Abstract
Untranslated regions (UTRs) of flaviviruses contain a large number of RNA structural elements involved in mediating the viral life cycle, including cyclisation, replication, and encapsidation. Here we report on a comparative genomics approach to characterize evolutionarily conserved RNAs in the 3UTR of tick-borne, insect-specific and no-known-vector flaviviruses in silico. Our data support the wide distribution of previously experimentally characterized exoribonuclease resistant RNAs (xrRNAs) within tick-borne and no-known-vector flaviviruses and provide evidence for the existence of a cascade of duplicated RNA structures within insect-specific flaviviruses. On a broader scale, our findings indicate that viral 3UTRs represent a flexible scaffold for evolution to come up with novel xrRNAs.
1. Introduction
Flaviviruses are small, single-stranded positive-sense RNA viruses that are typically transmitted between arthropod vectors and vertebrate hosts. They are endemic in tropic and sub-tropic regions and represent a global health threat, although humans are considered dead end hosts in many cases.
The genus Flavivirus within the Flaviviridae family comprises more than 70 species, which are organized into four groups, each with a specific host association: Mosquito-borne flaviviruses (MBFVs) and tick-borne flaviviruses (TBFVs) spread between vertebrate (mammals and birds) and invertebrate (mosquitoes and ticks) hosts, whereas insect-specific flaviviruses (ISFVs) replicate specifically in mosquitoes and no-known-vector flaviviruses (NKVs) have only been found in rodents and bats, respectively. This natural host-range-based classification is in good agreement with sequence-based phylogenetic clustering, mainly because all flaviviruses share a common genome organization [1]. Conversely, epidemiology, disease association [2] and transmission cycles [3] are fundamentally different among different flavivirus groups.
Emerging and re-emerging MBFVs such as Dengue virus (DENV), Japanese encephalitis virus (JEV), West Nile virus (WNV), Yellow fever virus (YFV) or Zika virus (ZIKV) are the causative agents of large-scale outbreaks that result in millions of human and veterinary infections every year [4]. Likewise, tick-borne encephalitis virus (TBEV), Powassan virus (POWV) and other members of the tick-borne serocomplex are neuropathogenic agents that cause a large number of infections every year, resulting in a massive incidence increase since the 1970ies [5]. Consequently, much research effort has gone into studying MBFV and TBFV biology, biochemistry and phylogeny [6]. The two remaining groups, ISFVs and NKVs, however, have received limited attention in the research community, mainly because they are generally not associated with human or veterinary disease and therefore are still underrepresented in the literature. The phylogenetic relationship among the four ecological flavivirus groups is shown in Figure 1. Table A1 lists all viral species considered in the present study.
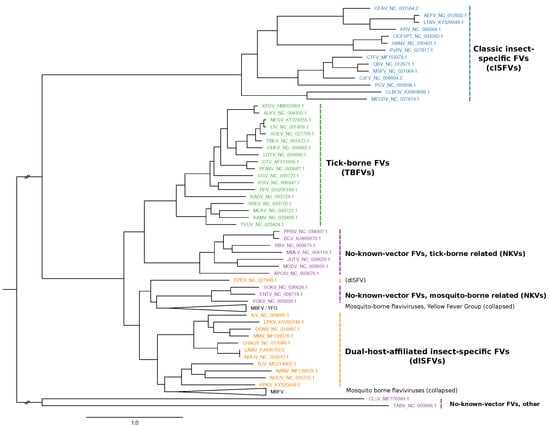
Figure 1.
Maximum-likelihood phylogenetic tree of the genus Flavivirus, highlighting the major groups ISFVs (blue), dISFVs(orange), TBFVs (green), and NKVs (magenta). The MBFV Yellow Fever virus group (YFG) and the main MBFV branch were not covered in this study and are both collapsed. The tree has been computed from a MAFFT alignment of complete polyprotein amino acid sequences with iq-tree. Figure rendered with FigTree.
TBFVs form a monophyletic group consisting of a single serocomplex, although pathology and clinical manifestations vary among different viruses. They comprise more than a dozen of recognized species and separate into three groups: Mammalian tick-borne flaviviruses (M-TBFV), seabird tick-borne flaviviruses (S-TBFV) and the Kadam virus group. See [7] for a comprehensive review.
ISFVs naturally infect hematophagous Diptera and are typically divided into two groups [8]: Classical insect-specific flaviviruses (cISFVs) naturally infect mosquitoes and excursively replicate in mosquito cells in vitro. They form a phylogenetically distinct clade among known flaviviruses, appearing at the root of the MBFV, TBFV and NKV branches. The cISFV group separates into two clades, one associated with Aedes spp. mosquitoes and the other associated with Culex spp. mosquitoes, respectively [9]. They lack the ability to infect vertebrates and to replicate in vertebrate cell lines and have not been in the research spotlight until very recently. The second group is comprised of arbovirus-related or dual-host affiliated insect-specific flaviviruses (dISFVs), which represent a non-monophyletic group which is phylogenetically and antigenically related to mosquito/vertebrate flaviviruses, although they do not appear to infect vertebrate cells [10]. Insect-specific viruses play a crucial role in the mosquito microbiome and have been shown to modulate the replication of other arboviruses [11]. In this line, they are currently considered as biological control agents and vaccine platforms [12].
NKVs represent an ecologically and phylogenetically diverse set of viruses which have been isolated exclusively from vertebrates (mainly bats and rodents), without evidence for transmission by arthropod vectors. They form a non-monophyletic group among flaviviruses and are typically divided into bat- (B-NKV) and rodent-associated (R-NKV) groups, see Table A1. B-NKVs can be further separated into Entebbe virus group, which is phylogenetically closer to MBFVs, and Rio Bravo virus group, which is a sister clade to TBFVs. Species in the R-NKV group form the Modoc virus group, which is phylogenetically close to the B-NKV Rio Bravo group [13]. While NKVs are poorly characterized they represent a valuable resource to study evolutionary traits related to host-switch capacity mediated by conserved genomic elements.
Conserved RNA Structures Mediate Pathogenesis
Conserved RNA structures in the untranslated regions (UTRs) of RNA viruses are of particular interest because they mediate the viral life cycle by promoting or enhancing replication, as proposed for elements in both 5UTRs [14] and 3UTRs [15,16,17,18]. Mosquito/vertebrate viruses must operate efficiently in vectors and hosts, phylogenetically distinct organisms with different cellular machineries. This requires a high level of flexibility of viral regulatory elements to evade various antiviral response strategies while assuring proper replication conditions required for maintaining a stable quasispecies population. To achieve this resilience in host adaptation, RNA duplication strategies have been proposed as an evolutionary trait for MBFVs [19]. Tandem RNA structures within DENV 3UTR are under different selective pressures in alternating hosts, suggesting the idea that duplicated RNA structures differentially evolved to accommodate specific functions in the two hosts [20]. Likewise, there is evidence for evolutionary pressure on maintaining the primary sequence of parts of duplicated RNA elements, as recently shown for flaviviral dumbbell (DB) elements in the context of finding a biophysical model for explaining a possible route for ZIKV-induced neurotropism [21].
Viral RNA genomes are different from procaryotic and eucaryotic mRNA. In addition to coding for and regulating the viral machinery, viral genomic RNA (gRNA) exhibits functional regions that act upon different stages of the viral life cycle. The 10-12kB flaviviral gRNA is capped, but non-polyadenylated and encodes a single open reading frame (ORF). Upon translation, a polyprotein is produced, which is then cleaved by viral and cellular enzymes into structural and non-structural proteins [22]. The ORF is flanked by highly structured untranslated regions (UTRs), which contain evolutionary conserved RNA elements that are crucially related to regulation of the viral life cycle, thereby inducing processes such as genome circularization, viral replication and packaging [23,24,25].
