Significance of Coronavirus Mutants in Feces and Diseased Tissues of Cats Suffering from Feline Infectious Peritonitis
Abstract
:1. Introduction
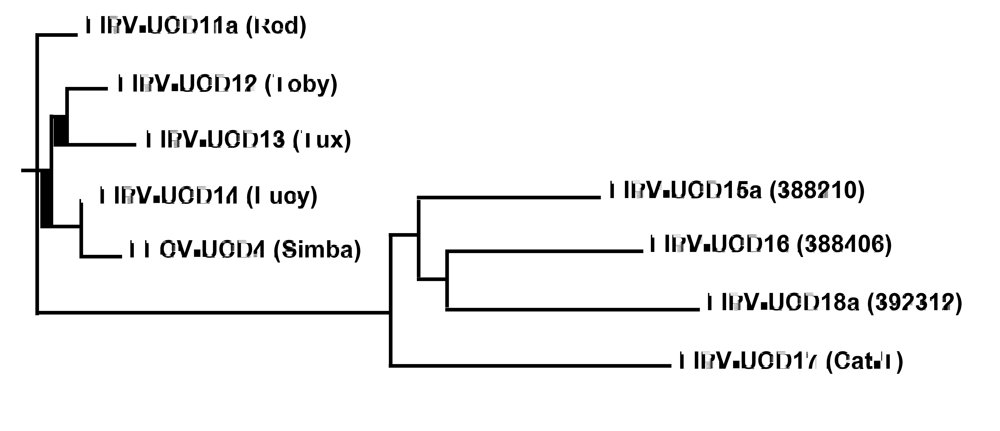
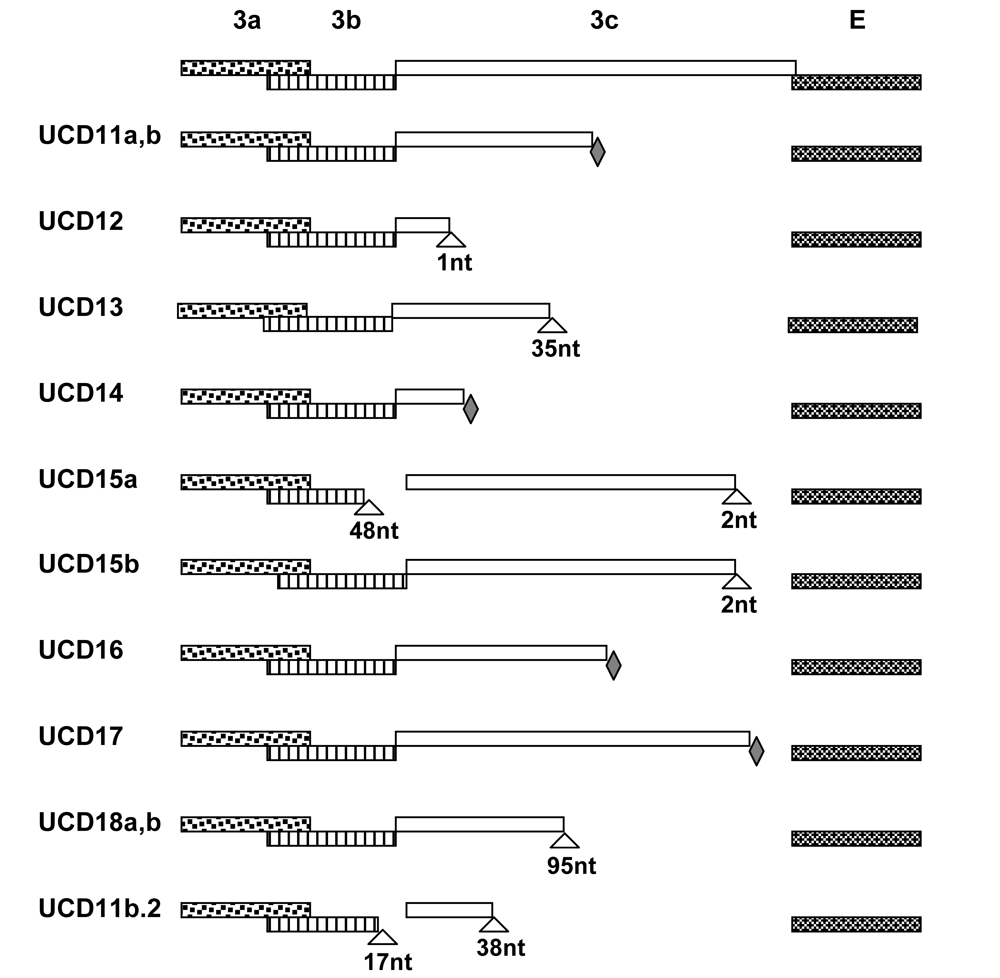
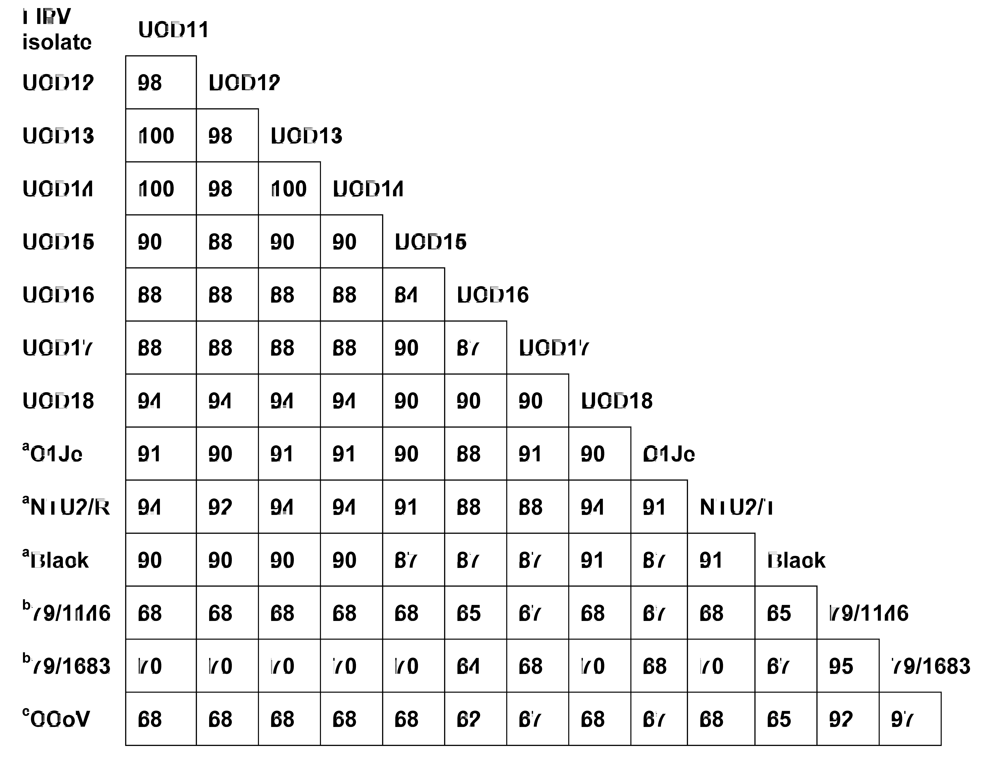
2. Results and Discussion
2.2. Experimental infection of laboratory cats establishes that FIPV-UCD11a, b possess the FIP biotype and that co-infection with both variants leads to infection with one or the other variant but not both
| Cat # | Isolate | Genes sequenced | #SNPs/nts sequenced a | Type of mutation in 3c | GenBank Accession # |
|---|---|---|---|---|---|
| 07-036 | FIPV-UCD11a.1ab | E,M,N,3a-c,7a,b | 5/6711 | Stop codon same as FIPV-UCD11a | FJ917530 |
| FIPV-UCD11a.1bc | 7/6711 | FJ917531 | |||
| 05-243 | FIPV-UCD11b.1ab | E,M,N,3a-c,7a,b | 6/4680 | Stop codon same as FIPV-UCD11b | FJ917532 |
| FIPV-UCD11b.1bc | 7/4680 | FJ917533 | |||
| 98-272 | FIPV-UCD11b.2ab | S,E,M,N,3a-c,7a,b | 5/8943 | Stop codon same as FIPV-UCD11b, plus deletions in 3b,c | FJ917534 |
| FIPV-UCD11b.2bc | 8/8943 | FJ917535 |
2.5. There was no evidence for cat-to-cat (i.e., horizontal) transmission of 3c gene mutants among cats in the same environment
2.6. Precedence and possible role for functional 3c gene mutations
