24.3, and 24.4 (both 99.8%) while less agreement was observed for specimens 24.1 (98.4%) and 26.5 (98.1%). There was some correlation between the level of agreement and sequence heterogeneity in the specimen: specimens 24.2, 24.3, and 24.4 had no or very few mixed bases in the VQA SS consensus sequence, while 24.1, 26.3, and 26.5 had mixtures at more than 2% of positions (Table 1 and Figure 3). Specimen 26.5 had the lowest concordance overall, as well as the lowest viral load and the highest proportion of mixtures in the VQA SS consensus.
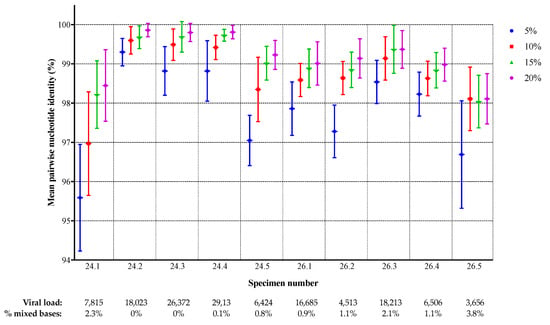
Figure 3.
Protease/reverse transcriptase nucleotide sequence concordance between laboratories. The mean percent identity with standard deviation is shown for each specimen and threshold.
3.3. Quality Assurance Anomalies
At low NGS mutation detection thresholds, it is more likely that sequence anomalies resulting from RT-PCR error and/or host DNA editing enzymes will be detected [29]. Sequence anomalies include frameshifts, stop codons, APOBEC mutations, and “unusual” mutations (amino acid changes that have only rarely or never been observed before in the Stanford HIV sequence database). This could be at least partly a result of artefactual codon sequences that can occur when two or more bases in the codon are mixed; for example, “YGR” may in fact be a mixture of TGG (tryptophan) and CGA (arginine), but could also be translated as TGA (stop) or CGG (arginine). The total number of sequence anomalies for all laboratories detected at 5, 10, 15, or 20% using the Stanford HIVdb sequence analysis tool was 51, 26, 15, and 12, respectively. The numbers of each type of anomaly are shown in Figure 4. All four types of QA anomalies were most common when the 5% threshold was used, and least common at 15% or 20%. No QA anomalies were found in the VQA SS consensus sequences, apart from those that resulted from the “N” where 80% consensus was not reached.
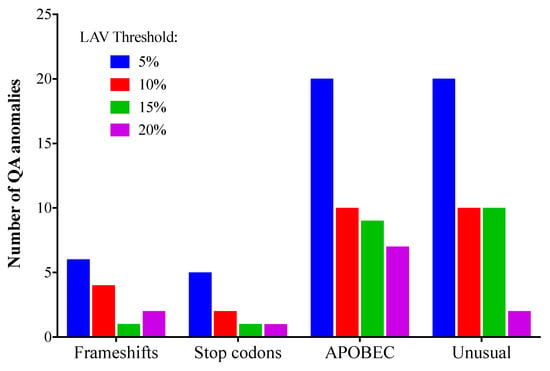
Figure 4.
Sequence quality assurance anomalies (total for all laboratories) at different low-abundance variant (LAV) thresholds. Sequence quality evaluation was performed with Stanford HIVdb (https://hivdb.stanford.edu/). HIVdb sequence analysis was performed using NGS sequences generated using the 5%, 10%, 15%, or 20% threshold levels.
4. Discussion
The global surveillance of HIVDR relies on high quality, standardized methods for detecting DRMs in specimens from survey participants. The current standard platform method is SS, and until NGS is accessible to all laboratories that are contributing sequence data for HIVDR surveys, those that adopt NGS-based methods must be able to produce sequences that have the same performance characteristics as laboratories using SS. It is recognized that this transitional approach may initially lead to the under-utilization of some potential advantages of NGS, including better sensitivity for LAV detection and de-convolution of complex mixtures.