Upon flavivirus infection, accumulation of both gRNA as well as viral long non-coding RNAs (lncRNAs) is observed. These lncRNAs, which have been referred to as subgenomic flaviviral RNAs (sfRNAs) [26] are stable decay intermediates produced by exploiting the host’s mRNA degradation machinery [27] and are associated with viral replication, pathogenesis and cytopathicity [28,29]. The production of sfRNA is induced by partial degradation of viral gRNA by the 5-3 exoribonuclease Xrn1, an enzyme associated with the cell’s RNA turnover machinery [30,31]. Mechanistically, sfRNAs are generated by stalling Xrn1 at conserved structural elements in the viral 3UTR, termed xrRNA (exoribonuclease-resistant RNA elements). These structures efficiently stall Xrn1 from progressing through from the 5 direction, thus protecting the downstream RNA from degradation, while pass-through in the 3-5 direction, as required for viral RNA-dependent RNA-polymerase is still possible [32]. In particular, different types of stem-loop (SL) and dumbbell (DB) elements found in many MBFVs and TBFVs have been related to quantitative protection of downstream virus RNA against exoribonuclease degradation [33].
Xrn1 stalling results in dysregulation of cellular function with the aim of promoting viral infections. In this regard, functions of sfRNA in modulating cellular mRNA decay and RNAi pathways [34] as well as modulating anti-viral interferon response [35,36] have been reported.
The genomic architecture of flaviviruses has been extensively studied to understand the molecular principles required for sfRNA production. Chemical and enzymatic probing methods [37], together with x-ray crystallography revealed the 3UTR structure of the MBFVs WNV [38], YFV [39], DENV [40], Murray Valley encephalitis virus (MVEV) [41], ZIKV [42] and recently different species of the TBFV and NKV groups [33], highlighting the possibility that exoribonuclease resistance might be a pervasive mechanism of the viral world. Interestingly, several conserved RNA structural elements in viral 3UTRs have been predicted in our group [43,44,45,46,47], some of which have later been attributed to xrRNA functionality [26]. To further expand the set of potential xrRNAs, we report here on a comparative genomics survey aimed at characterization of evolutionary conserved RNA structures in flavivirus 3UTRs, focusing on TBFVs and the hitherto understudied groups of ISFVs and NKVs. A detailed study on the evolutionary traits of conserved RNAs in MBFV 3UTRs will be published elsewhere.
2. Materials and Methods
Viral genome data for the present study were obtained from the public National Center for Biotechnology Information (NCBI) refseq (https://www.ncbi.nlm.nih.gov/refseq/) and genbank (https://www.ncbi.nlm.nih.gov/genbank/) databases on 28 May 2018. We downloaded all complete viral genomes under taxonomy ID 11051 (genus Flavivirus) and filtered for TBFV, ISFV and NKV species listed in Table A1. Whenever refseq annotation was not available for a species, we selected the longest complete genome from the genbank set as representative sequence. In total, the data set is comprised of 86 ISFV, 275 TBFV and 27 NKV isolates, respectively. The number of isolates with available 3UTR sequence data per species varies between 1 and 167.
2.1. Phylogeny Reconstruction
The polyprotein/coding sequence (CDS) regions of most flaviviruses can be aligned consistently, however, UTRs typically show large variance both in length and sequence composition, rendering them ill-suited for phylogeny reconstruction. A phylogeny of all members of the genus Flavivirus (Figure 1) was therefore reconstructed via a multiple sequence alignment (MSA) of the nucleotide sequences of the CDS regions only. The MSA was computed with MAFFT[48] and subsequent maximum-likelihood tree reconstruction was performed using iq-tree [49] using the GTR+F+R7 substitution model.
2.2. Structural Homology Search with Covariance Models
The present study is centered around structural homology of RNA elements among phylogenetically narrow subgroups. A straightforward approach to finding novel homologous RNA structures is to search RNA sequence databases with Covariance Models (CMs), i.e., statistical models of RNA structure that extend classic Hidden-Markov-Models (HMMs) to simultaneously represent sequence and secondary structure. CMs, as implemented in the infernal package [50] allow for rapid screening of large RNA sequence databases to find even weakly conserved sequence-only or structurally homologous RNAs. We have recently applied this approach to identify novel telomerase RNAs in Saccharomycetes [51].
Here, structural multiple sequence alignments of the viral 3UTR sequences were generated with locARNA [52] and CMs were built for known or experimentally verified xrRNAs [33]. All 3UTR sequences were then screened and novel candidate sequences were added to perform iterative refinement until convergence. Weak sequence conservation of putative xrRNA elements resulted in initially fragile results, indicating that infernal default parameters are typically not optimal. Adjusting parameters, in particular disabling both heuristic filtering and local end detection, however, allowed our CMs to find homologs with strongly conserved secondary structures in presence of large sequence deviation from the original sequence the CM was built from. Likewise, cmsearch E-values turned out unsuitable for assessing hit quality in case of major sequence divergence. We therefore employed a cutoff approach, requiring a hit to form at least 75% of all base pairs listed in the CM in order to be considered significant.
2.3. De novo Discovery of Conserved RNA Elements
Beside characterization of RNAs with homology to known structurally conserved elements, we aimed at identifying novel elements, considering both thermodynamic stability and sequence covariation as evolutionary traits. In this line, locARNA-generated structural alignments of full UTR sequences were cut manually into blocks corresponding to conserved secondary structures. Alternatively, we employed RNALalifold from the ViennaRNA package [53] to compute locally stable secondary structures for aligned UTR sequences. A CM was built for each structure and searched against all flavivirus 3UTRs, keeping only CMs that scored well multiple times per UTR. The rationale here is that the occurrence of multiple copies hints towards a possible functional role of a structural element, given that the ability of two or more independently evolving sequences to form a common structure is unlikely.
The above approach is implemented as a set of custom Perl and Python scripts for semi-automatic characterization and annotation of conserved RNAs in viral UTR sequences. Internally, these scripts build on the ViennaRNA scripting language interface for thermodynamics calculations, the ViennaNGS [54] suite for extraction of genomic loci, the RNAaliSplit package [55] for splitting alignments into subparts with common consensus structures (i.e., common structures formed by all individual sequences), R2R [56] for visualization, and the ETE3 framework [57] for tree annotation and visualization.
3. Results
Several flaviviruses have previously been studied in great detail, yielding a varied landscape of repeated RNA sequence and structure elements within the 3UTRs of these viruses, which are likely to have evolved from numerous duplications [19,58,59]. Many of these studies relied on single sequence predictions, which resulted in a good understanding of both structure and genomic position of conserved elements in individual species. A unified picture of homologous RNAs within the 3UTRs of flaviviruses, however, has not been available.
The comparative approach applied in the present study outperforms single sequence predictions by considering consensus structures formed by all sequences. This allows us not only to confirm previously described RNA structures but also to elucidate hitherto unrecognized tandem repeats in many species. In this line our results can help in understanding the complex evolution of flavivirus 3UTRs.
3.1. Construction of Seed Alignments
Based on recent experimental evidence for the existence of xrRNAs in TBFVs, ISFVs and NKVs, and previously characterized conserved RNA elements in flaviviral 3UTRs, we built seed alignments for initial CMs, which were then refined iteratively, i.e., subjected to multiple rounds of screening and incorporation of best hits into the CM. Likewise, candidate structures from RNALalifold calculations were used as seeds for identification of conserved RNA structures. Figure 2 shows an overview of refined consensus structures for each ecologic group of flaviviruses analyzed here.
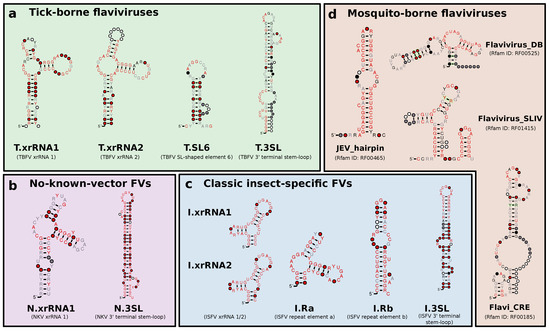
Figure 2.
Overview of consensus structures of all CMs used for the annotation of flavivirus 3UTRs. TBFV, ISFV, and NKV elements were refined from published experimental data (T.xrRNA1/2, I.xrRNA1/2, N.xrRNA) or identified computationally (T.SL6, I.Ra, I.Rb as well as all 3-terminal stem-loop structures). MBFV elements were obtained from Rfam. Throughout this paper, all CMs are referred to by the name written in bold. References to xrRNA-like structures refer to the generalized xrRNA CM (Section 3.6).
