Abstract
The RNA interference (RNAi) pathway is a potent antiviral defense mechanism in plants and invertebrates, in response to which viruses evolved suppressors of RNAi. In mammals, the first line of defense is mediated by the type I interferon system (IFN); however, the degree to which RNAi contributes to antiviral defense is still not completely understood. Recent work suggests that antiviral RNAi is active in undifferentiated stem cells and that antiviral RNAi can be uncovered in differentiated cells in which the IFN system is inactive or in infections with viruses lacking putative viral suppressors of RNAi. In this review, we describe the mechanism of RNAi and its antiviral functions in insects and mammals. We draw parallels and highlight differences between (antiviral) RNAi in these classes of animals and discuss open questions for future research.
1. Introduction
RNA interference (RNAi) or RNA silencing was first described in the model organism Caenorhabditis elegans [1] and following this ground-breaking discovery, studies in the field of small, noncoding RNAs have advanced tremendously. RNAi acts, with variations, in all eukaryotes ranging from unicellular organisms to complex species from the plant and animal kingdoms [2]. The key concept of all RNA silencing pathways is the association of single-stranded small RNAs of 20–30 nucleotides (nt) to a protein of the Argonaute superfamily [3,4]. In animals, three classes of small RNAs exist: small interfering RNAs (siRNAs), microRNAs (miRNAs) and PIWI-interacting RNAs (piRNAs) [2,5]. These RNAs guide Argonaute proteins onto target RNAs via Watson-Crick base pairing, usually resulting in gene silencing [6]. Whereas all three pathways adhere to the general concept of RNA silencing pathways, they differ in the mechanism for small RNA biogenesis and effector functions. For example, biogenesis of siRNAs and miRNAs depends on processing of double-stranded RNA (dsRNA) precursors into small RNAs by RNase-III Dicer enzymes [6], whereas piRNA biogenesis is Dicer independent.
Early on, it was recognized that RNAi could be a mechanism for antiviral defense, and, in fact, siRNAs were first detected in virus-infected plants [7,8,9]. It is now well established that RNAi is a major defense mechanism against parasitic nucleic acids in diverse organisms, including fungi, plants, and invertebrates [10,11,12]. Thus, recognition and processing of viral dsRNA into viral siRNAs (vsiRNAs) initiates a potent antiviral RNAi response that restricts virus accumulation. However, even though the mechanism of RNAi is evolutionarily conserved in mammals, the degree to which it contributes to antiviral defense has been a matter of debate. Positive and negative-sense RNA viruses were recently proposed to be a substrate for the RNAi pathway in several mammalian cell culture and animal models [13,14,15], yet conflicting evidence has also emerged in several studies that failed to detect vsiRNAs [16,17,18,19]. In vertebrates, RNAi coincides with the dsRNA-activated protein-based interferon response and recent findings suggest that mammalian RNAi is inhibited by the interferon response, suggestive of competition between both pathways [20,21].
In this review, we will discuss recent work on the antiviral function of RNAi in mammals, focusing on negative and positive-sense RNA viruses (excluding retroviruses). We will first describe the principal concepts of RNAi in insects and mammals (for a review on RNA silencing in plants, see [10]) and briefly discuss interferon-based antiviral immunity in mammals. Finally, we will discuss the antiviral activity of RNAi in insects and different mammalian experimental systems. Special attention will be given to stem cells, which seem to have specific characteristics, both in the interferon response and antiviral RNAi. To avoid ambiguity, we will only consider “classical” antiviral RNAi, in which viral dsRNA is processed into viral siRNAs to limit virus infection; we will not consider miRNA-dependent effects on virus replication.
2. The Mechanism of RNAi
Although RNA silencing pathways adhere to the same general concepts, paralogs of Dicer and Argonaute genes have emerged via duplications during eukaryotic evolution. This, along with the recruitment of different accessory proteins and co-factors, has led to functional diversification or specialization in different organisms [22]. For example, insects such as the fruit fly Drosophila melanogaster encode two Dicer genes, of which Dicer-1 mediates miRNA biogenesis, whereas Dicer-2 is responsible for siRNA biogenesis [6]. In contrast, mammals only encode a single Dicer that generates both miRNAs and siRNAs. Likewise, Argonaute-2 is responsible for siRNA-mediated target RNA cleavage in insects, whereas Argonaute-1 mediates miRNA-dependent gene silencing. Mammals, in contrast, encode four Argonaute genes, all of which engage in microRNA-guided gene silencing, and only Argonaute-2 is capable of cleaving target RNAs (also referred to as slicing) to mediate siRNA-dependent RNAi.
Below, we will discuss the siRNA and miRNA pathways of insects and mammals in more detail. Although the piRNA pathway has been suggested to mediate antiviral defense, especially in vector mosquitoes [23], piRNAs have not been studied in the context of viral infection in mammals and will not be discussed.
2.1. The siRNA Pathway in Insects
The “classical” RNAi mechanism, uncovered by Fire and Mello [1], is triggered by the presence of double-stranded RNA (dsRNA) in the cytoplasm. This initiates a series of processing steps that eventually results in the production of siRNAs that associate with an Argonaute protein (Figure 1). In insects, the RNase-III enzyme Dicer-2 recognizes cytoplasmic dsRNA and cleaves it into 21 nt siRNA duplexes with characteristic two-nucleotide overhangs at the 3’ ends of both strands (Figure 2) [24,25,26,27]. One of the two strands (the guide strand) is selectively incorporated into the RNA-induced silencing complex (RISC) with at its catalytic core the Argonaute-2 (Ago2) protein. The complementary strand (the passenger strand) is degraded in a process that requires Ago2 and the endonuclease Component 3 Promoter of RISC (C3PO) [28,29,30,31]. Selection of the guide and passenger strand is a non-stochastic process and involves the activity of the Dicer-2-associated co-factor R2D2 [32,33]. R2D2 probes the thermodynamic stability of the siRNA duplex and binds the more stable 5’ end, eventually defining the passenger strand. Dicer-2 selects the opposite strand that will be loaded as guide strand into Ago2 [34]. Dicer-2 processing and RISC loading is further promoted by the activity of co-factors including the dsRNA binding protein Loquacious (PD isoform, Loqs-PD), Ars2 and heat shock proteins [35,36,37,38]. These proteins enhance siRNA biogenesis by stabilizing the RNA-protein complexes or facilitating conformational changes during RISC loading. After the guide strand is stably bound by Ago2, it is 2’-O-methylated at the 3’ terminal nucleotide by the RNA methyl-transferase DmHen1 finalizing the maturation of an siRNA-loaded RISC [39].
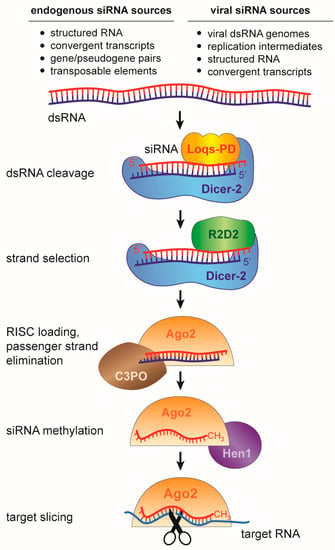
Figure 1.
The small interfering RNA (siRNA) pathway in Drosophila melanogaster. Double-stranded RNA precursors of different sources are processed by Dicer-2 into short interfering RNAs of ~21 nt in size. The siRNA duplex is loaded into an Argonaute2 containing RISC complex, where one strand (passenger) is degraded, and the guide strand is retained. The guide strand mediates target RNA recognition through Watson-Crick base pairing, followed by target cleavage (slicing) by Argonaute. Loqs-PD is required for endo-siRNA biogenesis, but dispensable for viral siRNA (vsiRNA) biogenesis.
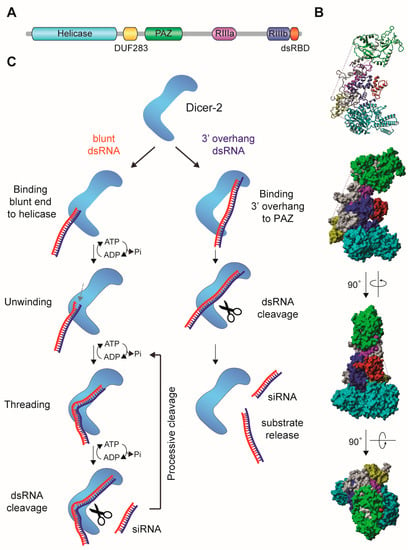
Figure 2.
Dicer proteins process double-stranded RNA (dsRNA) into small interfering RNA (siRNA). (A) Schematic representation of the domain organization of human Dicer protein [40]. RIIIa, RNase-IIIa; RIIIb, RNase-IIIb (B) Cryo-EM structure of human Dicer. Protein domains are colored in accordance to the scheme in A. The structure was determined by Liu et al. [41], and the published PDB file (5ZAM) was edited in Yasara View [42]. Drosophila Dicer-2 has a similar domain structure and L-shaped Cryo-EM structure as human Dicer [40]. (C) Schematic representation of the recognition and cleavage of dsRNA with a 3’ overhang and dsRNA with blunt termini by Drosophila Dicer-2, proposed by Sinha and colleagues [40]. Substrates with a 3’ overhang were proposed to bind the PAZ-Platform domains (referred to as PAZ in panel A) via the 3’ terminal overhang. Blunt-ended termini bind to the helicase domain and the dsRNA threads through this domain, after which cleavage occurs by the two RNaseIII domains. The latter mode results in processive, ATP-dependent cleavage of dsRNA and may contribute to efficient production of vsiRNAs for antiviral defense.
Two models of substrate processing depending on their termini have been proposed for Dicer-2 [40]. Substrates with 3’ overhangs are cleaved distributively by Dicer-2 in an ATP independent manner, releasing the dsRNA substrate after each cleavage. In contrast, dsRNA with blunt termini are locally unwound, with one of the strand threading through the helicase domain in an ATP dependent manner, after which the dsRNA re-anneals and becomes processively cleaved [40] (Figure 2C). Ago2-bound siRNAs recognize target RNAs via Watson-Crick base pairing and usually complementarity across the entire length of the siRNA/target duplex is required for efficient target cleavage. An exception is the first nucleotide of the siRNA, which is locked in a pocket of the Ago2 MID/PIWI domain [43] (Figure 3A,B). Upon formation of the siRNA/target RNA duplex, Ago2 cleaves the target RNA between nucleotide ten and eleven counted from the 5’ end of the siRNA (slicing, Figure 3C) [25,26,28,44]. This small RNA-mediated endonuclease activity (slicing) requires the catalytic DEDX tetrad (where X is D or H) in the PIWI domain of Argonaute proteins (Figure 3A) [45,46]. This motif is conserved amongst slicing-competent Argonaute proteins; nonetheless it is not sufficient for slicing activity since some slicing-incompetent Argonaute proteins also contain the motif [47]. After cleavage of target RNA, the slicing products are quickly degraded by cellular ribonucleases [48].