We evaluated several thresholds for reporting LAV from NGS data and demonstrated that the similarity to SS data was highest when a 20% threshold was applied. Furthermore, inter-laboratory comparability was also highest at this threshold. Previous studies that evaluated the sensitivity of SS for LAV detection or that compared sensitive point mutation assays and SS are consistent with the 20% threshold [30,31,32,33].
The decreased agreement between NGS and SS data at thresholds below 20% might have been considered predictable, based on the concept that additional mixtures are expected to be reported in the NGS sequences, as LAV present at low frequency are detected more frequently. However, we found that the observed decreased agreement below 20% is not solely the result of the better sensitivity of NGS, since the inter-laboratory agreement also decreased as the threshold was lowered. These observations suggest that the detection of LAV can be subject to stochastic effects that may not be robustly repeatable or reproducible between methods or laboratories. Importantly, our results raise concerns about accuracy and inter-laboratory (and perhaps also intra-laboratory) reproducibility at low thresholds such as 5%, and strongly suggest that if even lower thresholds were to be used, the reproducibility would continue to decline. In the future, if the clinical significance of drug resistant LAV is conclusively shown to increase the predictive value of HIVDR genotyping for clinical outcomes and a threshold below 20% is established, there may be enough impetus to transition laboratory assays that support the public health surveillance of HIVDR to NGS platforms. At that time, it will be important to gain a better understanding of the sources of inter-laboratory variability in sequence determination and implement ways to minimize their impact. Both processes would be greatly facilitated by the development and use of standardized reference and/or control material with relevant LAV at specific frequencies, for use in external QA programs and/or assay optimization and validation. Other challenges inherent in the capacity of laboratories in LMIC to perform NGS-based HIVDR genotyping (e.g., instrument cost, operator training, and the availability of technical support) will also require attention and significant resources.
The low inter-laboratory reproducibility of NGS sequences may also be at least partly related to input amplifiable copy number, specimen sequence heterogeneity, position in the genome (Figure 2 and Figure 3), and differences in the bioinformatics pipelines used. This complexity strongly suggests that clinical specimens with these characteristics should be included in external QA programs and inter-laboratory comparisons, rather than virus clones or reconstructed mixtures. With regard to differences in pipelines, Lee et al. [34] evaluated the data from six of the ten laboratories included in this study using five pipelines and reported that sensitivity was good (over 99%) using thresholds as low as 1%, but specificity was low (82.4%) at the 1% threshold; they therefore suggested that a 2% threshold would be more reliable than 1%.
Our study has several limitations. (1) One or more of the NGS methods used may include unique aspects that make them more accurate than others or than those used to generate the SS standard comparator sequences (for example, the use of unique molecular identifiers, different input RNA volumes, or bioinformatic analysis pipelines). In this case, that method might generate sequences that are very different from the gold standard, but in reality, closer to the correct result. (2) Because thresholds over 20% were not evaluated, it is possible that the optimal threshold is higher; it is expected that at very high thresholds, a decrease in concordance would be seen, as mixtures start to be under-called. (3) We have analyzed similarity to SS across the entire sequence uniformly; different optimal thresholds may exist for specific DRM positions, due to the context dependence of chromatogram peak height in SS raw data. For example, a LAV that involves a change from a “weak” A base to a “strong” G might be expected to reach maximum identity at thresholds lower than 20%. (4) It is possible that many of the sites where the variability between laboratories is introduced involve synonymous mutations that would not have any impact on the predicted amino acid sequence and DR interpretation. (5) All participating laboratories used the Illumina MiSeq platform, limiting the application of our conclusions to that platform. Finally, (6) several assay variables that could be hypothesized to have an impact on NGS assay reproducibility have not been explored, including PCR reaction input copy number, sampling bias related to procedural bottlenecks, PCR-associated errors, and analysis pipeline methodology.