The four ecologic groups of flaviviruses show a varied 3UTR architecture, however, the terminal 3 stem-loop structure (3SL, also referred to as 3 long stable hairpin, 3LSH) has been shown to be associated with panhandle-formation during virus replication and is therefore present in the terminal region of all flaviviruses [14]. The element is listed in Rfam as RF00185 (Flavivirus 3UTR cis-acting replication element, Flavi_CRE) and we could use it to consistently identify terminal regions within 3UTRs. Absence of this element from a UTR sequence is indicative of incomplete or truncated sequence data. The underlying sequences generally form a stable stem-loop structure upon structural alignment and single sequence folding. We built individual 3SL seed alignments and CMs for each ecologic group, termed T.3SL, N.3SL and I.3SL, respectively.
3.2. Tick-Borne Flaviviruses
MacFadden et al. [33] suggested two different exoribonuclease-resistant structures in TBFV 3UTRs. We used the proposed sequences from TBEV, POWV, Karshi virus (KSIV), Langat virus (LGTV), Louping ill virus (LIV), Omsk hemorrhagic fever virus (OHFV) and Alkhumra hemorrhagic fever virus (ALKV) as templates for a set of initial structural alignments and CMs. These models were then employed to search for high confidence hits within all TBFV 3UTRs to construct seed alignments of the two exoribonuclease resistant structures in TBFVs, termed T.xrRNA1 and T.xrRNA2. These models allowed us to construct highly specific CMs for both TBFV xrRNAs, which were subsequently used to annotate xrRNA instances in already studied and previously unstudied TBFV species (Figure 3a).
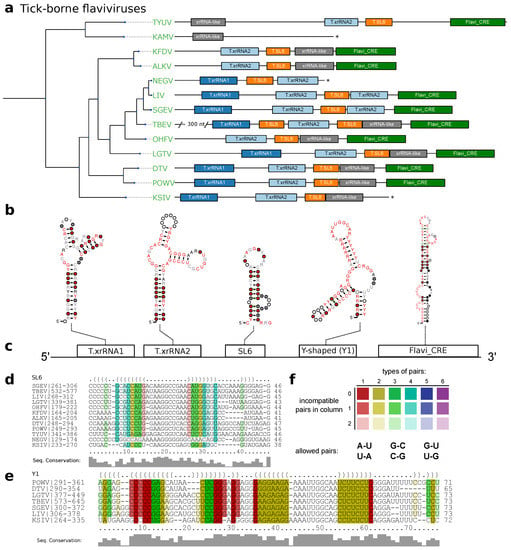
Figure 3.
(a) Annotated 3UTRs of TBFVs. The phylogenetic tree on the left has been computed from complete coding sequence nucleotide alignments and corresponds to the TBFV subtree in Figure 1. For each species with available 3UTR sequence a sketch of the 3UTR is drawn to scale next to the leaves of the tree. Colored boxes represent conserved RNA structural elements. Identifiers within the boxes indicate the CM which was used to infer homology at this position. Asterisks indicate incomplete 3UTR sequences. Species without available 3UTR are not shown. (b) Consensus structure plots of CM hits as calculated by mlocarna. (c) Schematic depiction of the common structural architecture of TBFV 3UTRs. (d,e) Structural alignments of elements SL6 and Y1. (f) RNAalifold coloring scheme for paired columns in alignments. Colors indicate the number of basepair combinations found in pair of columns. Fainter colors indicate that some sequences cannot form a base pair.
The full structural alignment of the 3UTRs of selected tick-borne species moreover suggests a short stem-loop element in several species, which is characterized by high sequence heterogeneity but heavily conserved structure supported by multiple covariations. Evidence for this element, termed stem-loop 6 (SL6), has been reported earlier for at least TBEV, LGTV and OHFV [58,59]. We kept this nomenclature and identified the exact position in each TBFV 3UTR (Figure 3d).
Our data further shows that both TBFV xrRNA CMs (Figure 2a), as well as NKV xrRNA CMs (Figure 2b and Section 3.5) consistently yield plausible hits with a high degree of structure conservation immediately upstream of the strongly conserved terminal stem-loop element. Existence of a Y-shaped element (termed Y1) and putative similarity to NKVs has been proposed earlier based on single sequence structure predictions [58]. Structural locARNA alignment and subsequent RNAalifold consensus structure prediction indicates strong secondary structure conservation with frequent structure-conserving sequence covariations. Taken together, this suggests good evidence that respective regions in TBFVs harbor a putatively structured and functional xrRNA-like RNA (Y1, Figure 3d).
Despite the differences in length and sequence composition, the 3UTRs of most species in the TBFV group share a common architecture. Similar to MBFV SL-elements [19], two copies of xrRNAs are found in almost every species of this ecologic group, generally succeeded by one instance of SL6 and Y1. Likewise, the terminal 3 stem loop is conserved in all TBFVs and can be reliably annotated by both our CM, T.3SL, and the Rfam Flavi_CRE model, which is used in Figure 3. Among all investigated species, only ALKV, OHFV and Kyasanur forest disease virus (KFDV) do not have a copy of xrRNA1, indicating that these viruses may have previously lost this element. Conversely, the two seabird-associated TBFVs with available 3UTR data, Tyuleniy virus (TYUV) and Kama virus (KAMV) do not fit into this general scheme. Likewise, we were not able to annotate additional homologous or conserved structures with any CM used in this screen in the variable region of the 3UTR of TBEV [5], despite the substantially longer UTR (+300 nts).
3.3. Classic Insect-Specific Flaviviruses
Classic insect-specific flaviviruses present diverged 3UTR architectures, which likely result from the association of different species to Aedes spp. and Culex spp. vectors, respectively, which are also reflected by clade separation in the ISFV phylogenetic tree. Previous studies employed single sequence predictions to propose a varied set of homologous RNA structures in combination with an unusually large number of duplicated sequence signals [59]. Recent experimental evidence, however, suggests the presence of xrRNAs that have a similar fold to those known from MBFVs in cISFVs. Consequently, we set out to independently characterize conserved RNA elements for different subclades.
3.3.1. Exoribonuclease-Resistant RNAs in Aedes-Associated cISFVs
MacFadden et al. [33] used SHAPE structure probing to report the presence of two exoribonuclease-resistant RNAs in Cell fusing agent virus (CFAV), and provided evidence for a duplicated set of homologous structures in Aedes flavivirus (AEFV) and Kamiti river virus (KRV). We constructed initial alignments from the reported sequences in this clade in Aedes spp. associated viruses, resulting in two seed alignments, termed I.xrRNA1 and I.xrRNA2 (Figure 2c). For both elements, seed CMs were iteratively built from structural locARNA alignments. Minor manual adjustments to the alignments were required here, since the predicted consensus structures diverged slightly from the published SHAPE-guided prediction. Both models were then employed to search for additional high confidence hits within other isolates of CFAV, AEFV and KRV which were subsequently added to the seed alignments.
Screening the entire set of flavivirus 3UTRs revealed that both ISFV xrRNA elements, I.xrRNA1 and I.xrRNA2, are only found in CFAV, AEFV and KRV, i.e., species initially used for the construction of the respective CMs. Furthermore, also in terms of pure structural conservation, no reliable hits in any other ISFV species could be obtained with any of these CMs (Figure 4a). This suggests that both ISFV xrRNAs may represent a specialized class of xrRNA elements only present in CFAV, AEFV and KRV. The 3UTR of KRV is unique among all known flaviviruses because it harbors an additional copy of the terminal 3 stem-loop element 600 nts upstream of the actual 3-terminus, supporting previous reports that the KRV 3UTR has undergone a full duplication during its evolution [60].