3. Experimental Section
3.2. Subjects
3.3. FIPV transmission studies
3.6. Cycle sequencing
4. Conclusions
Acknowledgments
References
- Holzworth, J.E. Some important disorders of cats. Cornell Vet. 1963, 53, 157–160. [Google Scholar] [PubMed]
- Feldman, B.F.; Jortner, B.S. Clinico-pathology conference. J. Am. Vet. Med. Assoc. 1964, 144, 1409–1411. [Google Scholar] [PubMed]
- Rohrbach, B.W.; Legendre, A.M.; Baldwin, C.A.; Lein, D.H.; Reed, W.M.; Wilson R.B. Epidemiology of feline infectious peritonitis among cats examined at veterinary medical teaching hospitals. J. Am. Vet. Med. Assoc. 2001, 218, 1111–1115. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, L.G.; Griesemer, R.A. Feline infectious peritonitis. Pathol. Vet. 1966, 3, 255–270. [Google Scholar] [PubMed]
- Zook, B.C.; King, N.W.; Robinson. R.L.; McCombs H.L. Ultrastructural evidence for the viral etiology of feline infectious peritonitis. Pathol. Vet. 1968, 5, 91–95. [Google Scholar]
- Ward, J. Morphogenesis of a virus in cats with experimental feline infectious peritonitis. Virol. 1970, 41, 191–194. [Google Scholar] [CrossRef]
- Montali, R.J.; Strandberg, J.D. Extraperitoneal lesions in feline infectious peritonitis. Vet. Pathol. 1972, 9, 109–121. [Google Scholar] [PubMed]