5. Conclusions
Of the LAV thresholds tested here, 20% led to PR-RT NGS consensus sequences that matched SS most closely and had the highest level of inter-laboratory agreement. Using the 20% threshold, we observed excellent agreement between NGS and SS, but significant differences at lower thresholds that may limit their use for global surveillance of HIVDR. Understanding how variation in NGS methods influences sequence quality is essential for NGS-based HIV-1 drug resistance genotyping and other applications where LAV detection reproducibility is important.
Author Contributions
Conceptualization, N.T.P. and C.J.; methodology, N.T.P. and C.J.; formal analysis, N.T.P. and D.J.Z.; writing—original draft preparation, N.T.P.; writing—review and editing, all co-authors; visualization, N.T.P.; project administration, N.T.P. and C.J. All authors have read and agreed to the published version of the manuscript.
Funding
S.A.-R. was supported by the Mexican Government (Comisión de Equidad y Género de las Legislaturas LX-LXI y Comisión de Igualdad de Género de la Legislatura LXII de la H. Cámara de Diputados de la República Mexicana), and Consejo Nacional de Ciencia y Tecnología (CONACyT SALUD-2017-01-289725). H.J. was supported by the Canadian Federal Initiative to Address HIV and AIDS and the Public Health Agency of Canada. R.K. and M.H. were supported in part by grants from the NIH R01AI136058, R01AI120792, K24AI134359 and P30AI042853. V.R.-A. was supported by the National Institutes of Minority Health and Health Disparities, grant number U54 MD007579. R.S. was supported by R01AI140970 and P30 AI50410. S.Z. was supported by NIAID grant 5R01 AI140970. S.H.E. and Y.Z. were supported by UM1AI069613. VQA activities were performed under contract HHSN272201200023C from NIAID.
Acknowledgments
This study was presented at the 27th International Workshop on HIV Drug Resistance and Treatment Strategies in Johannesburg, South Africa, on October 23, 2018. We thank the High-throughput Sequencing Facility at University of North Carolina, Chapel Hill; Mia Coetzer, Brown University; Adriana Alvarez, Hodan Mohamed and Carmen Manso from the Antiviral Unit, Public Health England; the members of the BCCfE clinical HIV genotyping team for specimen processing and data collection, and Don Kirkby and Jeff Knaggs for their contributions to software development and data analysis; Andrea Arias and Raphael Sánchez (Ponce Health Sciences University-Ponce Research Institute) for their contributions in specimen processing and data analysis; Silvia Bertagnolio, WHO, Geneva, Switzerland and Paul Sandstrom (PHAC) for administrative support.
Conflicts of Interest
N.P. is a consultant to the WHO HIVDR surveillance team and has performed contract work for Abbott Molecular, Aldatu Biosciences, Gilead Sciences, Roche Molecular Systems, Stanford University, and ThermoFisher Scientific. C.J.B. has received honoraria from Gilead Canada paid to his institution. The University of North Carolina is pursuing IP protection for Primer ID, and R.S. is listed as a co-inventor and has received nominal royalties.
Appendix A

Table A1.
Assay details.
Table A1.
Assay details.