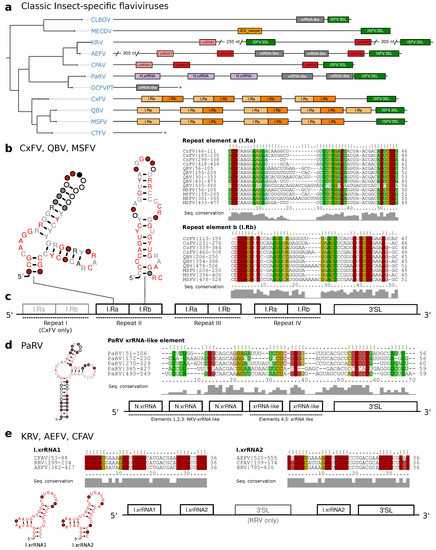
Figure 4.
(a) Annotated Tree of cISFV 3UTRs. Asterisks denote incomplete 3UTR sequences. Species without available 3UTR are not shown. (b) Consensus secondary structure plots and structural alignments of CM hits of Repeat a/b elements in CxFV, QBV, and MSFV. (c) Schematic of the common architecture of CxFV, QBV, and MSFV. Element I-IV refers to the respective repeat of elements. (d) Consensus structure plot and structural alignment of all CM hits of xrRNA-like elements in PaRV. (e) Proposed 3UTR architecture of KRV, AEFV, and CFAV with consensus structure plots and structural alignments of I.xrRNA1 and I.xrRNA2.
3.3.2. Conserved Structures in Culex-Associated cISFVs
The second distinct clade of cISFVs includes Culex flavivirus (CxFV), Quang Binh virus (QBV), Mosquito flavivirus (MSFV), Palm Creek virus (PCV), Culex theileri flavivirus (CTFV) as well as a few other species with only partial genome sequence availability [9] and is associated with Culex spp. vectors. An interesting observation in this clade is that no other CM from any of the four ecologic flavivirus groups shows a hit, not even with remote sequence or structure conservation. We therefore set out to produce a a high quality structural alignment of the complete 3UTRs of CxFV, QBV and MSFV. Consensus structure folding of the full alignment revealed each species to harbor 3–4 repeats of two highly conserved elements supported by multiple co-varying base pairs (Figure 4b,c). We termed these “Repeat element a/b”, respectively (Ra and Rb). Both elements, while strongly conserving their folds, show highly variable loop regions as well as weak sequence conservation in the case of the Ra element. Structure conservation and occurrence in multiple copies, as typically seen with other exoribonuclease-stalling elements, hints towards possible functional importance. These results complement earlier reports of sequence repeats in the 3UTR of CxFV [61] with the identification of a quadruplicated pair of conserved structures.
3.3.3. Diverged 3UTR Architecture in Many cISFVs
Interestingly, a screen of all available CMs in Parramatta River virus (PaRV) revealed five xrRNA-like elements (Figure 4a), with elements 1–3 sharing sequence and structure properties with NKV xrRNAs (Section 3.5), while elements 4 and 5 only conserve N.xrRNAs structure. All five hits can be structurally aligned into a consistent consensus structure (Figure 4d), despite the overall weak sequence consensus.
Conversely, the 3UTRs of Calbertado virus (CLBOV) and Mercadeo virus (MECDV) appear structurally different from the other cISFVs. A general lack of characteristic CM hits lets these species appear more like an outgroup among cISFVs. In particular, we could only find significant hits for the omnipresent terminal 3stem-loop structure, a putative xrRNA-like element in CLBOV and a single instance of a structure homologous to the Rfam model RF00465 (Japanese encephalitis virus hairpin structure) in MECDV. Still, limited availability of 3UTR sequence data renders the characterization of conserved elements and interpretation difficult here.
Our data suggests that the 3UTRs of cISFVs, in contrast to TBFVs (Section 3.2 and dISFVs (Section 3.4), do not appear to have a consistent architectural organization. In agreement with the cISFV phylogenetic subtree (Figure 1) we constitute three diverged groups with common 3UTR organization that conform to their respective sub-clades: (i) CFAV-AEFV-KRV, each with two instances of xrRNAs, (ii) CxFV-QBV-MSFV with 3–4 copies of I.Ra/I.Rb elements and (iii) PaRV with 4–5 copies of xrRNA-like structures. Although no full 3UTR sequences are available for the phylogenetically closest relatives of PaRV, HANV and OCFVPT, an xrRNA-like element in the small available fragment (syntenic to PaRV UTR) of OCFVPT 3UTR suggests that both viruses might be organized in a similar manner, as supported by earlier reports that these viruses should be classified within the same species [1]. For CLBOV and MECDV, no clear pattern of conserved elements can be identified with our CMs. Both viruses either employ an entirely different class of elements or might not require capability for exoribonuclease stalling at all. The only element shared universally among all cISFVs is the 3-terminal stem-loop, although cISFVs seem to diverge from other flaviviruses here, indicated by the inability of Rfam model RF00185 (Flavivirus CRE) to reliably annotate any cISFV 3UTR.
3.4. Dual-Host Affiliated Insect-Specific Flaviviruses
Isolated almost exclusively from mosquitoes, dISFVs do not seem to infect vertebrate cells, despite their phylogenetic proximity to MBFVs (Figure 1). This association is reflected by good hits of the Rfam covariance models RF00525 (Flavivirus DB element) and RF00465 (Japanese encephalitis virus hairpin structure) in all dISFV isolates studied here (Figure 5). Interestingly, we could not find evidence for any sequences or structures homologous to tick-borne or other insect-specific flaviviruses. In this line, our data is in good agreement with the phylogenetic location of these viruses, which share ancestral roots with MBFVs [10].
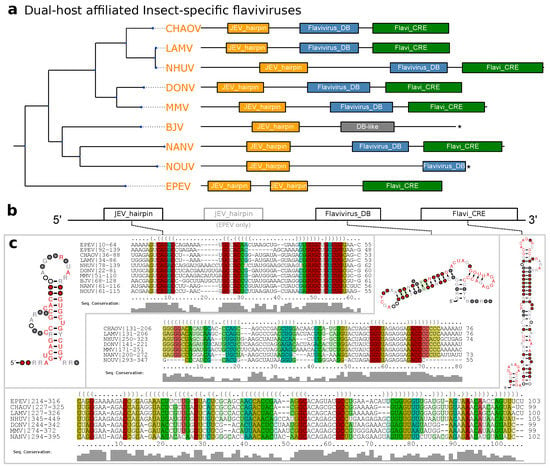
Figure 5.
(a) Annotated Tree of dISFV 3UTRs. Asterisks denote incomplete 3UTR sequences. Species without available 3UTR are not shown. (b) Schematic Architecture of the dISFV 3UTR. (c) Structural alignments and consensus structure plots of dISFV elements.
An unusual species within this group is Ecuador Paraiso Escondido virus (EPEV), which has been isolated from New World sandflies and has been classified as insect-specific virus. EPEV phylogenetically appears at the root of the Entebbe bat virus group (ENTVG), a clade comprised of the three NKVs Entebbe bat virus (ENTV), Sokoluk virus (SOKV) and Yokose virus (YOKV). While all of these viruses contain homologs of conserved stem-loop (SL) and dumbbell (DB) elements found in MBFVs, ENTVG species may have lost their vector dependence [1].
3.5. No-Known-Vector Flaviviruses
Rather than forming a monophyletic group, the no-known-vector flaviviruses can be separated into two distinct lineages, which are closely related to either TBFVs or MBFVs, respectively (Figure 1). Two additional NKVs, Tamana bat virus (TABV) and Cyclopterus lumpus virus (CLuV), are phylogenetically distant and serve as an outgroup to all flaviviruses. In analogy to the procedure outlined above for TBFVs (Section 3.2) and ISFVs (Section 3.3), we built a CM for experimentally verified xrRNAs in tick-borne related NKVs, termed N.xrRNA.
We found multiple hits of this CM at various loci within the 3UTRs of tick-borne related NKVs, indicating that these species, in contrast to TBFVs, do not conserve a common 3UTR architecture (Figure 6a). Surprisingly, we could identify several high-quality hits of the Rfam model RF00525 (Flavivirus DB element), an element typically found in MBFVs, in Rio Bravo virus (RBV), Montana myotis leukoencephalitis virus (MMLV) and Modoc virus (MODV). This is in so far remarkable as there is no evidence for conservation of this element in TBFVs, which phylogenetically cluster with this clade of NKVs. This element might have been introduced by an ancestral recombination event. Alternatively, conservation of an MBFV element in NKVs might be indicative of an association with an unknown vector, in agreement with the hypothesis that vector specificity is mediated by characteristic 3UTR elements [19].