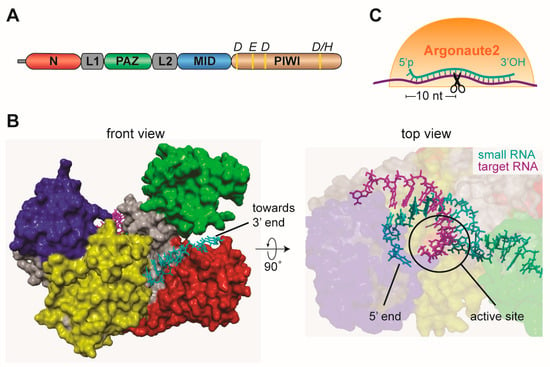
Figure 3.
Argonaute proteins are at the core of small RNA silencing pathways. (A) Schematic representation of the domain organization of mammalian Argonaute and the conserved residues required for slicer activity. (B) Crystal structure of human AGO2 in association with a guide RNA and a target RNA base pairing from nucleotide 2 to 8. Protein domains are colored in accordance to the scheme in A. The structure was determined by Schirle and colleagues [49] and the published PDB file (4W5Q) was edited in Yasara View. (C) Schematic representation of target slicing by Argonaute proteins.
Endogenous sources of dsRNA are long inverted repeats that fold into perfectly complementary hairpins or transcripts that are derived from convergent transcription. In addition, gene-pseudogene pairs and transposon insertions are potential sources of dsRNA when they express transcripts with full or partial complementarity (Figure 1). These genome-encoded dsRNA molecules are processed into endogenous siRNAs (endo-siRNA) that have been implicated in transposon control and anecdotally in the regulation of gene expression [50,51,52,53,54]. Yet, dsRNA is usually not very abundant in healthy, uninfected cells and the major function of this pathway seems to be defense against foreign dsRNA of viral origin [55] (discussed in Section 4).
2.2. The miRNA Pathway in Insects
miRNAs are an endogenous class of small RNAs, expressed by plants, animals, protists and even viruses [2]. Biogenesis of animal miRNAs resembles siRNA biogenesis, with some differences including the origin of precursor RNAs. miRNAs are processed from genome-encoded hairpins, called primary-miRNAs (pri-miRNAs) that are transcribed by RNA polymerase II and, less frequently, by RNA polymerase III [56,57,58]. Pri-miRNAs are typically a few kb in length [59] and harbor either single or multiple local stem-loop structures that undergo a series of maturation steps to generate an Argonaute-associated miRNA [60]. Typically, these stem-loops are ~80 nt in size and consist of two imperfectly base pairing arms, separated by a single-stranded loop region [61]. They are released from the pri-miRNA transcript in the nucleus by the microprocessor complex, consisting of the RNase-III enzyme Drosha and its co-factor Pasha [62,63,64,65,66]. Endonucleolytic cleavage by Drosha near the base of the hairpin produces the precursor miRNA (pre-miRNA), a ~60 nt small RNA hairpin with a two-nucleotide overhang at the 3’ end, indicative of RNase-III processing [66]. Subsequently, the pre-miRNA is exported from the nucleus via the Ran-GTP dependent nuclear exporter Exportin-5 [67,68,69,70]. In the cytoplasm, another RNase-III enzyme, Dicer-1, in a complex with the PB isoform of Loqs cleaves off the loop of the pre-miRNA resulting in an RNA heteroduplex with two-nucleotide overhangs at both 3’ ends [24,71,72]. One of the two strands is selectively incorporated into the Argonaute-1 containing miRNA induced silencing complex (miRISC) [73,74]. Strand selection is thought to be primarily based on the thermodynamic properties of the heteroduplex; usually the strand with the weaker stability at its 5’ end is incorporated into Ago1 [75,76]. The miRNA guides miRISC to target sites in the 3’ untranslated regions (UTR) of mRNAs, akin to target recognition in mammals [77] (described in Section 2.3).
2.3. RNAi Pathway in Mammals
Whereas the miRNA and siRNA pathways are largely independent in insects, siRNA and miRNA biogenesis and function in mammals depend on shared components (Figure 4), which results in crosstalk between these pathways. Like in insects, miRNAs in mammals are an abundant class of small RNAs of 21–22 nt in length [78] that are primarily produced from RNA polymerase II synthesized pri-miRNAs. These pri-miRNAs are processed into pre-miRNAs (pre-miRNAs) by the Microprocessor complex, consisting of the RNaseIII Drosha along with DGCR8 (DiGeorge Syndrome Critical Region 8) [79]. Pre-miRNAs are transported to the cytoplasm, where they are cleaved by Dicer into miRNA duplexes. These duplexes are loaded by Dicer and its co-factors TRBP (TAR RNA binding protein) and PACT (Protein kinase RNA activator) into an Argonaute (AGO) containing RISC complex, from which the passenger strand is eliminated. The RISC-associated mature miRNA base pairs with cognate messenger RNAs (mRNA), resulting in destabilization of target mRNAs or blocking their translation [24,61,80,81]. All four ubiquitously expressed mammalian AGO proteins mediate miRNA-mRNA interactions with approximately equivalent affinities [82,83,84] and overexpression experiments indicate that their miRNA binding patterns are similar [85,86].
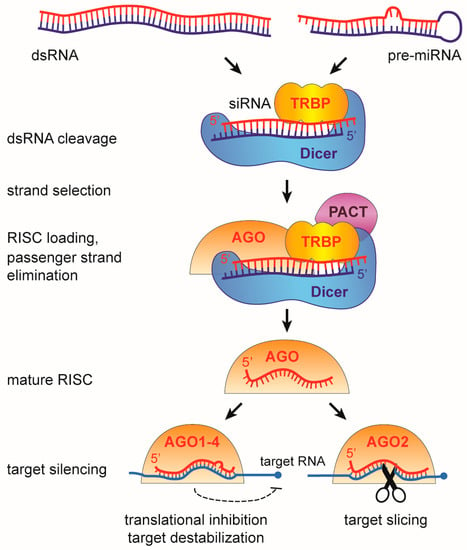
Figure 4.
The RNA interference (RNAi) pathway in mammals. A single Dicer protein processes long dsRNA into siRNAs and pre-miRNAs into miRNA duplexes. These small RNAs are loaded into an Argonaute containing RISC complex, from which one of the strands is eliminated and degraded. The other strand, referred to as guide strand (for siRNAs) or the mature miRNA (for miRNAs), is retained and used to guide Argonaute onto target RNAs, resulting in cleavage (siRNA) or translational inhibition or target RNA destabilization (miRNA). The scheme shows the cytoplasmic stage of the miRNA pathway; the nuclear stage (pri-miRNA transcription, processing, and pre-miRNA nuclear export) is not shown.
In contrast to canonical Dicer-dependent miRNAs, non-canonical miRNAs bypass processing by Dicer or the Microprocessor complex. These non-canonical miRNAs can be derived from introns, small nucleolar RNAs (snoRNAs), and tRNAs [87,88,89,90,91,92,93]. For example, the mirtron pathway, which is also found in D. melanogaster and C. elegans, produces pre-miRNAs by the processing of introns by spliceosomes and debranching enzymes in the nucleus [94]. Another non-canonical miRNA is produced by processing of snoRNA ACA45 in a Drosha/DGCR8 independent, but Dicer dependent manner [88].
The miRISC complex is guided by the miRNA to target sites typically located in the 3’ UTRs of mRNAs [77]. Target recognition is initiated by a short nucleotide stretch at the 5’ end of the miRNA (position 2–8), the so-called seed sequence, accompanied with various degree of base pairing at the 3’ end [77,95,96,97]. Mechanisms for miRNA-mediated gene silencing include translational repression, de-adenylation, and enhancement of mRNA decay [77,98,99]. The majority of mRNAs is estimated to be regulated by miRNAs [100], and post-transcriptional regulation by miRNAs is thus implicated in almost all cell biological processes.
Although miRNA-mediated gene regulation seems to be the dominant function of mammalian RNAi, early evidence has already indicated that the siRNA pathway is functional in mammals. Transfection of synthetic siRNAs or expression of short-hairpin RNAs (shRNAs) with complementarity to a gene of interest was found to induce robust and sequence-specific RNAi, without activation of the interferon response as siRNAs are too short to be detected by dsRNA sensors (discussed in Section 3.1) [25,101]. Moreover, long dsRNA was reported to be functional in gene knockdown in embryonal teratocarcinoma cell lines that are interferon defective [102,103,104].
RNAi in mammals is characterized by processing of dsRNA by Dicer into 21–23 nt short interfering RNAs (siRNAs) [105]. Subsequently, siRNAs are preferentially loaded onto AGO1 or AGO2, of which only AGO2 possesses slicing activity in mammals [83,86]. After elimination of the passenger strand, the guide strand directs AGO2 onto complementary mRNA through base pairing. In contrast to the seed-based target recognition of miRNAs, siRNA targeting requires base pairing of the entire small RNA, resulting in target RNA cleavage by AGO2. As in insects, target cleavage occurs between nucleotide ten and eleven, counted from the 5’ end of the siRNA [106].