| Laboratory ID | RNA Extraction Method (Specimen Volume) | RT-PCR Amplification Strategy | Negative Control | % of Extracted RNA Used | Coverage (PR, RT aa) | Minimum Read Depth a | Minimum Variant Count b | Analysis Pipeline |
|---|---|---|---|---|---|---|---|---|
| 1 | QIAamp Viral RNA Mini kit (0.14 mL) | RT with primerID, then nested PCR | Water | 50% | PR 1–99, RT 34–122 and 152–236 | Varies | NA c | TCS pipeline in house |
| 2 | ViroSeq RNA extraction kit (0.5 mL) | RT then nested PCR | Water | 10% | PR 1–99, RT 1–440 | 1000 | 1000 | CLC Genomics Workbench and In-house |
| 3 | QIAamp Viral RNA Mini kit (1 mL) | One-step RT-PCR then nested PCR | Water | 10% | PR 6–99, RT 1–251 | 1000 | 50 | HyDRA [35] |
| 4 | MagnaPure LC (0.5 mL) | One-step RT-PCR, then nested PCR | Water | 30% | PR 1–99, RT 1–250 | 330 | NA c | Geneious |
| 5 | QIAamp Viral RNA Mini kit (0.14 mL) | One-step RT-PCR then nested PCR | Water | 10% | PR 1–99 RT 1–300 | 100 | 5 | Trim Galore!, HydDRA [35] |
| 6 | NucliSENS easyMAG (0.4 mL) | One-step RT-PCR, then nested PCR | Water | ~9% | PR 1–99, RT 1–250 | 100 | 5 | HyDRA [35] |
| 7 | QIAamp UltraSens Virus kit (0.5 mL) | Primary RT-PCR, then nested PCR | Fetal bovine serum | 16.7% | PR 5–99, RT 1–320 | 1000 | NA c | In-house [36] |
| 8 | QIAamp Viral RNA Mini kit (0.14 mL) | One-step RT-PCR then nested PCR | Water | 25 % | PR 1–99, RT 1–440 | 1000 | 10 | PASeq.org [35] |
| 9 | EZ1 Advance XL (variable) d | RT then nested PCR | Water | 16.7% | PR 1–99, RT 1–240 | 1000 | 10 e | Hivmmer [9] |
| 10 | NucliSENS easyMAG (0.5 mL) | RT then nested PCR | Water | 6.7% | PR 1–99, RT 1–400 or 1–240 | 100 | NA c | MiCall [35] |
a minimum number of reads required for data quality assurance. b minimum number of individual variants required for reporting. c in some analysis pipelines, the minimum variant count is not specified, although in practice is defined by the minimum coverage and variant proportion. d volume adjusted based on viral load to contain at least 5000 copies. e and ≥1% of total coverage at site.

Table A2.
Nucleotide sequence identity vs. VQA Sanger consensus at different variant thresholds.
Table A2.
Nucleotide sequence identity vs. VQA Sanger consensus at different variant thresholds.
| 5% | 10% | 15% | 20% | |
|---|---|---|---|---|
| Number | 94 | 94 | 94 | 85 |
| Minimum | 95.0 | 95.7 | 98.2 | 98.3 |
| Median | 98.9 | 99.6 | 99.7 | 99.9 |
| Mean | 98.7 | 99.4 | 99.6 | 99.7 |
| Std. Deviation | 0.95 | 0.63 | 0.41 | 0.40 |
| Lower 95% CI of mean | 98.5 | 99.2 | 99.5 | 99.6 |
| Upper 95% CI of mean | 98.9 | 99.5 | 99.7 | 99.8 |

Table A3.
Random Effects model testing differences between thresholds.
Table A3.
Random Effects model testing differences between thresholds.