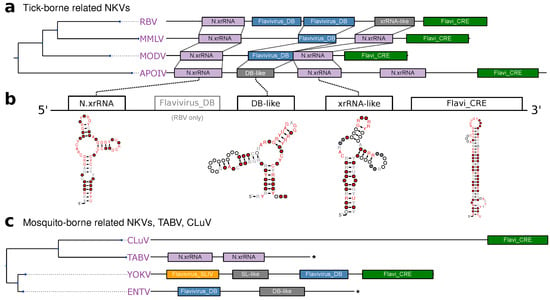
Figure 6.
(a,c) Annotated 3UTRs of NKVs. Asterisks denote incomplete 3UTR sequences. (b) Schematic of TBFV-associated NKV-FV UTR architecture with consensus structures of NKV structure elements.
Conversely, there seems to be no generally conserved 3UTR architecture among members of the mosquito-borne related NKVs (Figure 6c). While sequence data has not been available for Sokoluk virus (SOKV), we could annotate typical MBFV elements in the next relatives Entebbe bat virus (ENTV) and Yokose virus (YOKV), as proposed previously [32].
3.6. A Generalized xrRNA Structure
Earlier work suggested that xrRNAs from TBFVs and tick-borne related NKVs fall into a more general structural class of xrRNAs [33]. Following this line of reasoning, we investigated whether all high confidence hits obtained with our TBFV and NKV CMs could be assembled into one coherent CM that conserves the xrRNA-typical fold. A further advantage of a generalized CM would be higher sensitivity, allowing for identification of common features and eventually lead to annotation of previously unannotated xrRNAs.
Structural alignment and consensus structure prediction revealed all high confidence hits to fold into a common secondary structure (Figure 7a,b). While most of the consensus structure is characterized by low sequence conservation, stem 3 (S3) and loop 1 (L1) show medium to high degree of sequence conservation. The length of all stems is well conserved, although both major loop regions L2 and L3 show large fluctuations, with the length of L3 being de facto constant and L2 showing a high degree of flexibility.
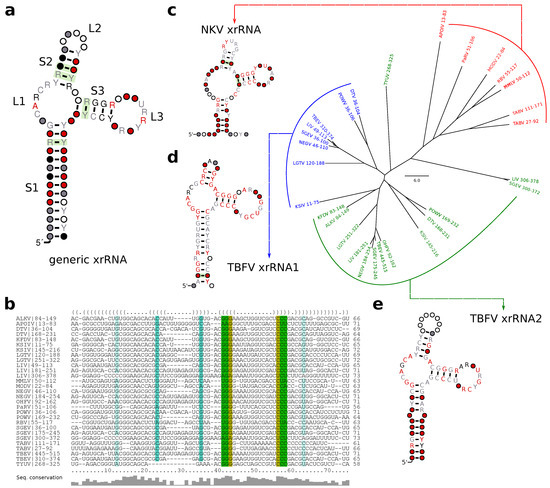
Figure 7.
Generalized structure of all high confidence (cmsearch evalue ) hits of T.xrRNA1, T.xrRNA2 and N.xrRNA. (a) Consensus structure prediction and (b) structural alignment of all high confidence hits. (c–e) Neighbor-joining tree of all high confidence hits of N.xrRNA (c), T.xrRNA1 (d), and T.xrRNA2 (e). Leaves are grouped and colored by the CM used for annotation, coordinates correspond to the position in the respective 3UTR. For each group a separate structural alignment was computed, the consensus structures are shown.
We further investigated whether any high confidence hits from I.xrRNA1/2 in cISFVs could be aligned to the generalized xrRNA model. Although both cISFV xrRNAs (Figure 2c) bear some similarity to the generalized model, in particular to S3 and L3, we were not able to build a common alignment or consensus structure. Despite seemingly similar shape, our data also suggests that cISFV xrRNAs form a separate xrRNA subclass, unrelated to MBFV xrRNAs. In particular, we could not obtain hits of Rfam CMs (which can be seen as representatives of MBFV elements) in cISFVs, nor could we confirm any hits of cISFV-specific elements in MBFVs.
In addition to learning xrRNA features, a more generalized CM enabled us to detect xrRNA-like structures (indicated as such in all annotation plots) that could not be found previously.
4. Discussion
Mediated gRNA decay in the form of exoribonuclease resistance seems to be a pervasive strategy employed by viruses to circumvent host immune responses. Evidence of sfRNA production following incomplete Xrn1 degradation has not only been observed in different members of the Flavivirus genus [62], but also in other species of the Flaviviridae family, however, with major differences in xrRNA structure and sfRNA characteristics. While MBFV produce a 300-500nt sfRNA that corresponds to degradation products of the gRNA 3UTR, hepaciviruses and pestiviruses produce a long subgenomic RNA whose 5 end is located within the first 130nt of the viral gRNA [63].
Moreover, recent studies have identified xrRNA functionality in several phylogenetically distant RNA viruses, such as animal-infecting, segmented viruses of the Bunyaviridae and Arenavividae [64] families, as well as plant-infecting viruses of the Tombusuviridae and Luteoviridae families [65,66]. The interesting question whether exoribonucleases other than Xrn1 would be blocked as well has recently been answered. MacFadden et al. [33] could show that both RNAse J1 and Dxo1 are stalled by MBFV xrRNAs, thereby demonstrating the general nature of this structure-induced blocking mechanism. These novel findings, together with previous knowledge of Xrn1 stalling in segmented plant viruses [67,68] provide evidence for a convergent evolution scenario where xrRNAs depend on a specific folded RNA structure and form a distinct class of functional RNAs.
Repeated RNA elements appear to be a hallmark of flavivirus 3UTR architecture. While there seems to be a plethora of conserved structure classes, our data emphasizes the consistent trend that these elements typically do not occur as single copies. Rather, duplicate or even multiple occurrences of these elements hint towards functional relevance. This is further underlined by the evolutionary conservation of both patterns and elements among different species. Since, exoribonuclease stalling is presumably never perfect, it makes sense that viruses might employ multiple copies of such elements.
In this contribution we set out to identify homologs of known exoribonuclease stalling elements and novel conserved structures. To this end, we computationally characterized homologs of experimentally verified xrRNA in tick-borne and no-known vector viruses that seem to form a coherent class of RNA structures with capability to stall exoribonucleases (T.xrRNA1/2, N.xrRNA). Likewise, we identified another class of xrRNAs in classic insect-specific flaviviruses (I.xrRNA1/2) which appears to be only distantly related to the former class. In the same line, we predicted a set of novel conserved elements in cISFVs that appear in quadruples and do not coincide with other insect-specific elements (I.Ra,I.Rb).
While we did not focus on studying the evolutionary history of these groups of elements in detail, our data suggests that many elements share ancestral roots. This is supported by the observation that at least the tick-borne, no-known-vector and Aedes spp. associated xrRNAs fold into a similar Y-shaped substructure, although the exact fold varies significantly among individual species.
We compiled a set of covariance models that can be used for rapid screening assays in the identification and characterization of novel flaviviruses. All models are available from GitHub via https://github.com/mtw/ITNFV-Data.
A major problem is the limited availability of diverse 3UTR sequence data for many viruses analyzed here, particularly within cISFVs. Many novel ISFVs have previously been discovered, but 3UTR sequences were only available for a subset of them. Future studies are required to shed more light on the evolutionary history of 3UTR evolution in flaviviruses.
Author Contributions
M.T.W. conceived the study. R.O. and M.T.W. analyzed the data, characterized conserved RNA elements and performed phylogenetic studies. All authors contributed to writing the paper and approved of the submitted version.
Funding
This work was funded in part by the Doktoratskolleg RNA Biology at Univ. Vienna and the Austrian Science Fund (SFB F43, I-1303).
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A. Data Set Analyzed in This Study

Table A1.