The evolutionary conservation of AGO2-mediated target cleavage in mammals suggests important functions for this activity. AGO2 efficiently mediates target repression independent of its slicer activity, as miRNA-mediated gene silencing in AGO1, AGO3, and AGO4 deficient embryonic stem cells was comparable to control cells [83]. Yet, biogenesis of the non-canonical miRNA miR-451, implicated in the regulation of erythroid development, is Dicer-independent and instead depends on AGO2 catalysis [107]. In this case, the short length of the stem of only 17 bp likely explains why miR-451 fails to be processed by Dicer [108]. Besides being indispensable for miR-451 biogenesis, inactivation of AGO2 by insertional mutagenesis in mice results in a lethal phenotype as only wild-type and heterozygous offspring are observed [86]. In addition, loss of AGO2 results in a severe developmental phenotype, including a defect or failure in neural tube closure and mispatterning of brain structures [86]. The fact that AGO2 inactivation leads to these phenotypes in a background of wildtype AGO1, AGO3, and AGO4, which act redundantly in the miRNA pathway, suggests that slicing activity of AGO2 is important in development. Yet, biochemical or genetic evidence that slicing is required for the observed phenotypes is currently lacking. Evolutionary conservation of slicer activity would also be consistent with an antiviral function of AGO2 in mammals; this will be discussed in Section 5.
6. Summary and Open Questions
Vertebrates rely on the protein-based IFN response to combat viral infections, whereas the RNAi machinery, known for its potent antiviral activity in invertebrates, is conserved but primarily functions in gene regulation. Mammals encode a single Dicer protein and four AGO proteins, of which only AGO2 is slicer competent. The function of these proteins in both the miRNA and siRNA pathways makes it difficult to genetically dissect the role of the RNAi pathway in inhibiting viral replication. The notion that both the IFN response and RNAi rely on dsRNA to initiate the antiviral response adds another level of complexity.
With the advancement of next generation sequencing technologies, a growing body of evidence has emerged that supports a role for RNAi in antiviral defense in mammals. Detection of canonical vsiRNAs in ESCs infected with EMCV or Nodamura virus ΔB2 provided the first compelling evidence for a role of antiviral RNAi in mammals [13,14]. ESCs possess an attenuated immune response [145], caused by reduced gene expression of IFN-pathway components or, in some instances, failure to respond to dsRNA triggers [146]. It is now apparent that RNAi is suppressed by the IFN pathway, likely due to the action of one or more ISGs [21] and through the interaction of Dicer and LGP2 [20].
VSRs seem to play an important role in differentiated cells, demonstrated by the accumulation of vsiRNAs during Nodamura virus ΔB2 and HEV71 3A mutant virus infections [13,14,206]. These findings were complemented by the detection of AGO2-associated siRNAs in somatic cells infected with Influenza A virus ΔNS1 [15]. These studies indicate that VSRs may mask the antiviral RNAi response in mammals. This situation is markedly different from the situation in plants and insects, in which vsiRNAs are readily detected with most, if not all wildtype viruses analyzed, hinting at differences in processivity of Dicer enzymes or differences in accessibility of viral dsRNA in mammals and insects.
Antiviral RNAi thus seems to be affected by the cellular context, IFN responses, and viral counter-defense mechanisms (Figure 6). Important questions still remain for each of these aspects. (i) How does the cellular context affect the antiviral immune response? Are there tissue and cell type specific differences in antiviral RNAi? Why is the antiviral RNAi pathway functional in stem cells and why is this activity lost upon differentiation? For example, are there specific determinants in stem cells that favor RNAi over the IFN response? How does cell potency (e.g., toti-, pluri-, and multipotency) affect the dominant antiviral immune response, and do tissue stem cells use RNAi for antiviral defense? (ii) Which factors, beyond LGP2, contribute to the inhibition of RNAi in differentiated cells? What are the relative contributions of the IFN and RNAi responses to host defense. (iii) How widespread is RNAi suppression among mammalian viruses? Do mammalian viruses encode VSRs that suppress AGO2, and what is the course of infection of virus mutants lacking this activity? Answers to these questions will shed light on the sophisticated RNAi pathway and its functions in antiviral defense.
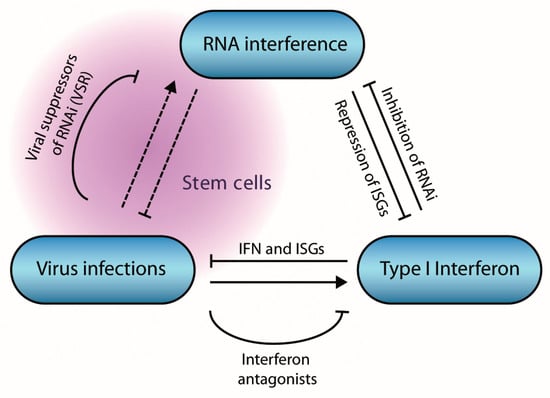
Figure 6.
Interactions between viruses, RNA interference (RNAi), and the interferon pathway in mammals. Virus infection induces the expression of type I interferons, leading to the expression of Interferon stimulated genes (ISGs) that collectively restrict virus infection. The interferon pathway inhibits RNAi via multiple mechanisms, whereas miRNAs inhibit expression of ISGs. Virus infection induces an antiviral RNAi response under specific conditions, in stem cells or in absence of viral suppressors of RNAi.
Acknowledgments
We thank members of the laboratory for fruitful discussions. This work was financially supported by a PhD fellowship from the Radboud Institute for Molecular Life Sciences (to S.S.), funded by the Graduate Programme of the Netherlands Organisation for Scientific Research (NWO), a VICI grant from the Netherlands Organization for Scientific Research (grant number 016.VICI.170.090), and a European Research Council Consolidator Grant under the European Union’s Seventh Framework Programme (grant number ERC CoG 615680) to R.P.v.R.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Ghildiyal, M.; Zamore, P.D. Small silencing RNAs: An expanding universe. Nat. Rev. Genet. 2009, 10, 94–108. [Google Scholar] [CrossRef] [PubMed]
- Schirle, N.T.; Kinberger, G.A.; Murray, H.F.; Lima, W.F.; Prakash, T.P.; MacRae, I.J. Structural analysis of human Argonaute-2 bound to a modified siRNA guide. J. Am. Chem. Soc. 2016, 138, 8694–8697. [Google Scholar] [CrossRef] [PubMed]
- Elkayam, E.; Kuhn, C.D.; Tocilj, A.; Haase, A.D.; Greene, E.M.; Hannon, G.J.; Joshua-Tor, L. The structure of human argonaute-2 in complex with miR-20a. Cell 2012, 150, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Ketting, R.F. The many faces of RNAi. Dev. Cell 2011, 20, 148–161. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Sontheimer, E.J. Origins and mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef]
- Hamilton, A.J.; Baulcombe, D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 1999, 286, 950–952. [Google Scholar] [CrossRef]
- Ratcliff, F.; Harrison, B.D.; Baulcombe, D.C. A similarity between viral defense and gene silencing in plants. Science 1997, 276, 1558–1560. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.W.; Voinnet, O. Antiviral immunity directed by small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Bronkhorst, A.W.; van Rij, R.P. The long and short of antiviral defense: Small RNA-based immunity in insects. Curr. Opin. Virol. 2014, 7, 19–28. [Google Scholar] [CrossRef]
- Dang, Y.; Yang, Q.; Xue, Z.; Liu, Y. RNA interference in fungi: Pathways, functions, and applications. Eukaryot. Cell 2011, 10, 1148–1155. [Google Scholar] [CrossRef]
- Maillard, P.V.; Ciaudo, C.; Marchais, A.; Li, Y.; Jay, F.; Ding, S.W.; Voinnet, O. Antiviral RNA interference in mammalian cells. Science 2013, 342, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lu, J.; Han, Y.; Fan, X.; Ding, S.W. RNA interference functions as an antiviral immunity mechanism in mammals. Science 2013, 342, 231–234. [Google Scholar] [CrossRef]
- Li, Y.; Basavappa, M.; Lu, J.; Dong, S.; Cronkite, D.A.; Prior, J.T.; Reinecker, H.C.; Hertzog, P.; Han, Y.; Li, W.X.; et al. Induction and suppression of antiviral RNA interference by influenza A virus in mammalian cells. Nat. Microbiol. 2016, 2, 16250. [Google Scholar] [CrossRef]
- Lin, Y.T.; Kincaid, R.P.; Arasappan, D.; Dowd, S.E.; Hunicke-Smith, S.P.; Sullivan, C.S. Small RNA profiling reveals antisense transcription throughout the KSHV genome and novel small RNAs. RNA 2010, 16, 1540–1558. [Google Scholar] [CrossRef] [PubMed]
- Schuster, S.; Tholen, L.E.; Overheul, G.J.; van Kuppeveld, F.J.M.; van Rij, R.P. Deletion of cytoplasmic double-stranded RNA sensors does not uncover viral small interfering rna production in human cells. mSphere 2017, 2, e00333-17. [Google Scholar] [CrossRef]
- Pfeffer, S.; Sewer, A.; Lagos-Quintana, M.; Sheridan, R.; Sander, C.; Grasser, F.A.; van Dyk, L.F.; Ho, C.K.; Shuman, S.; Chien, M.; et al. Identification of microRNAs of the herpesvirus family. Nat. Methods 2005, 2, 269–276. [Google Scholar] [CrossRef]
- Umbach, J.L.; Cullen, B.R. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009, 23, 1151–1164. [Google Scholar] [CrossRef]