| Comparison | N | Rand Eff Model Mean (SEM) | Rand Eff Model p-value Test = 0 | Mean Diff | Median Diff | SD Diff | Min Diff | Max Diff | Paired t p value | Sign Test p value | Sign Rank p value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 10% vs 5% | 94 | 0.0062 (0.0016) | 0.004 | 0.0065 | 0.0044 | 0.0071 | −0.0056 | 0.0359 | 0 | 0 | 0 |
| 15% vs 5% | 94 | 0.0087 (0.0020) | 0.002 | 0.009 | 0.0056 | 0.0094 | −0.0052 | 0.0466 | 0 | 0 | 0 |
| 15% vs 10% | 94 | 0.0025 (0.00076) | 0.01 | 0.0025 | 0.0011 | 0.0048 | −0.0028 | 0.04 | 0.0000022 | 0 | 0 |
| 20% vs 5% | 85 | 0.0094 (0.0022) | 0.003 | 0.0097 | 0.0055 | 0.01 | −0.0034 | 0.0477 | 0 | 0 | 0 |
| 20% vs 10% | 85 | 0.0033 (0.00091) | 0.007 | 0.0034 | 0.0022 | 0.0055 | −0.0063 | 0.0411 | 0.0000002 | 0 | 0 |
| 20% vs 15% | 85 | 0.00084 (0.0002) | 0.003 | 0.0008 | 0 | 0.0017 | −0.0075 | 0.0056 | 0.0000265 | 0 | 0.0000012 |
References
- Parikh, U.M.; McCormick, K.; van Zyl, G.; Mellors, J.W. Future technologies for monitoring HIV drug resistance and cure. Curr. Opin. HIV AIDS 2017, 12, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Casadella, M.; Paredes, R. Deep sequencing for HIV-1 clinical management. Virus Res. 2017, 239, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Van Laethem, K.; Theys, K.; Vandamme, A.M. HIV-1 genotypic drug resistance testing: digging deep, reaching wide? Curr. Opin. Virol. 2015, 14, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Barzon, L.; Lavezzo, E.; Costanzi, G.; Franchin, E.; Toppo, S.; Palu, G. Next-generation sequencing technologies in diagnostic virology. J. Clin. Virol. 2013, 58, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Z.; Paredes, R.; Ribaudo, H.J.; Svarovskaia, E.S.; Metzner, K.J.; Kozal, M.J.; Hullsiek, K.H.; Balduin, M.; Jakobsen, M.R.; Geretti, A.M.; et al. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA 2011, 305, 1327–1335. [Google Scholar] [CrossRef]
- Paredes, R.; Lalama, C.M.; Ribaudo, H.J.; Schackman, B.R.; Shikuma, C.; Giguel, F.; Meyer, W.A., 3rd; Johnson, V.A.; Fiscus, S.A.; D’Aquila, R.T.; et al. Pre-existing minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J. Infect. Dis. 2010, 201, 662–671. [Google Scholar] [CrossRef]
- Ji, H.; Enns, E.; Brumme, C.J.; Parkin, N.; Howison, M.; Lee, E.R.; Capina, R.; Marinier, E.; Avila-Rios, S.; Sandstrom, P.; et al. Bioinformatic data processing pipelines in support of next-generation sequencing-based HIV drug resistance testing: the Winnipeg Consensus. J. Int. AIDS Soc. 2018, 21, e25193. [Google Scholar] [CrossRef]
- Taylor, T.; Lee, E.R.; Nykoluk, M.; Enns, E.; Liang, B.; Capina, R.; Gauthier, M.K.; Domselaar, G.V.; Sandstrom, P.; Brooks, J.; et al. A MiSeq-HyDRA platform for enhanced HIV drug resistance genotyping and surveillance. Sci Rep. 2019, 9, 8970. [Google Scholar] [CrossRef]
- Howison, M.; Coetzer, M.; Kantor, R. Measurement error and variant-calling in deep Illumina sequencing of HIV. Bioinformatics 2019, 35, 2029–2035. [Google Scholar] [CrossRef]
- Raymond, S.; Nicot, F.; Carcenac, R.; Lefebvre, C.; Jeanne, N.; Saune, K.; Delobel, P.; Izopet, J. HIV-1 genotypic resistance testing using the Vela automated next-generation sequencing platform. J. Antimicrob. Chemother. 2018, 73, 1152–1157. [Google Scholar] [CrossRef]