Viral genomes considered in this study. Flaviviruses are categorized into the groups tick-borne flaviviruses (TBFV), insect-specific flaviviruses (ISFV) and no-known-vector flaviviruses (NKV). The length of the 3UTR is listed for each isolate. Representative accession number from the refseq database. Whenever a refseq genome was not available, the isolate with the longest 3UTR was selected as representative species. Mammalian TBFVs. Seabird TBFVs. Classic ISFVs. Dual-host affiliated ISFVs. e Rodent-associated NKVs. Bat-associated NKVs. 3UTR partial or not available in the refseq data set.
Table A1.
Viral genomes considered in this study. Flaviviruses are categorized into the groups tick-borne flaviviruses (TBFV), insect-specific flaviviruses (ISFV) and no-known-vector flaviviruses (NKV). The length of the 3UTR is listed for each isolate. Representative accession number from the refseq database. Whenever a refseq genome was not available, the isolate with the longest 3UTR was selected as representative species. Mammalian TBFVs. Seabird TBFVs. Classic ISFVs. Dual-host affiliated ISFVs. e Rodent-associated NKVs. Bat-associated NKVs. 3UTR partial or not available in the refseq data set.
| Group | Accession Number | Acronym | Scientific Name | 3UTR Length (nt) | Isolates |
|---|---|---|---|---|---|
| ISFV | NC_012932.1 | AEFV | Aedes flavivirus | 942 | 3 |
| ISFV | NC_001564.2 | CFAV | Cell fusing agent virus | 553 | 3 |
| ISFV | KX669689.1 | CLBOV | Calbertado virus | 546 | 8 |
| ISFV | MF153378.1 | CTFV | Culex theileri flavivirus | 112 | 1 |
| ISFV | NC_008604.2 | CxFV | Culex flavivirus | 654 | 13 |
| ISFV | NC_030401.1 | HANV | Hanko virus | ||
| ISFV | NC_005064.1 | KRV | Kamiti River virus | 1208 | 3 |
| ISFV | NC_027819.1 | MECDV | Mercadeo virus | 638 | 3 |
| ISFV | NC_021069.1 | MSFV | Mosquito flavivirus | 674 | 6 |
| ISFV | NC_034242.1 | OCFVPT | Ochlerotatus caspius flavivirus | 148 | 2 |
| ISFV | NC_027817.1 | PaRV | Parramatta River virus | 629 | 2 |
| ISFV | NC_033694.1 | PCV | Palm Creek virus | ||
| ISFV | NC_012671.1 | QBV | Quang Binh virus | 673 | 2 |
| ISFV | MG214905.1 | BJV | Barkedji virus | 335 | 1 |
| ISFV | NC_017086.1 | CHAOV | Chaoyang virus | 326 | 2 |
| ISFV | NC_016997.1 | DONV | Donggang virus | 343 | 2 |
| ISFV | NC_027999.1 | EPEV | Paraiso Escondido virus | 316 | 2 |
| ISFV | NC_024805.1 | ILV | Ilomantsi virus | ||
| ISFV | KY320648.1 | KPKV | Kampung Karu virus | ||
| ISFV | FJ606789.2 | LAMV | Lammi virus | 326 | 1 |
| ISFV | KY290249.1 | LPKV | Long Pine Key virus | ||
| ISFV | KY320649.1 | LTNV | La Tina virus | ||
| ISFV | MF139576.1 | MMV | Marisma mosquito virus | 376 | 1 |
| ISFV | MF139575.1 | NANV | Nanay virus | 399 | 1 |
| ISFV | NC_024017.1 | NHUV | Nhumirim virus | 451 | 1 |
| ISFV | NC_033715.1 | NOUV | Nounane virus | 347 | 3 |
| NKV e | NC_003676.1 | APOIV | Apoi virus | 576 | 1 |
| NKV | KJ469370.1 | BCV | Batu Cave virus | ||
| NKV | MF776369.1 | CLuV | Cyclopterus lumpus virus | 601 | 1 |
| NKV | NC_008718.1 | ENTV | Entebbe bat virus | 308 | 3 |
| NKV e | NC_026620.1 | JUTV | Jutiapa virus | ||
| NKV | NC_004119.1 | MMLV | Montana myotis leukoencephalitis virus | 460 | 1 |
| NKV e | NC_003635.1 | MODV | Modoc virus | 366 | 1 |
| NKV | NC_034007.1 | PPBV | Phnom Penh bat virus | ||
| NKV | NC_003675.1 | RBV | Rio Bravo virus | 486 | 2 |
| NKV | NC_026624.1 | SOKV | Sokoluk virus | ||
| NKV | NC_003996.1 | TABV | Tamana bat virus | 241 | 1 |
| NKV | NC_005039.1 | YOKV | Yokose virus | 429 | 1 |
| TBFV | NC_004355.1 | ALKV | Alkhumra hemorrhagic fever virus | 393 | 21 |
| TBFV | AF311056.1 | DTV | Deer tick virus | 459 | 1 |
| TBFV | NC_033723.1 | GGV | Gadgets Gully virus | ||
| TBFV | NC_033724.1 | KADV | Kadam virus | ||
| TBFV | NC_023439.1 | KAMV | Kama virus | 282 | 2 |
| TBFV | HM055369.1 | KFDV | Kyasanur forest disease virus | 392 | 6 |
| TBFV | NC_006947.1 | KSIV | Karshi virus | 381 | 3 |
| TBFV | NC_003690.1 | LGTV | Langat virus | 568 | 5 |
| TBFV | NC_001809.1 | LIV | Louping ill virus | 500 | 5 |
| TBFV | NC_033721.1 | MEAV | Meaban virus | ||
| TBFV | KT224355.1 | NEGV | Negishi virus | 266 | 1 |
| TBFV | NC_005062.1 | OHFV | Omsk hemorrhagic fever virus | 410 | 4 |
| TBFV | NC_003687.1 | POWV | Powassan virus | 480 | 23 |
| TBFV | DQ235149.1 | RFV | Royal Farm virus | ||
| TBFV | NC_027709.1 | SGEV | Spanish goat encephalitis virus | 493 | 2 |
| TBFV | NC_033726.1 | SREV | Saumarez Reef virus | ||
| TBFV | NC_001672.1 | TBEV | Tick-borne encephalitis virus | 764 | 167 |
| TBFV | NC_023424.1 | TYUV | Tyuleniy virus | 591 | 3 |
References
- Kuno, G.; Chang, G.J.J.; Tsuchiya, K.R.; Karabatsos, N.; Cropp, C.B. Phylogeny of the Genus Flavivirus. J. Virol. 1998, 72, 73–83. [Google Scholar]
- Gaunt, M.W.; Sall, A.A.; de Lamballerie, X.; Falconar, A.K.; Dzhivanian, T.I.; Gould, E.A. Phylogenetic relationships of flaviviruses correlate with their epidemiology, disease association and biogeography. J. Gen. Virol. 2001, 82, 1867–1876. [Google Scholar] [CrossRef]
- Vasilakis, N.; Weaver, S.C. Flavivirus transmission focusing on Zika. Curr. Opin. Virol. 2017, 22, 30–35. [Google Scholar] [CrossRef]
- Barrows, N.J.; Campos, R.K.; Liao, K.C.; Prasanth, K.R.; Soto-Acosta, R.; Yeh, S.C.; Schott-Lerner, G.; Pompon, J.; Sessions, O.M.; Bradrick, S.S.; et al. Biochemistry and Molecular Biology of Flaviviruses. Chem. Rev. 2018, 118, 4448–4482. [Google Scholar] [CrossRef]
- Kellman, E.; Offerdahl, D.; Melik, W.; Bloom, M. Viral Determinants of Virulence in Tick-Borne Flaviviruses. Viruses 2018, 10, 329. [Google Scholar] [CrossRef] [PubMed]
- Braack, L.; de Almeida, A.P.G.; Cornel, A.J.; Swanepoel, R.; De Jager, C. Mosquito-borne arboviruses of African origin: Review of key viruses and vectors. Parasite Vector 2018, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- Gritsun, T.; Lashkevich, V.; Gould, E. Tick-borne encephalitis. Antivir. Res. 2003, 57, 129–146. [Google Scholar] [CrossRef]
- Guzman, H.; Contreras-Gutierrez, M.A.; da Rosa, A.P.T.; Nunes, M.R.; Cardoso, J.F.; Popov, V.L.; Young, K.I.; Savit, C.; Wood, T.G.; Widen, S.G.; et al. Characterization of three new insect-specific flaviviruses: Their relationship to the mosquito-borne flavivirus pathogens. Am. J. Trop. Med. Hyg. 2018, 98, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Blitvich, B.; Firth, A. Insect-Specific Flaviviruses: A Systematic Review of Their Discovery, Host Range, Mode of Transmission, Superinfection Exclusion Potential and Genomic Organization. Viruses 2015, 7, 1927–1959. [Google Scholar] [CrossRef] [PubMed]