- Van der Veen, A.G.; Maillard, P.V.; Schmidt, J.M.; Lee, S.A.; Deddouche-Grass, S.; Borg, A.; Kjaer, S.; Snijders, A.P.; Reis e Sousa, C. The RIG-I-like receptor LGP2 inhibits Dicer-dependent processing of long double-stranded RNA and blocks RNA interference in mammalian cells. EMBO J. 2018, 37, e97479. [Google Scholar] [CrossRef] [PubMed]
- Maillard, P.V.; Van der Veen, A.G.; Deddouche-Grass, S.; Rogers, N.C.; Merits, A.; Reis e Sousa, C. Inactivation of the type I interferon pathway reveals long double-stranded RNA-mediated RNA interference in mammalian cells. EMBO J. 2016, 35, 2505–2518. [Google Scholar] [CrossRef] [PubMed]
- Cerutti, H.; Casas-Mollano, J.A. On the origin and functions of RNA-mediated silencing: From protists to man. Curr. Genet. 2006, 50, 81–99. [Google Scholar] [CrossRef]
- Miesen, P.; Joosten, J.; van Rij, R.P. PIWIs go viral: Arbovirus-derived piRNAs in vector mosquitoes. PLoS Pathog. 2016, 12, e1006017. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Lendeckel, W.; Tuschl, T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001, 15, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Elbashir, S.M.; Martinez, J.; Patkaniowska, A.; Lendeckel, W.; Tuschl, T. Functional anatomy of siRNAs for mediating efficient rnai in Drosophila melanogaster embryo lysate. EMBO J. 2001, 20, 6877–6888. [Google Scholar] [CrossRef]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single processing center models for human Dicer and bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef]
- Miyoshi, K.; Tsukumo, H.; Nagami, T.; Siomi, H.; Siomi, M.C. Slicer function of Drosophila argonautes and its involvement in RISC formation. Genes Dev. 2005, 19, 2837–2848. [Google Scholar] [CrossRef]
- Matranga, C.; Tomari, Y.; Shin, C.; Bartel, D.P.; Zamore, P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 2005, 123, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Rand, T.A.; Petersen, S.; Du, F.; Wang, X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 2005, 123, 621–629. [Google Scholar] [CrossRef]
- Liu, Y.; Ye, X.; Jiang, F.; Liang, C.; Chen, D.; Peng, J.; Kinch, L.N.; Grishin, N.V.; Liu, Q. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science 2009, 325, 750–753. [Google Scholar] [CrossRef]
- Liu, Q.; Rand, T.A.; Kalidas, S.; Du, F.; Kim, H.E.; Smith, D.P.; Wang, X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 2003, 301, 1921–1925. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jiang, F.; Kalidas, S.; Smith, D.; Liu, Q. Dicer-2; R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA 2006, 12, 1514–1520. [Google Scholar] [CrossRef]
- Tomari, Y.; Matranga, C.; Haley, B.; Martinez, N.; Zamore, P.D. A protein sensor for siRNA asymmetry. Science 2004, 306, 1377–1380. [Google Scholar] [CrossRef]
- Marques, J.T.; Kim, K.; Wu, P.H.; Alleyne, T.M.; Jafari, N.; Carthew, R.W. Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat. Struct. Mol. Biol. 2010, 17, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Sabin, L.R.; Zhou, R.; Gruber, J.J.; Lukinova, N.; Bambina, S.; Berman, A.; Lau, C.K.; Thompson, C.B.; Cherry, S. Ars2 regulates both miRNA- and siRNA- dependent silencing and suppresses RNA virus infection in Drosophila. Cell 2009, 138, 340–351. [Google Scholar] [CrossRef]
- Iwasaki, S.; Kobayashi, M.; Yoda, M.; Sakaguchi, Y.; Katsuma, S.; Suzuki, T.; Tomari, Y. Hsc70/hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell 2010, 39, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, T.; Takeuchi, A.; Siomi, H.; Siomi, M.C. A direct role for HSP90 in pre-RISC formation in Drosophila. Nat. Struct. Mol. Biol. 2010, 17, 1024–1026. [Google Scholar] [CrossRef]
- Horwich, M.D.; Li, C.; Matranga, C.; Vagin, V.; Farley, G.; Wang, P.; Zamore, P.D. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007, 17, 1265–1272. [Google Scholar] [CrossRef] [PubMed]
- Sinha, N.K.; Iwasa, J.; Shen, P.S.; Bass, B.L. Dicer uses distinct modules for recognizing dsRNA termini. Science 2018, 359, 329–334. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, J.; Cheng, H.; Ke, X.; Sun, L.; Zhang, Q.C.; Wang, H.W. Cryo-EM structure of human dicer and its complexes with a pre-miRNA substrate. Cell 2018, 173, 1549–1550. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. Yasara view-molecular graphics for all devices-from smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef]
- Ma, J.B.; Yuan, Y.R.; Meister, G.; Pei, Y.; Tuschl, T.; Patel, D.J. Structural basis for 5’-end-specific recognition of guide RNA by the A. fulgidus PIWI protein. Nature 2005, 434, 666–670. [Google Scholar] [CrossRef]
- Rand, T.A.; Ginalski, K.; Grishin, N.V.; Wang, X. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc. Natl. Acad. Sci. USA 2004, 101, 14385–14389. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, K.; Weinberg, D.E.; Bartel, D.P.; Patel, D.J. Structure of yeast argonaute with guide RNA. Nature 2012, 486, 368–374. [Google Scholar] [CrossRef]
- Nowotny, M.; Gaidamakov, S.A.; Crouch, R.J.; Yang, W. Crystal structures of RNase H bound to an RNA/DNA hybrid: Substrate specificity and metal-dependent catalysis. Cell 2005, 121, 1005–1016. [Google Scholar] [CrossRef]
- Faehnle, C.R.; Elkayam, E.; Haase, A.D.; Hannon, G.J.; Joshua-Tor, L. The making of a slicer: Activation of human Argonaute-1. Cell Rep. 2013, 3, 1901–1909. [Google Scholar] [CrossRef] [PubMed]
- Orban, T.I.; Izaurralde, E. Decay of mRNAs targeted by RISC requires XRN1, the ski complex, and the exosome. RNA 2005, 11, 459–469. [Google Scholar] [CrossRef]
- Schirle, N.T.; MacRae, I.J. The crystal structure of human argonaute2. Science 2012, 336, 1037–1040. [Google Scholar] [CrossRef]
- Czech, B.; Malone, C.D.; Zhou, R.; Stark, A.; Schlingeheyde, C.; Dus, M.; Perrimon, N.; Kellis, M.; Wohlschlegel, J.A.; Sachidanandam, R.; et al. An endogenous small interfering RNA pathway in Drosophila. Nature 2008, 453, 798–802. [Google Scholar] [CrossRef]
- Ghildiyal, M.; Seitz, H.; Horwich, M.D.; Li, C.; Du, T.; Lee, S.; Xu, J.; Kittler, E.L.; Zapp, M.L.; Weng, Z.; et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 2008, 320, 1077–1081. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, Y.; Saito, K.; Kin, T.; Ono, Y.; Asai, K.; Sunohara, T.; Okada, T.N.; Siomi, M.C.; Siomi, H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 2008, 453, 793–797. [Google Scholar] [CrossRef]
- Chung, W.J.; Okamura, K.; Martin, R.; Lai, E.C. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr. Biol. 2008, 18, 795–802. [Google Scholar] [CrossRef]
- Okamura, K.; Chung, W.J.; Ruby, J.G.; Guo, H.; Bartel, D.P.; Lai, E.C. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 2008, 453, 803–806. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Aliyari, R.; Li, W.X.; Li, H.W.; Kim, K.; Carthew, R.; Atkinson, P.; Ding, S.W. RNA interference directs innate immunity against viruses in adult Drosophila. Science 2006, 312, 452–454. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [CrossRef]
- Saini, H.K.; Griffiths-Jones, S.; Enright, A.J. Genomic analysis of human microRNA transcripts. Proc. Natl. Acad. Sci. USA 2007, 104, 17719–17724. [Google Scholar] [CrossRef]
- Finnegan, E.F.; Pasquinelli, A.E. MicroRNA biogenesis: Regulating the regulators. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 51–68. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Landthaler, M.; Yalcin, A.; Tuschl, T. The human DiGeorge syndrome critical region gene 8 and its d. Melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004, 14, 2162–2167. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef]
- Lund, E.; Guttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef]
- Zeng, Y.; Cullen, B.R. Structural requirements for pre-microRNA binding and nuclear export by exportin 5. Nucleic Acids Res. 2004, 32, 4776–4785. [Google Scholar] [CrossRef]
- Hutvagner, G.; McLachlan, J.; Pasquinelli, A.E.; Balint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Nakahara, K.; Pham, J.W.; Kim, K.; He, Z.; Sontheimer, E.J.; Carthew, R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 2004, 117, 69–81. [Google Scholar] [CrossRef]
- Forstemann, K.; Horwich, M.D.; Wee, L.; Tomari, Y.; Zamore, P.D. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell 2007, 130, 287–297. [Google Scholar] [CrossRef]
- Tomari, Y.; Du, T.; Zamore, P.D. Sorting of Drosophila small silencing RNAs. Cell 2007, 130, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.S.; Hutvagner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Ohrt, T.; Muetze, J.; Svoboda, P.; Schwille, P. Intracellular localization and routing of miRNA and RNAi pathway components. Curr. Top. Med. Chem. 2012, 12, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant. Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef] [PubMed]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef]
- Ender, C.; Meister, G. Argonaute proteins at a glance. J. Cell Sci. 2010, 123, 1819–1823. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Trombly, M.I.; Chen, J.; Wang, X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009, 23, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Riley, K.J.; Yario, T.A.; Steitz, J.A. Association of Argonaute proteins and microRNAs can occur after cell lysis. RNA 2012, 18, 1581–1585. [Google Scholar] [CrossRef]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef]