- Weber, J.; Volkova, I.; Sahoo, M.K.; Tzou, P.L.; Shafer, R.W.; Pinsky, B.A. Prospective evaluation of the vela diagnostics next-generation sequencing platform for HIV-1 genotypic resistance testing. J. Mol. Diagn. 2019, 21, 961–970. [Google Scholar] [CrossRef] [PubMed]
- May, S.; Adamska, E.; Tang, J. Evaluation of Vela Diagnostics HIV-1 genotyping assay on an automated next generation sequencing platform. J. Clin. Virol. 2020, 127, 104376. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Diaz, A.; McCormick, A.; Booth, C.; Gonzalez, D.; Sayada, C.; Haque, T.; Johnson, M.; Webster, D. Analysis of transmitted HIV-1 drug resistance using 454 ultra-deep-sequencing and the DeepChek®-HIV system. J. Int. AIDS Soc. 2014, 17 (4 Suppl 3), 19752. [Google Scholar] [CrossRef]
- Li, J.Z.; Kuritzkes, D.R. Clinical implications of HIV-1 minority variants. Clin. Infect. Dis. 2013, 56, 1667–1674. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, J.Z.; Paredes, R.; Ribaudo, H.J.; Kozal, M.J.; Svarovskaia, E.S.; Johnson, J.A.; Geretti, A.M.; Metzner, K.J.; Jakobsen, M.R.; Hullsiek, K.H.; et al. Impact of minority nonnucleoside reverse transcriptase inhibitor resistance mutations on resistance genotype after virologic failure. J. Infect. Dis. 2013, 207, 893–897. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Z.; Paredes, R.; Ribaudo, H.J.; Svarovskaia, E.S.; Kozal, M.J.; Hullsiek, K.H.; Miller, M.D.; Bangsberg, D.R.; Kuritzkes, D.R. Relationship between minority nonnucleoside reverse transcriptase inhibitor resistance mutations, adherence, and the risk of virologic failure. AIDS 2012, 26, 185–192. [Google Scholar] [CrossRef]
- Inzaule, S.C.; Hamers, R.L.; Noguera-Julian, M.; Casadella, M.; Parera, M.; Kityo, C.; Steegen, K.; Naniche, D.; Clotet, B.; Rinke de Wit, T.F.; et al. Clinically relevant thresholds for ultrasensitive HIV drug resistance testing: a multi-country nested case-control study. Lancet HIV 2018, 5, e638–e646. [Google Scholar] [CrossRef]
- Mbunkah, H.A.; Bertagnolio, S.; Hamers, R.L.; Hunt, G.; Inzaule, S.; Rinke De Wit, T.F.; Paredes, R.; Parkin, N.T.; Jordan, M.R.; Metzner, K.J.; et al. Low-abundance drug-resistant HIV-1 variants in antiretroviral drug-naive individuals: A systematic review of detection methods, prevalence, and clinical impact. J. Infect. Dis. 2020, 221, 1584–1597. [Google Scholar] [CrossRef]
- Avila-Rios, S.; Garcia-Morales, C.; Matias-Florentino, M.; Romero-Mora, K.A.; Tapia-Trejo, D.; Quiroz-Morales, V.S.; Reyes-Gopar, H.; Ji, H.; Sandstrom, P.; Casillas-Rodriguez, J.; et al. Pretreatment HIV-drug resistance in Mexico and its impact on the effectiveness of first-line antiretroviral therapy: a nationally representative 2015 WHO survey. Lancet HIV 2016, 3, e579–e591. [Google Scholar] [CrossRef]
- World Health Organization. WHO HIV Drug Resistance Report 2012; World Health Organization: Geneva, Switzerland, 2012. [Google Scholar]
- World Health Organization. WHO HIV Drug Resistance Report 2017; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- World Health Organization. WHO HIV Drug Resistance Report 2019; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Parkin, N.; de Mendoza, C.; Schuurman, R.; Jennings, C.; Bremer, J.; Jordan, M.R.; Bertagnolio, S. Evaluation of in-house genotyping assay performance using dried blood spot specimens in the Global World Health Organization laboratory network. Clin. Infect. Dis. 2012, 54 (Suppl 4), S273–S279. [Google Scholar] [CrossRef]