- Alkan, C.; Zapata, S.; Bichaud, L.; Moureau, G.; Lemey, P.; Firth, A.E.; Gritsun, T.S.; Gould, E.A.; de Lamballerie, X.; Depaquit, J.; et al. Ecuador Paraiso Escondido virus, a new flavivirus isolated from New World sandflies in Ecuador is the first representative of a novel clade in the genus Flavivirus. J. Virol. 2015, 89, 11773–11785. [Google Scholar] [CrossRef] [PubMed]
- Halbach, R.; Junglen, S.; van Rij, R.P. Mosquito-specific and mosquito-borne viruses: Evolution, infection, and host defense. Curr. Opin. Insect. Sci. 2017, 22, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Bolling, B.; Weaver, S.; Tesh, R.; Vasilakis, N. Insect-Specific Virus Discovery: Significance for the Arbovirus Community. Viruses 2015, 7, 4911–4928. [Google Scholar] [CrossRef]
- Blitvich, B.; Firth, A. A Review of Flaviviruses that Have No Known Arthropod Vector. Viruses 2017, 9, 154. [Google Scholar] [CrossRef] [PubMed]
- Filomatori, C.V.; Lodeiro, M.F.; Alvarez, D.E.; Samsa, M.M.; Pietrasanta, L.; Gamarnik, A.V. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Gene Dev. 2006, 20, 2238–2249. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, D.E.; Ezcurra, A.L.D.L.; Fucito, S.; Gamarnik, A.V. Role of RNA structures present at the 3′ UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology 2005, 339, 200–212. [Google Scholar] [CrossRef] [PubMed]
- Manzano, M.; Reichert, E.D.; Polo, S.; Falgout, B.; Kasprzak, W.; Shapiro, B.A.; Padmanabhan, R. Identification of Cis-Acting Elements in the 3′-Untranslated Region of the Dengue Virus Type 2 RNA That Modulate Translation and Replication. J Biol. Chem. 2011, 286, 22521–22534. [Google Scholar] [CrossRef]
- Brinton, M.A.; Basu, M. Functions of the 3′ and 5′ genome RNA regions of members of the genus Flavivirus. Virus Res. 2015, 206, 108–119. [Google Scholar] [CrossRef]
- Ng, W.; Soto-Acosta, R.; Bradrick, S.; Garcia-Blanco, M.; Ooi, E. The 5′ and 3′ untranslated regions of the flaviviral genome. Viruses 2017, 9, 137. [Google Scholar] [CrossRef] [PubMed]
- Villordo, S.M.; Carballeda, J.M.; Filomatori, C.V.; Gamarnik, A.V. RNA Structure Duplications and Flavivirus Host Adaptation. Trends Microbiol. 2016, 24, 270–283. [Google Scholar] [CrossRef] [PubMed]
- de Borba, L.; Villordo, S.M.; Marsico, F.L.; Carballeda, J.M.; Filomatori, C.V.; Gebhard, L.G.; Pallarés, H.M.; Lequime, S.; Lambrechts, L.; Vargas, I.S.; et al. RNA Structure Duplication in the Dengue Virus 3′ UTR: Redundancy or Host Specificity? mBio 2019, 10, e02506–18. [Google Scholar] [CrossRef]
- de Bernardi Schneider, A.; Wolfinger, M.T. Musashi binding elements in Zika and related Flavivirus 3′UTRs: A comparative study in silico. bioRxiv 2018. [Google Scholar] [CrossRef]
- Rice, C.M.; Lenches, E.M.; Eddy, S.R.; Shin, S.; Sheets, R.; Strauss, J. Nucleotide sequence of yellow fever virus: Implications for flavivirus gene expression and evolution. Science 1985, 229, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Hahn, C.S.; Hahn, Y.S.; Rice, C.M.; Lee, E.; Dalgarno, L.; Strauss, E.G.; Strauss, J.H. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 1987, 198, 33–41. [Google Scholar] [CrossRef]
- Villordo, S.M.; Alvarez, D.E.; Gamarnik, A.V. A balance between circular and linear forms of the dengue virus genome is crucial for viral replication. RNA 2010, 16, 2325–2335. [Google Scholar] [CrossRef] [PubMed]
- de Borba, L.; Villordo, S.M.; Iglesias, N.G.; Filomatori, C.V.; Gebhard, L.G.; Gamarnik, A.V. Overlapping local and long-range RNA-RNA interactions modulate dengue virus genome cyclization and replication. J. Virol. 2015, 89, 3430–3437. [Google Scholar] [CrossRef] [PubMed]
- Pijlman, G.P.; Funk, A.; Kondratieva, N.; Leung, J.; Torres, S.; Van der Aa, L.; Liu, W.J.; Palmenberg, A.C.; Shi, P.Y.; Hall, R.A.; et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 2008, 4, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, B.M.; Eiler, D.; Kieft, J.S. Structured RNAs that evade or confound exonucleases: Function follows form. Curr. Opin. Struct. Biol. 2016, 36, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.L.; Anderson, J.R.; Kumagai, Y.; Wilusz, C.J.; Akira, S.; Khromykh, A.A.; Wilusz, J. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA 2012, 18, 2029–2040. [Google Scholar] [CrossRef]
- Roby, J.A.; Pijlman, G.P.; Wilusz, J.; Khromykh, A.A. Noncoding subgenomic flavivirus RNA: Multiple functions in West Nile virus pathogenesis and modulation of host responses. Viruses 2014, 6, 404–427. [Google Scholar] [CrossRef]
- Jones, C.I.; Zabolotskaya, M.V.; Newbury, S.F. The 5′-> 3′ exoribonuclease Xrn1/Pacman and its functions in cellular processes and development. Wiley Interdiscip. Rev. RNA 2012, 3, 455–468. [Google Scholar] [CrossRef]
- Antic, S.; Wolfinger, M.T.; Skucha, A.; Hosiner, S.; Dorner, S. General and miRNA-mediated mRNA degradation occurs on ribosome complexes in Drosophila cells. Mol. Cell Biol. 2015. [Google Scholar] [CrossRef]
- Kieft, J.S.; Rabe, J.L.; Chapman, E.G. New hypotheses derived from the structure of a flaviviral Xrn1-resistant RNA: Conservation, folding, and host adaptation. RNA Biol. 2015, 12, 1169–1177. [Google Scholar] [CrossRef]
- MacFadden, A.; O’Donoghue, Z.; Silva, P.A.; Chapman, E.G.; Olsthoorn, R.C.; Sterken, M.G.; Pijlman, G.P.; Bredenbeek, P.J.; Kieft, J.S. Mechanism and structural diversity of exoribonuclease-resistant RNA structures in flaviviral RNAs. Nat. Commun. 2018, 9, 119. [Google Scholar] [CrossRef]
- Hussain, M.; Torres, S.; Schnettler, E.; Funk, A.; Grundhoff, A.; Pijlman, G.P.; Khromykh, A.A.; Asgari, S. West Nile virus encodes a microRNA-like small RNA in the 3′ untranslated region which up-regulates GATA4 mRNA and facilitates virus replication in mosquito cells. Nucleic Acids Res. 2012, 40, 2210–2223. [Google Scholar] [CrossRef] [PubMed]
- Schuessler, A.; Funk, A.; Lazear, H.M.; Cooper, D.A.; Torres, S.; Daffis, S.; Jha, B.K.; Kumagai, Y.; Takeuchi, O.; Hertzog, P.; et al. West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J. Virol. 2012, 86, 5708–5718. [Google Scholar] [CrossRef] [PubMed]