- Liu, J.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.J.; Hammond, S.M.; Joshua-Tor, L.; Hannon, G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004, 305, 1437–1441. [Google Scholar] [CrossRef]
- Berezikov, E.; Chung, W.J.; Willis, J.; Cuppen, E.; Lai, E.C. Mammalian mirtron genes. Mol. Cell 2007, 28, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Ender, C.; Krek, A.; Friedlander, M.R.; Beitzinger, M.; Weinmann, L.; Chen, W.; Pfeffer, S.; Rajewsky, N.; Meister, G. A human snoRNA with microRNA-like functions. Mol. Cell 2008, 32, 519–528. [Google Scholar] [CrossRef]
- Babiarz, J.E.; Ruby, J.G.; Wang, Y.; Bartel, D.P.; Blelloch, R. Mouse es cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008, 22, 2773–2785. [Google Scholar] [CrossRef] [PubMed]
- Cole, C.; Sobala, A.; Lu, C.; Thatcher, S.R.; Bowman, A.; Brown, J.W.; Green, P.J.; Barton, G.J.; Hutvagner, G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 2009, 15, 2147–2160. [Google Scholar] [CrossRef]
- Brameier, M.; Herwig, A.; Reinhardt, R.; Walter, L.; Gruber, J. Human box C/D snoRNAs with miRNA like functions: Expanding the range of regulatory RNAs. Nucleic Acids Res. 2011, 39, 675–686. [Google Scholar] [CrossRef]
- Ono, M.; Scott, M.S.; Yamada, K.; Avolio, F.; Barton, G.J.; Lamond, A.I. Identification of human miRNA precursors that resemble box c/d snoRNAs. Nucleic Acids Res. 2011, 39, 3879–3891. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.S.; Avolio, F.; Ono, M.; Lamond, A.I.; Barton, G.J. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput. Biol. 2009, 5, e1000507. [Google Scholar] [CrossRef] [PubMed]
- Mourelatos, Z.; Dostie, J.; Paushkin, S.; Sharma, A.; Charroux, B.; Abel, L.; Rappsilber, J.; Mann, M.; Dreyfuss, G. Mirnps: A novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002, 16, 720–728. [Google Scholar] [CrossRef]
- Farh, K.K.; Grimson, A.; Jan, C.; Lewis, B.P.; Johnston, W.K.; Lim, L.P.; Burge, C.B.; Bartel, D.P. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science 2005, 310, 1817–1821. [Google Scholar] [CrossRef]
- Grimson, A.; Farh, K.K.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef]
- Brennecke, J.; Stark, A.; Russell, R.B.; Cohen, S.M. Principles of microRNA-target recognition. PLoS Biol. 2005, 3, e85. [Google Scholar] [CrossRef]
- Eichhorn, S.W.; Guo, H.; McGeary, S.E.; Rodriguez-Mias, R.A.; Shin, C.; Baek, D.; Hsu, S.H.; Ghoshal, K.; Villen, J.; Bartel, D.P. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell 2014, 56, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Ameres, S.L.; Zamore, P.D. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 2013, 14, 475–488. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Brummelkamp, T.R.; Bernards, R.; Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science 2002, 296, 550–553. [Google Scholar] [CrossRef]
- Paddison, P.J.; Caudy, A.A.; Hannon, G.J. Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl. Acad. Sci. USA 2002, 99, 1443–1448. [Google Scholar] [CrossRef]
- Yang, S.; Tutton, S.; Pierce, E.; Yoon, K. Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol. Cell. Biol. 2001, 21, 7807–7816. [Google Scholar] [CrossRef]
- Billy, E.; Brondani, V.; Zhang, H.; Muller, U.; Filipowicz, W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl. Acad. Sci. USA 2001, 98, 14428–14433. [Google Scholar] [CrossRef]
- Zamore, P.D.; Tuschl, T.; Sharp, P.A.; Bartel, D.P. Rnai: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 2000, 101, 25–33. [Google Scholar] [CrossRef]
- Martinez, J.; Patkaniowska, A.; Urlaub, H.; Luhrmann, R.; Tuschl, T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 2002, 110, 563–574. [Google Scholar] [CrossRef]
- Dueck, A.; Ziegler, C.; Eichner, A.; Berezikov, E.; Meister, G. MicroRNAs associated with the different human Argonaute proteins. Nucleic Acids Res. 2012, 40, 9850–9862. [Google Scholar] [CrossRef]
- Siolas, D.; Lerner, C.; Burchard, J.; Ge, W.; Linsley, P.S.; Paddison, P.J.; Hannon, G.J.; Cleary, M.A. Synthetic shRNAs as potent RNAi triggers. Nat. Biotechnol. 2005, 23, 227–231. [Google Scholar] [CrossRef]
- Isaacs, A.; Cox, R.A.; Rotem, Z. Foreign nucleic acids as the stimulus to make interferon. Lancet 1963, 2, 113–116. [Google Scholar] [CrossRef]
- Hardy, M.P.; Owczarek, C.M.; Trajanovska, S.; Liu, X.; Kola, I.; Hertzog, P.J. The soluble murine type I interferon receptor Ifnar-2 is present in serum, is independently regulated, and has both agonistic and antagonistic properties. Blood 2001, 97, 473–482. [Google Scholar] [CrossRef]
- Hardy, M.P.; Sanij, E.P.; Hertzog, P.J.; Owczarek, C.M. Characterization and transcriptional analysis of the mouse chromosome 16 cytokine receptor gene cluster. Mamm. Genome 2003, 14, 105–118. [Google Scholar] [CrossRef]
- Hardy, M.P.; Hertzog, P.J.; Owczarek, C.M. Multiple regions within the promoter of the murine Ifnar-2 gene confer basal and inducible expression. Biochem. J. 2002, 365, 355–367. [Google Scholar] [CrossRef]
- Goubau, D.; Deddouche, S.; Reis e Sousa, C. Cytosolic sensing of viruses. Immunity 2013, 38, 855–869. [Google Scholar] [CrossRef]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef]
- Schoggins, J.W.; Rice, C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011, 1, 519–525. [Google Scholar] [CrossRef]
- Li, G.; Xiang, Y.; Sabapathy, K.; Silverman, R.H. An apoptotic signaling pathway in the interferon antiviral response mediated by RNase l and c-Jun NH2-terminal kinase. J. Biol. Chem. 2004, 279, 1123–1131. [Google Scholar] [CrossRef]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef]
- Schlee, M.; Hartmann, G. Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 2016, 16, 566–580. [Google Scholar] [CrossRef]
- Levin, D.; London, I.M. Regulation of protein synthesis: Activation by double-stranded RNA of a protein kinase that phosphorylates eukaryotic initiation factor 2. Proc. Natl. Acad. Sci. USA 1978, 75, 1121–1125. [Google Scholar] [CrossRef]
- Zilberstein, A.; Kimchi, A.; Schmidt, A.; Revel, M. Isolation of two interferon-induced translational inhibitors: A protein kinase and an oligo-isoadenylate synthetase. Proc. Natl. Acad. Sci. USA 1978, 75, 4734–4738. [Google Scholar] [CrossRef]
- Hovanessian, A.G.; Brown, R.E.; Kerr, I.M. Synthesis of low molecular weight inhibitor of protein synthesis with enzyme from interferon-treated cells. Nature 1977, 268, 537–540. [Google Scholar] [CrossRef]
- Zhou, A.; Hassel, B.A.; Silverman, R.H. Expression cloning of 2-5a-dependent RNAase: A uniquely regulated mediator of interferon action. Cell 1993, 72, 753–765. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef]
- Luo, D.; Kohlway, A.; Pyle, A.M. Duplex RNA activated atpases (dras): Platforms for RNA sensing, signaling and processing. RNA Biol. 2013, 10, 111–120. [Google Scholar] [CrossRef]
- Cui, S.; Eisenacher, K.; Kirchhofer, A.; Brzozka, K.; Lammens, A.; Lammens, K.; Fujita, T.; Conzelmann, K.K.; Krug, A.; Hopfner, K.P. The C-terminal regulatory domain is the RNA 5’-triphosphate sensor of RIG-I. Mol. Cell 2008, 29, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Takahasi, K.; Yoneyama, M.; Nishihori, T.; Hirai, R.; Kumeta, H.; Narita, R.; Gale, M., Jr.; Inagaki, F.; Fujita, T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell 2008, 29, 428–440. [Google Scholar] [CrossRef]
- Reikine, S.; Nguyen, J.B.; Modis, Y. Pattern recognition and signaling mechanisms of RIG-I and MDA5. Front. Immunol. 2014, 5, 342. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Chen, Z.J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014, 32, 461–488. [Google Scholar] [CrossRef]
- Sohn, J.; Hur, S. Filament assemblies in foreign nucleic acid sensors. Curr. Opin. Struct. Biol. 2016, 37, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Bruns, A.M.; Leser, G.P.; Lamb, R.A.; Horvath, C.M. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Mol. Cell 2014, 55, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Bruns, A.M.; Horvath, C.M. LGP2 synergy with MDA5 in rlr-mediated RNA recognition and antiviral signaling. Cytokine 2015, 74, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, T.; Valdes, M.; Elsby, R.; Kakuta, S.; Caceres, G.; Saijo, S.; Iwakura, Y.; Barber, G.N. Loss of dexd/h box RNA helicase lGP2 manifests disparate antiviral responses. J. Immunol. 2007, 178, 6444–6455. [Google Scholar] [CrossRef]
- Satoh, T.; Kato, H.; Kumagai, Y.; Yoneyama, M.; Sato, S.; Matsushita, K.; Tsujimura, T.; Fujita, T.; Akira, S.; Takeuchi, O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc. Natl. Acad. Sci. USA 2010, 107, 1512–1517. [Google Scholar] [CrossRef]
- Rothenfusser, S.; Goutagny, N.; DiPerna, G.; Gong, M.; Monks, B.G.; Schoenemeyer, A.; Yamamoto, M.; Akira, S.; Fitzgerald, K.A. The RNA helicase LGP2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 2005, 175, 5260–5268. [Google Scholar] [CrossRef]