- Parkin, N.; Bremer, J.; Bertagnolio, S. Genotyping external quality assurance in the World Health Organization HIV drug resistance laboratory network during 2007–2010. Clin. Infect. Dis. 2012, 54 (Suppl 4), S266–S272. [Google Scholar] [CrossRef]
- World Health Organization. WHO/HIVResNet HIV Drug Resistance Laboratory Operational Framework; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Huang, D.D.; Bremer, J.W.; Brambilla, D.J.; Palumbo, P.E.; Aldrovandi, G.; Eshleman, S.; Brown, C.; Fiscus, S.; Frenkel, L.; Hamdan, H.; et al. Model for assessment of proficiency of human immunodeficiency virus type 1 sequencing-based genotypic antiretroviral assays. J. Clin. Microbiol. 2005, 43, 3963–3970. [Google Scholar] [CrossRef] [PubMed]
- Jabara, C.B.; Jones, C.D.; Roach, J.; Anderson, J.A.; Swanstrom, R. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc. Natl. Acad. Sci. USA 2011, 108, 20166–20171. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Jones, C.; Mieczkowski, P.; Swanstrom, R. Primer ID validates template sampling depth and greatly reduces the error rate of next-generation sequencing of HIV-1 genomic RNA populations. J. Virol. 2015, 89, 8540–8555. [Google Scholar] [CrossRef] [PubMed]
- Tzou, P.L.; Kosakovsky Pond, S.L.; Avila-Rios, S.; Holmes, S.P.; Kantor, R.; Shafer, R.W. Analysis of unusual and signature APOBEC-mutations in HIV-1 pol next-generation sequences. PLoS ONE 2020, 15, e0225352. [Google Scholar] [CrossRef] [PubMed]
- Church, J.D.; Jones, D.; Flys, T.; Hoover, D.; Marlowe, N.; Chen, S.; Shi, C.; Eshleman, J.R.; Guay, L.A.; Jackson, J.B.; et al. Sensitivity of the ViroSeq HIV-1 genotyping system for detection of the K103N resistance mutation in HIV-1 subtypes A, C, and D. J. Mol. Diagn. 2006, 8, 430–432; [Google Scholar] [CrossRef] [PubMed]
- Leitner, T.; Halapi, E.; Scarlatti, G.; Rossi, P.; Albert, J.; Fenyo, E.M.; Uhlen, M. Analysis of heterogeneous viral populations by direct DNA sequencing. Biotechniques 1993, 15, 120–127. [Google Scholar]
- Halvas, E.K.; Aldrovandi, G.M.; Balfe, P.; Beck, I.A.; Boltz, V.F.; Coffin, J.M.; Frenkel, L.M.; Hazelwood, J.D.; Johnson, V.A.; Kearney, M.; et al. Blinded, multicenter comparison of methods to detect a drug-resistant mutant of human immunodeficiency virus type 1 at low frequency. J. Clin. Microbiol. 2006, 44, 2612–2614. [Google Scholar] [CrossRef]
- Larder, B.A.; Kohli, A.; Kellam, P.; Kemp, S.D.; Kronick, M.; Henfrey, R.D. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature 1993, 365, 671–673. [Google Scholar] [CrossRef]
- Lee, E.R.; Parkin, N.; Jennings, C.; Brumme, C.J.; Enns, E.; Casadella, M.; Howison, M.; Coetzer, M.; Avila-Rios, S.; Capina, R.; et al. Performance comparison of next generation sequencing analysis pipelines for HIV-1 drug resistance testing. Sci. Rep. 2020, 10, 1634. [Google Scholar] [CrossRef]
- Noguera-Julian, M.; Edgil, D.; Harrigan, P.R.; Sandstrom, P.; Godfrey, C.; Paredes, R. Next-Generation Human Immunodeficiency Virus sequencing for patient management and drug resistance surveillance. J. Infect. Dis. 2017, 216 (suppl_9), S829–s833. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, E.; Chan, Y.T.; Aghaizu, A.; Bibby, D.F.; Murphy, G.; Tosswill, J.; Harris, R.J.; Myers, R.; Field, N.; Delpech, V.; et al. Enhanced surveillance of HIV-1 drug resistance in recently infected MSM in the UK. J. Antimicrob. Chemother. 2017, 72, 227–234. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).