- Bidet, K.; Dadlani, D.; Garcia-Blanco, M.A. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 2014, 10, e1004242. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Wolfinger, M.T.; Tanzer, A.; Hofacker, I.L. Predicting RNA Structures from Sequence and Probing Data. Methods 2016, 103, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Funk, A.; Truong, K.; Nagasaki, T.; Torres, S.; Floden, N.; Melian, E.B.; Edmonds, J.; Dong, H.; Shi, P.Y.; Khromykh, A.A. RNA structures required for production of subgenomic flavivirus RNA. J. Virol. 2010, 84, 11407–11417. [Google Scholar] [CrossRef]
- Silva, P.A.; Pereira, C.F.; Dalebout, T.J.; Spaan, W.J.; Bredenbeek, P.J. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J. Virol. 2010, 84, 11395–11406. [Google Scholar] [CrossRef] [PubMed]
- Sztuba-Solinska, J.; Teramoto, T.; Rausch, J.W.; Shapiro, B.A.; Padmanabhan, R.; Le Grice, S.F. Structural complexity of Dengue virus untranslated regions: cis-acting RNA motifs and pseudoknot interactions modulating functionality of the viral genome. Nucleic Acids Res. 2013, 41, 5075–5089. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.G.; Costantino, D.A.; Rabe, J.L.; Moon, S.L.; Wilusz, J.; Nix, J.C.; Kieft, J.S. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science 2014, 344, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, B.M.; Laurence, H.M.; Massey, A.R.; Costantino, D.A.; Xie, X.; Yang, Y.; Shi, P.Y.; Nix, J.C.; Beckham, J.D.; Kieft, J.S. Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science 2016, 354, 1148–1152. [Google Scholar] [CrossRef]
- Rauscher, S.; Flamm, C.; Mandl, C.W.; Heinz, F.X.; Stadler, P.F. Secondary structure of the 3′-noncoding region of flavivirus genomes: comparative analysis of base pairing probabilities. RNA 1997, 3, 779–791. [Google Scholar] [PubMed]
- Hofacker, I.L.; Fekete, M.; Flamm, C.; Huynen, M.A.; Rauscher, S.; Stolorz, P.E.; Stadler, P.F. Automatic detection of conserved RNA structure elements in complete RNA virus genomes. Nucleic Acid Res. 1998, 26, 3825–3836. [Google Scholar] [CrossRef] [PubMed]
- Witwer, C.; Rauscher, S.; Hofacker, I.L.; Stadler, P.F. Conserved RNA secondary structures in Picornaviridae genomes. Nucleic Acid Res. 2001, 29, 5079–5089. [Google Scholar] [CrossRef]
- Hofacker, I.L.; Stadler, P.F.; Stocsits, R.R. Conserved RNA secondary structures in viral genomes: A survey. Bioinformatics 2004, 20, 1495–1499. [Google Scholar] [CrossRef]
- Thurner, C.; Witwer, C.; Hofacker, I.L.; Stadler, P.F. Conserved RNA secondary structures in Flaviviridae genomes. J. Gen. Virol. 2004, 85, 1113–1124. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold Faster RNA Homology Searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef]
- Waldl, M.; Thiel, B.C.; Ochsenreiter, R.; Holzenleiter, A.; de Araujo Oliveira, J.V.; Walter, M.E.M.; Wolfinger, M.T.; Stadler, P.F. TERribly Difficult: Searching for Telomerase RNAs in Saccharomycetes. Genes 2018, 9, 372. [Google Scholar] [CrossRef] [PubMed]
- Will, S.; Reiche, K.; Hofacker, I.L.; Stadler, P.F.; Backofen, R. Inferring noncoding RNA families and classes by means of genome-scale structure-based clustering. PLoS Comp. Biol. 2007, 3, e65. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Zu Siederdissen, C.H.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithm Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Wolfinger, M.T.; Fallmann, J.; Eggenhofer, F.; Amman, F. ViennaNGS: A toolbox for building efficient next-generation sequencing analysis pipelines. F1000Research 2015, 4. [Google Scholar] [CrossRef]
- Wolfinger, M.T. Bio::RNA::RNAaliSplit 0.09. 2019. Available online: https://github.com/mtw/Bio-RNA-RNAaliSplit (accessed on 23 March 2019).
- Weinberg, Z.; Breaker, R.R. R2R-software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinform. 2011, 12, 3. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Gritsun, T.; Gould, E. Origin and Evolution of 3′UTR of Flaviviruses: Long Direct Repeats as A Basis for the Formation of Secondary Structures and Their Significance for Virus Transmission. Adv. Virus Res. 2006, 69, 203–248. [Google Scholar]
- Gritsun, D.J.; Jones, I.M.; Gould, E.A.; Gritsun, T.S. Molecular Archaeology of Flaviviridae Untranslated Regions: Duplicated RNA Structures in the Replication Enhancer of Flaviviruses and Pestiviruses Emerged via Convergent Evolution. PLoS ONE 2014, 9, e92056. [Google Scholar] [CrossRef]
- Gritsun, T.; Gould, E. The 3′ untranslated regions of Kamiti River virus and Cell fusing agent virus originated by self-duplication. J. Gen. Virol. 2006, 87, 2615–2619. [Google Scholar] [CrossRef]
- Hoshino, K.; Isawa, H.; Tsuda, Y.; Yano, K.; Sasaki, T.; Yuda, M.; Takasaki, T.; Kobayashi, M.; Sawabe, K. Genetic characterization of a new insect flavivirus isolated from Culex pipiens mosquito in Japan. Virology 2007, 359, 405–414. [Google Scholar] [CrossRef]
- Clarke, B.; Roby, J.; Slonchak, A.; Khromykh, A. Functional non-coding RNAs derived from the flavivirus 3′ untranslated region. Virus Res. 2015, 206, 53–61. [Google Scholar] [CrossRef]
- Moon, S.L.; Blackinton, J.G.; Anderson, J.R.; Dozier, M.K.; Dodd, B.J.; Keene, J.D.; Wilusz, C.J.; Bradrick, S.S.; Wilusz, J. X1 Stalling in the 5’UTR of Hepatitis C Virus and Bovine Viral Diarrhea Virus Is Associated with Dysregulated Host mRNA Stability. PLoS Pathog. 2015, 11, e1004708. [Google Scholar] [CrossRef]
- Charley, P.A.; Wilusz, C.J.; Wilusz, J. Identification of Phlebovirus and Arenavirus RNA sequences that stall and repress the exoribonuclease XRN1. J. Biol. Chem. 2018, 293, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Steckelberg, A.L.; Akiyama, B.M.; Costantino, D.A.; Sit, T.L.; Nix, J.C.; Kieft, J.S. A folded viral noncoding RNA blocks host cell exoribonucleases through a conformationally dynamic RNA structure. Proc. Natl. Acad. Sci. USA 2018, 115, 6404–6409. [Google Scholar] [CrossRef] [PubMed]
- Steckelberg, A.L.; Vicens, Q.; Kieft, J.S. Exoribonuclease-Resistant RNAs Exist within both Coding and Noncoding Subgenomic RNAs. mBio 2018, 9, e02461-18. [Google Scholar] [CrossRef] [PubMed]
- Iwakawa, H.o.; Mizumoto, H.; Nagano, H.; Imoto, Y.; Takigawa, K.; Sarawaneeyaruk, S.; Kaido, M.; Mise, K.; Okuno, T. A viral noncoding RNA generated by cis-element-mediated protection against 5′- 3′ RNA decay represses both cap-independent and cap-dependent translation. J. Virol. 2008, 82, 10162–10174. [Google Scholar] [CrossRef] [PubMed]
- Flobinus, A.; Hleibieh, K.; Klein, E.; Ratti, C.; Bouzoubaa, S.; Gilmer, D. A viral noncoding RNA complements a weakened viral RNA silencing suppressor and promotes efficient systemic host infection. Viruses 2016, 8, 272. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).