- Yoneyama, M.; Kikuchi, M.; Matsumoto, K.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Foy, E.; Loo, Y.M.; Gale, M., Jr.; Akira, S.; et al. Shared and unique functions of the dexd/h-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005, 175, 2851–2858. [Google Scholar] [CrossRef]
- Si-Tahar, M.; Blanc, F.; Furio, L.; Chopy, D.; Balloy, V.; Lafon, M.; Chignard, M.; Fiette, L.; Langa, F.; Charneau, P.; et al. Protective role of LGP2 in influenza virus pathogenesis. J. Infect. Dis. 2014, 210, 214–223. [Google Scholar] [CrossRef]
- Chopy, D.; Pothlichet, J.; Lafage, M.; Megret, F.; Fiette, L.; Si-Tahar, M.; Lafon, M. Ambivalent role of the innate immune response in rabies virus pathogenesis. J. Virol. 2011, 85, 6657–6668. [Google Scholar] [CrossRef] [PubMed]
- Suthar, M.S.; Ramos, H.J.; Brassil, M.M.; Netland, J.; Chappell, C.P.; Blahnik, G.; McMillan, A.; Diamond, M.S.; Clark, E.A.; Bevan, M.J.; et al. The RIG-I-like receptor LGP2 controls CD8(+) T cell survival and fitness. Immunity 2012, 37, 235–248. [Google Scholar] [CrossRef]
- Sato, S.; Li, K.; Kameyama, T.; Hayashi, T.; Ishida, Y.; Murakami, S.; Watanabe, T.; Iijima, S.; Sakurai, Y.; Watashi, K.; et al. The RNA sensor RIG-I dually functions as an innate sensor and direct antiviral factor for hepatitis B virus. Immunity 2015, 42, 123–132. [Google Scholar] [CrossRef]
- Weber, M.; Sediri, H.; Felgenhauer, U.; Binzen, I.; Banfer, S.; Jacob, R.; Brunotte, L.; Garcia-Sastre, A.; Schmid-Burgk, J.L.; Schmidt, T.; et al. Influenza virus adaptation PB2-627K modulates nucleocapsid inhibition by the pathogen sensor RIG-I. Cell Host Microbe 2015, 17, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Weissman, I.L. Stem cells: Units of development, units of regeneration, and units in evolution. Cell 2000, 100, 157–168. [Google Scholar] [CrossRef]
- Doetsch, F.; Caille, I.; Lim, D.A.; Garcia-Verdugo, J.M.; Alvarez-Buylla, A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 1999, 97, 703–716. [Google Scholar] [CrossRef]
- Kuznetsov, S.A.; Mankani, M.H.; Gronthos, S.; Satomura, K.; Bianco, P.; Robey, P.G. Circulating skeletal stem cells. J. Cell Biol. 2001, 153, 1133–1140. [Google Scholar] [CrossRef] [PubMed]
- Saiura, A.; Sata, M.; Hirata, Y.; Nagai, R.; Makuuchi, M. Circulating smooth muscle progenitor cells contribute to atherosclerosis. Nat. Med. 2001, 7, 382–383. [Google Scholar] [CrossRef]
- Burke, D.C.; Graham, C.F.; Lehman, J.M. Appearance of interferon inducibility and sensitivity during differentiation of murine teratocarcinoma cells in vitro. Cell 1978, 13, 243–248. [Google Scholar] [CrossRef]
- Chen, L.L.; Yang, L.; Carmichael, G.G. Molecular basis for an attenuated cytoplasmic dsRNA response in human embryonic stem cells. Cell Cycle 2010, 9, 3552–3564. [Google Scholar] [CrossRef]
- Witteveldt, J.; Knol, L.I.; Macias, S. MicroRNA-deficient mouse embryonic stem cells acquire a functional interferon response. eLife 2019, 8. [Google Scholar] [CrossRef]
- Wu, X.; Dao Thi, V.L.; Huang, Y.; Billerbeck, E.; Saha, D.; Hoffmann, H.H.; Wang, Y.; Silva, L.A.V.; Sarbanes, S.; Sun, T.; et al. Intrinsic immunity shapes viral resistance of stem cells. Cell 2018, 172, 423–438.e25. [Google Scholar] [CrossRef] [PubMed]
- Bailey, C.C.; Zhong, G.; Huang, I.C.; Farzan, M. IFITM-family proteins: The cell’s first line of antiviral defense. Annu. Rev. Virol. 2014, 1, 261–283. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Robotham, J.M.; Lee, E.; Dalton, S.; Kneteman, N.M.; Gilbert, D.M.; Tang, H. Productive hepatitis C virus infection of stem cell-derived hepatocytes reveals a critical transition to viral permissiveness during differentiation. PLoS Pathog. 2012, 8, e1002617. [Google Scholar] [CrossRef]
- Weber, F.; Kochs, G.; Haller, O.; Staeheli, P. Viral evasion of the interferon system: Old viruses, new tricks. J. Interferon Cytokine Res. 2003, 23, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Sastre, A. Ten strategies of interferon evasion by viruses. Cell Host Microbe 2017, 22, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Talon, J.; Horvath, C.M.; Polley, R.; Basler, C.F.; Muster, T.; Palese, P.; Garcia-Sastre, A. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 2000, 74, 7989–7996. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Latham, A.G.; Krug, R.M. Human influenza viruses activate an interferon-independent transcription of cellular antiviral genes: Outcome with influenza a virus is unique. Proc. Natl. Acad. Sci. USA 2002, 99, 10096–10101. [Google Scholar] [CrossRef] [PubMed]
- Versteeg, G.A.; Garcia-Sastre, A. Viral tricks to grid-lock the type I interferon system. Curr. Opin. Microbiol. 2010, 13, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Laurent-Rolle, M.; Morrison, J.; Rajsbaum, R.; Macleod, J.M.L.; Pisanelli, G.; Pham, A.; Ayllon, J.; Miorin, L.; Martinez, C.; tenOever, B.R.; et al. The interferon signaling antagonist function of yellow fever virus NS5 protein is activated by type I interferon. Cell Host Microbe 2014, 16, 314–327. [Google Scholar] [CrossRef]
- Hammond, S.M. Dicing and slicing: The core machinery of the RNA interference pathway. FEBS Lett. 2005, 579, 5822–5829. [Google Scholar] [CrossRef] [PubMed]
- Weber, F.; Wagner, V.; Rasmussen, S.B.; Hartmann, R.; Paludan, S.R. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 2006, 80, 5059–5064. [Google Scholar] [CrossRef] [PubMed]
- Bronkhorst, A.W.; van Cleef, K.W.; Vodovar, N.; Ince, I.A.; Blanc, H.; Vlak, J.M.; Saleh, M.C.; van Rij, R.P. The DNA virus invertebrate iridescent virus 6 is a target of the Drosophila RNAi machinery. Proc. Natl. Acad. Sci. USA 2012, 109, E3604–E3613. [Google Scholar] [CrossRef]
- Deddouche, S.; Matt, N.; Budd, A.; Mueller, S.; Kemp, C.; Galiana-Arnoux, D.; Dostert, C.; Antoniewski, C.; Hoffmann, J.A.; Imler, J.L. The dexd/h-box helicase dicer-2 mediates the induction of antiviral activity in Drosophila. Nat. Immunol. 2008, 9, 1425–1432. [Google Scholar] [CrossRef]
- Galiana-Arnoux, D.; Dostert, C.; Schneemann, A.; Hoffmann, J.A.; Imler, J.L. Essential function in vivo for dicer-2 in host defense against RNA viruses in Drosophila. Nat. Immunol. 2006, 7, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Sabin, L.R.; Zheng, Q.; Thekkat, P.; Yang, J.; Hannon, G.J.; Gregory, B.D.; Tudor, M.; Cherry, S. Dicer-2 processes diverse viral RNA species. PLoS ONE 2013, 8, e55458. [Google Scholar] [CrossRef]
- Marques, J.T.; Wang, J.P.; Wang, X.; de Oliveira, K.P.; Gao, C.; Aguiar, E.R.; Jafari, N.; Carthew, R.W. Functional specialization of the small interfering RNA pathway in response to virus infection. PLoS Pathog. 2013, 9, e1003579. [Google Scholar] [CrossRef]
- Han, Y.H.; Luo, Y.J.; Wu, Q.; Jovel, J.; Wang, X.H.; Aliyari, R.; Han, C.; Li, W.X.; Ding, S.W. Rna-based immunity terminates viral infection in adult Drosophila in the absence of viral suppression of RNA interference: Characterization of viral small interfering RNA populations in wild-type and mutant flies. J. Virol. 2011, 85, 13153–13163. [Google Scholar] [CrossRef]
- Mueller, S.; Gausson, V.; Vodovar, N.; Deddouche, S.; Troxler, L.; Perot, J.; Pfeffer, S.; Hoffmann, J.A.; Saleh, M.C.; Imler, J.L. RNAi-mediated immunity provides strong protection against the negative-strand RNA vesicular stomatitis virus in Drosophila. Proc. Natl. Acad. Sci. USA 2010, 107, 19390–19395. [Google Scholar] [CrossRef] [PubMed]
- van Rij, R.P.; Saleh, M.C.; Berry, B.; Foo, C.; Houk, A.; Antoniewski, C.; Andino, R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006, 20, 2985–2995. [Google Scholar] [CrossRef] [PubMed]
- Kemp, C.; Mueller, S.; Goto, A.; Barbier, V.; Paro, S.; Bonnay, F.; Dostert, C.; Troxler, L.; Hetru, C.; Meignin, C.; et al. Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J. Immunol. 2013, 190, 650–658. [Google Scholar] [CrossRef]
- Campbell, C.L.; Keene, K.M.; Brackney, D.E.; Olson, K.E.; Blair, C.D.; Wilusz, J.; Foy, B.D. Aedes aegypti uses RNA interference in defense against sindbis virus infection. BMC Microbiol. 2008, 8, 47. [Google Scholar] [CrossRef]
- Sanchez-Vargas, I.; Scott, J.C.; Poole-Smith, B.K.; Franz, A.W.; Barbosa-Solomieu, V.; Wilusz, J.; Olson, K.E.; Blair, C.D. Dengue virus type 2 infections of aedes aegypti are modulated by the mosquito’s RNA interference pathway. PLoS Pathog. 2009, 5, e1000299. [Google Scholar] [CrossRef] [PubMed]
- Keene, K.M.; Foy, B.D.; Sanchez-Vargas, I.; Beaty, B.J.; Blair, C.D.; Olson, K.E. RNA interference acts as a natural antiviral response to O’nyong-nyong virus (alphavirus; togaviridae) infection of Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2004, 101, 17240–17245. [Google Scholar] [CrossRef]
- Samuel, G.H.; Wiley, M.R.; Badawi, A.; Adelman, Z.N.; Myles, K.M. Yellow fever virus capsid protein is a potent suppressor of RNA silencing that binds double-stranded RNA. Proc. Natl. Acad. Sci. USA 2016, 113, 13863–13868. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, C.; Dishongh, R.; Moore, S.C.; Whitt, M.A.; Chow, M.; Machaca, K. RNA interference is an antiviral defence mechanism in caenorhabditis elegans. Nature 2005, 436, 1044–1047. [Google Scholar] [CrossRef] [PubMed]
- Felix, M.A.; Ashe, A.; Piffaretti, J.; Wu, G.; Nuez, I.; Belicard, T.; Jiang, Y.; Zhao, G.; Franz, C.J.; Goldstein, L.D.; et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011, 9, e1000586. [Google Scholar] [CrossRef]
- Lu, R.; Maduro, M.; Li, F.; Li, H.W.; Broitman-Maduro, G.; Li, W.X.; Ding, S.W. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature 2005, 436, 1040–1043. [Google Scholar] [CrossRef]
- Wassenegger, M.; Krczal, G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant. Sci. 2006, 11, 142–151. [Google Scholar] [CrossRef]
- Saleh, M.C.; Tassetto, M.; van Rij, R.P.; Goic, B.; Gausson, V.; Berry, B.; Jacquier, C.; Antoniewski, C.; Andino, R. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature 2009, 458, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Goic, B.; Vodovar, N.; Mondotte, J.A.; Monot, C.; Frangeul, L.; Blanc, H.; Gausson, V.; Vera-Otarola, J.; Cristofari, G.; Saleh, M.C. RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nat. Immunol. 2013, 14, 396–403. [Google Scholar] [CrossRef]
- Goic, B.; Stapleford, K.A.; Frangeul, L.; Doucet, A.J.; Gausson, V.; Blanc, H.; Schemmel-Jofre, N.; Cristofari, G.; Lambrechts, L.; Vignuzzi, M.; et al. Virus-derived DNA drives mosquito vector tolerance to arboviral infection. Nat. Commun. 2016, 7, 12410. [Google Scholar] [CrossRef]
- Tassetto, M.; Kunitomi, M.; Andino, R. Circulating immune cells mediate a systemic RNAi-based adaptive antiviral response in Drosophila. Cell 2017, 169, 314–325.e13. [Google Scholar] [CrossRef] [PubMed]
- Gammon, D.B.; Mello, C.C. RNA interference-mediated antiviral defense in insects. Curr. Opin. Insect Sci. 2015, 8, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Nayak, A.; Tassetto, M.; Kunitomi, M.; Andino, R. RNA interference-mediated intrinsic antiviral immunity in invertebrates. Curr. Top. Microbiol. Immunol. 2013, 371, 183–200. [Google Scholar]
- Li, H.; Li, W.X.; Ding, S.W. Induction and suppression of RNA silencing by an animal virus. Science 2002, 296, 1319–1321. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.A.; Lee, J.H.; Chapados, B.R.; Debler, E.W.; Schneemann, A.; Williamson, J.R. Dual modes of RNA-silencing suppression by flock house virus protein B2. Nat. Struct. Mol. Biol. 2005, 12, 952–957. [Google Scholar] [CrossRef] [PubMed]
- Lingel, A.; Simon, B.; Izaurralde, E.; Sattler, M. The structure of the flock house virus b2 protein, a viral suppressor of RNA interference, shows a novel mode of double-stranded RNA recognition. EMBO Rep. 2005, 6, 1149–1155. [Google Scholar] [CrossRef]
- Singh, G.; Popli, S.; Hari, Y.; Malhotra, P.; Mukherjee, S.; Bhatnagar, R.K. Suppression of RNA silencing by Flock house virus B2 protein is mediated through its interaction with the PAZ domain of Dicer. FASEB J. 2009, 23, 1845–1857. [Google Scholar] [CrossRef] [PubMed]
- Aliyari, R.; Wu, Q.; Li, H.W.; Wang, X.H.; Li, F.; Green, L.D.; Han, C.S.; Li, W.X.; Ding, S.W. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe 2008, 4, 387–397. [Google Scholar] [CrossRef] [PubMed]
- van Cleef, K.W.; van Mierlo, J.T.; Miesen, P.; Overheul, G.J.; Fros, J.J.; Schuster, S.; Marklewitz, M.; Pijlman, G.P.; Junglen, S.; van Rij, R.P. Mosquito and Drosophila entomobirnaviruses suppress dsRNA- and siRNA-induced RNAi. Nucleic Acids Res. 2014, 42, 8732–8744. [Google Scholar] [CrossRef] [PubMed]
- van Mierlo, J.T.; Bronkhorst, A.W.; Overheul, G.J.; Sadanandan, S.A.; Ekstrom, J.O.; Heestermans, M.; Hultmark, D.; Antoniewski, C.; van Rij, R.P. Convergent evolution of Argonaute-2 slicer antagonism in two distinct insect RNA viruses. PLoS Pathog. 2012, 8, e1002872. [Google Scholar] [CrossRef]
- Nayak, A.; Berry, B.; Tassetto, M.; Kunitomi, M.; Acevedo, A.; Deng, C.; Krutchinsky, A.; Gross, J.; Antoniewski, C.; Andino, R. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat. Struct. Mol. Biol. 2010, 17, 547–554. [Google Scholar] [CrossRef] [PubMed]
- van Mierlo, J.T.; Overheul, G.J.; Obadia, B.; van Cleef, K.W.; Webster, C.L.; Saleh, M.C.; Obbard, D.J.; van Rij, R.P. Novel Drosophila viruses encode host-specific suppressors of RNAi. PLoS Pathog. 2014, 10, e1004256. [Google Scholar] [CrossRef] [PubMed]
- Obbard, D.J.; Jiggins, F.M.; Halligan, D.L.; Little, T.J. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr. Biol. 2006, 16, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Svobodova, E.; Kubikova, J.; Svoboda, P. Production of small RNAs by mammalian Dicer. Pflugers Arch. 2016, 468, 1089–1102. [Google Scholar] [CrossRef]
- Carmell, M.A.; Xuan, Z.; Zhang, M.Q.; Hannon, G.J. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002, 16, 2733–2742. [Google Scholar] [CrossRef]
- Murchison, E.P.; Partridge, J.F.; Tam, O.H.; Cheloufi, S.; Hannon, G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA 2005, 102, 12135–12140. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, J.M.; Seila, A.C.; Yeo, G.W.; Sharp, P.A. RNA sequence analysis defines Dicer’s role in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 18097–18102. [Google Scholar] [CrossRef] [PubMed]
- Chong, M.M.; Zhang, G.; Cheloufi, S.; Neubert, T.A.; Hannon, G.J.; Littman, D.R. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010, 24, 1951–1960. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Kim, S.Y.; Carmell, M.A.; Murchison, E.P.; Alcorn, H.; Li, M.Z.; Mills, A.A.; Elledge, S.J.; Anderson, K.V.; Hannon, G.J. Dicer is essential for mouse development. Nat. Genet. 2003, 35, 215–217. [Google Scholar] [CrossRef]
- Kanellopoulou, C.; Muljo, S.A.; Kung, A.L.; Ganesan, S.; Drapkin, R.; Jenuwein, T.; Livingston, D.M.; Rajewsky, K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005, 19, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Frohn, A.; Eberl, H.C.; Stohr, J.; Glasmacher, E.; Rudel, S.; Heissmeyer, V.; Mann, M.; Meister, G. Dicer-dependent and -independent Argonaute2 protein interaction networks in mammalian cells. Mol. Cell. Proteom. 2012, 11, 1442–1456. [Google Scholar] [CrossRef] [PubMed]
- Smibert, P.; Yang, J.S.; Azzam, G.; Liu, J.L.; Lai, E.C. Homeostatic control of Argonaute stability by microRNA availability. Nat. Struct. Mol. Biol. 2013, 20, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Bogerd, H.P.; Whisnant, A.W.; Kennedy, E.M.; Flores, O.; Cullen, B.R. Derivation and characterization of dicer- and microRNA-deficient human cells. RNA 2014, 20, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Backes, S.; Langlois, R.A.; Schmid, S.; Varble, A.; Shim, J.V.; Sachs, D.; tenOever, B.R. The mammalian response to virus infection is independent of small RNA silencing. Cell Rep. 2014, 8, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Bogerd, H.P.; Skalsky, R.L.; Kennedy, E.M.; Furuse, Y.; Whisnant, A.W.; Flores, O.; Schultz, K.L.; Putnam, N.; Barrows, N.J.; Sherry, B.; et al. Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs. J. Virol. 2014, 88, 8065–8076. [Google Scholar] [CrossRef] [PubMed]
- Parameswaran, P.; Sklan, E.; Wilkins, C.; Burgon, T.; Samuel, M.A.; Lu, R.; Ansel, K.M.; Heissmeyer, V.; Einav, S.; Jackson, W.; et al. Six RNA viruses and forty-one hosts: Viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 2010, 6, e1000764. [Google Scholar] [CrossRef]
- Girardi, E.; Chane-Woon-Ming, B.; Messmer, M.; Kaukinen, P.; Pfeffer, S. Identification of RNase L-dependent, 3’-end-modified, viral small RNAs in Sindbis virus-infected mammalian cells. MBio 2013, 4, e00698-00613. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Xu, Y.; Zhang, Y.; Zhou, H.; Deng, Y.Q.; Li, X.F.; Miao, M.; Zhang, Q.; Zhong, B.; Hu, Y.; et al. Human virus-derived small RNAs can confer antiviral immunity in mammals. Immunity 2017, 46, 992–1004.e5. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.; Courtney, D.G.; Kennedy, E.M.; Cullen, B.R. Influenza a virus-derived siRNAs increase in the absence of NS1 yet fail to inhibit virus replication. RNA 2018, 24, 1172–1182. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, E.M.; Whisnant, A.W.; Kornepati, A.V.; Marshall, J.B.; Bogerd, H.P.; Cullen, B.R. Production of functional small interfering RNAs by an amino-terminal deletion mutant of human Dicer. Proc. Natl. Acad. Sci. USA 2015, 112, E6945–E6954. [Google Scholar] [CrossRef]
- Donaszi-Ivanov, A.; Mohorianu, I.; Dalmay, T.; Powell, P.P. Small RNA analysis in Sindbis virus infected human HEK293 cells. PLoS ONE 2013, 8, e84070. [Google Scholar] [CrossRef]
- Xu, Y.P.; Qiu, Y.; Zhang, B.; Chen, G.; Chen, Q.; Wang, M.; Mo, F.; Xu, J.; Wu, J.; Zhang, R.R.; et al. Zika virus infection induces RNAi-mediated antiviral immunity in human neural progenitors and brain organoids. Cell Res. 2019, 29, 265–273. [Google Scholar] [CrossRef]
- Haasnoot, J.; de Vries, W.; Geutjes, E.J.; Prins, M.; de Haan, P.; Berkhout, B. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 2007, 3, e86. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Wang, H.; Ji, Y.; Yang, J.; Xu, S.; Huang, X.; Wang, Z.; Qin, L.; Tien, P.; Zhou, X.; et al. The nucleocapsid protein of coronaviruses acts as a viral suppressor of RNA silencing in mammalian cells. J. Virol. 2015, 89, 9029–9043. [Google Scholar] [CrossRef] [PubMed]
- Fabozzi, G.; Nabel, C.S.; Dolan, M.A.; Sullivan, N.J. Ebolavirus proteins suppress the effects of small interfering RNA by direct interaction with the mammalian RNA interference pathway. J. Virol. 2011, 85, 2512–2523. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Sastre, A.; Egorov, A.; Matassov, D.; Brandt, S.; Levy, D.E.; Durbin, J.E.; Palese, P.; Muster, T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 1998, 252, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Li, W.X.; Li, H.; Lu, R.; Li, F.; Dus, M.; Atkinson, P.; Brydon, E.W.; Johnson, K.L.; Garcia-Sastre, A.; Ball, L.A.; et al. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. USA 2004, 101, 1350–1355. [Google Scholar] [CrossRef]
- Prins, K.C.; Delpeut, S.; Leung, D.W.; Reynard, O.; Volchkova, V.A.; Reid, S.P.; Ramanan, P.; Cardenas, W.B.; Amarasinghe, G.K.; Volchkov, V.E.; et al. Mutations abrogating VP35 interaction with double-stranded RNA render ebola virus avirulent in guinea pigs. J. Virol. 2010, 84, 3004–3015. [Google Scholar] [CrossRef]
- Pijlman, G.P.; Funk, A.; Kondratieva, N.; Leung, J.; Torres, S.; van der Aa, L.; Liu, W.J.; Palmenberg, A.C.; Shi, P.Y.; Hall, R.A.; et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 2008, 4, 579–591. [Google Scholar] [CrossRef]
- Schnettler, E.; Sterken, M.G.; Leung, J.Y.; Metz, S.W.; Geertsema, C.; Goldbach, R.W.; Vlak, J.M.; Kohl, A.; Khromykh, A.A.; Pijlman, G.P. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and mammalian cells. J. Virol. 2012, 86, 13486–13500. [Google Scholar] [CrossRef]
- Bidet, K.; Dadlani, D.; Garcia-Blanco, M.A. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 2014, 10, e1004242. [Google Scholar] [CrossRef]
- Backes, S.; Shapiro, J.S.; Sabin, L.R.; Pham, A.M.; Reyes, I.; Moss, B.; Cherry, S.; tenOever, B.R. Degradation of host microRNAs by poxvirus poly(A) polymerase reveals terminal RNA methylation as a protective antiviral mechanism. Cell Host Microbe 2012, 12, 200–210. [Google Scholar] [CrossRef]
- Johnson, K.L.; Price, B.D.; Eckerle, L.D.; Ball, L.A. Nodamura virus nonstructural protein B2 can enhance viral RNA accumulation in both mammalian and insect cells. J. Virol. 2004, 78, 6698–6704. [Google Scholar] [CrossRef] [PubMed]
- Luthra, P.; Ramanan, P.; Mire, C.E.; Weisend, C.; Tsuda, Y.; Yen, B.; Liu, G.; Leung, D.W.; Geisbert, T.W.; Ebihara, H.; et al. Mutual antagonism between the Ebola virus VP35 protein and the RIG-I activator pact determines infection outcome. Cell Host Microbe 2013, 14, 74–84. [Google Scholar] [CrossRef]
- Hartman, A.L.; Towner, J.S.; Nichol, S.T. A C-terminal basic amino acid motif of Zaire ebolavirus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza a virus. Virology 2004, 328, 177–184. [Google Scholar] [CrossRef]
- Hu, Y.; Li, W.; Gao, T.; Cui, Y.; Jin, Y.; Li, P.; Ma, Q.; Liu, X.; Cao, C. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Min, J.Y.; Krug, R.M. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2’-5’ oligo (a) synthetase/RNAse l pathway. Proc. Natl. Acad. Sci. USA 2006, 103, 7100–7105. [Google Scholar] [CrossRef]
- Guo, Z.; Chen, L.M.; Zeng, H.; Gomez, J.A.; Plowden, J.; Fujita, T.; Katz, J.M.; Donis, R.O.; Sambhara, S. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am. J. Respir. Cell Mol. Biol. 2007, 36, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Gack, M.U.; Albrecht, R.A.; Urano, T.; Inn, K.S.; Huang, I.C.; Carnero, E.; Farzan, M.; Inoue, S.; Jung, J.U.; Garcia-Sastre, A. Influenza A virus ns1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 2009, 5, 439–449. [Google Scholar] [CrossRef]
- Sullivan, C.S.; Ganem, D. A virus-encoded inhibitor that blocks RNA interference in mammalian cells. J. Virol. 2005, 79, 7371–7379. [Google Scholar] [CrossRef]
- Bergmann, M.; Garcia-Sastre, A.; Carnero, E.; Pehamberger, H.; Wolff, K.; Palese, P.; Muster, T. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 2000, 74, 6203–6206. [Google Scholar] [CrossRef] [PubMed]
- Benitez, A.A.; Spanko, L.A.; Bouhaddou, M.; Sachs, D.; tenOever, B.R. Engineered mammalian RNAi can elicit antiviral protection that negates the requirement for the interferon response. Cell Rep. 2015, 13, 1456–1466. [Google Scholar] [CrossRef] [PubMed]
- Kok, K.H.; Jin, D.Y. Influenza A virus NS1 protein does not suppress RNA interference in mammalian cells. J. Gen. Virol. 2006, 87, 2639–2644. [Google Scholar] [CrossRef] [PubMed]
- Perez, J.T.; Pham, A.M.; Lorini, M.H.; Chua, M.A.; Steel, J.; tenOever, B.R. MicroRNA-mediated species-specific attenuation of influenza A virus. Nat. Biotechnol. 2009, 27, 572–576. [Google Scholar] [CrossRef] [PubMed]
- Langlois, R.A.; Albrecht, R.A.; Kimble, B.; Sutton, T.; Shapiro, J.S.; Finch, C.; Angel, M.; Chua, M.A.; Gonzalez-Reiche, A.S.; Xu, K.; et al. MicroRNA-based strategy to mitigate the risk of gain-of-function influenza studies. Nat. Biotechnol. 2013, 31, 844–847. [Google Scholar] [CrossRef] [PubMed]
- Langlois, R.A.; Varble, A.; Chua, M.A.; Garcia-Sastre, A.; tenOever, B.R. Hematopoietic-specific targeting of influenza A virus reveals replication requirements for induction of antiviral immune responses. Proc. Natl. Acad. Sci. USA 2012, 109, 12117–12122. [Google Scholar] [CrossRef]
- Varble, A.; Chua, M.A.; Perez, J.T.; Manicassamy, B.; Garcia-Sastre, A.; tenOever, B.R. Engineered RNA viral synthesis of microRNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 11519–11524. [Google Scholar] [CrossRef]
- Pare, J.M.; Sullivan, C.S. Distinct antiviral responses in pluripotent versus differentiated cells. PLoS Pathog. 2014, 10, e1003865. [Google Scholar] [CrossRef]
- Tam, O.H.; Aravin, A.A.; Stein, P.; Girard, A.; Murchison, E.P.; Cheloufi, S.; Hodges, E.; Anger, M.; Sachidanandam, R.; Schultz, R.M.; et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 2008, 453, 534–538. [Google Scholar] [CrossRef]
- Hertzog, P.J.; Hwang, S.Y.; Kola, I. Role of interferons in the regulation of cell proliferation, differentiation, and development. Mol. Reprod. Dev. 1994, 39, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Yu, J.Y.; Shcherbata, H.R.; Mathieu, J.; Wang, A.J.; Seal, S.; Zhou, W.; Stadler, B.M.; Bourgin, D.; Wang, L.; et al. MicroRNAs regulate human embryonic stem cell division. Cell Cycle 2009, 8, 3729–3741. [Google Scholar] [CrossRef]
- Garcia-Perez, J.L.; Widmann, T.J.; Adams, I.R. The impact of transposable elements on mammalian development. Development 2016, 143, 4101–4114. [Google Scholar] [CrossRef] [PubMed]
- Ma, E.; MacRae, I.J.; Kirsch, J.F.; Doudna, J.A. Autoinhibition of human Dicer by its internal helicase domain. J. Mol. Biol. 2008, 380, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Provost, P.; Dishart, D.; Doucet, J.; Frendewey, D.; Samuelsson, B.; Radmark, O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002, 21, 5864–5874. [Google Scholar] [CrossRef] [PubMed]
- Flemr, M.; Malik, R.; Franke, V.; Nejepinska, J.; Sedlacek, R.; Vlahovicek, K.; Svoboda, P. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell 2013, 155, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Peaston, A.E.; Evsikov, A.V.; Graber, J.H.; de Vries, W.N.; Holbrook, A.E.; Solter, D.; Knowles, B.B. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell 2004, 7, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, L.; Berman, M.; Kong, Y.Y.; Dorf, M.E. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity 2011, 35, 426–440. [Google Scholar] [CrossRef]
- Takahashi, T.; Nakano, Y.; Onomoto, K.; Murakami, F.; Komori, C.; Suzuki, Y.; Yoneyama, M.; Ui-Tei, K. LGP2 virus sensor regulates gene expression network mediated by trbp-bound microRNAs. Nucleic Acids Res. 2018, 46, 9134–9147. [Google Scholar] [CrossRef]
- Seo, G.J.; Kincaid, R.P.; Phanaksri, T.; Burke, J.M.; Pare, J.M.; Cox, J.E.; Hsiang, T.Y.; Krug, R.M.; Sullivan, C.S. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe 2013, 14, 435–445. [Google Scholar] [CrossRef]
- Cloonan, N.; Brown, M.K.; Steptoe, A.L.; Wani, S.; Chan, W.L.; Forrest, A.R.; Kolle, G.; Gabrielli, B.; Grimmond, S.M. The miR-17-5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 2008, 9, R127. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, L.H.; Jacobsen, A.B.; Frankel, L.B.; Wen, J.; Krogh, A.; Lund, A.H. MicroRNA-145 targets YES and STAT1 in colon cancer cells. PLoS ONE 2010, 5, e8836. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.Y.; Ezelle, H.J.; Venkataraman, T.; Lapidus, R.G.; Scheibner, K.A.; Hassel, B.A. Regulation of human RNase-L by the miR-29 family reveals a novel oncogenic role in chronic myelogenous leukemia. J. Interferon Cytokine Res. 2013, 33, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Ostermann, E.; Tuddenham, L.; Macquin, C.; Alsaleh, G.; Schreiber-Becker, J.; Tanguy, M.; Bahram, S.; Pfeffer, S.; Georgel, P. Deregulation of type I IFN-dependent genes correlates with increased susceptibility to cytomegalovirus acute infection of Dicer mutant mice. PLoS ONE 2012, 7, e43744. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).