- Pedersen, N.C.; Ward, W. Antigenic relationship of the feline infections peritonitis virus to coronaviruses of other species. Arch. Virol. 1978, 58, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, N. C.; Black, J. W.; Boyle, J. F.; Evermann, J. F.; McKeirnan, A. J.; Ott, R. L. Pathogenic differences between various feline coronavirus isolates. Coronaviruses; molecular biology and pathogenesis. Adv. Exp. Med. Biol. 1984, 173, 365–380. [Google Scholar] [PubMed]
- Pedersen, N.C.; Boyle, J.F.; Floyd, K.; Fudge, A.; Barker J. An enteric coronavirus infection of cats and its relationship to feline infectious peritonitis. Am. J. Vet. Res. 1981, 42, 368–377. [Google Scholar] [PubMed]
- Pedersen, N.C. A review of feline infectious peritonitis virus infection: 1963-2008. J. Feline Med. Surg. 2009, 11, 225–258. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, N.C.; Allen, C.E.; Lyons L.A. Pathogenesis of feline enteric coronavirus infection. J. Feline Med. Surg. 2008, 10, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, N.C. Virologic and immunologic aspects of feline infectious peritonitis virus infection. Adv. Exp. Med. Biol. 1987, 218, 529–550. [Google Scholar] [PubMed]
- Horzinek, M.C.; Herrewegh, A.; de Groot, R.J. Perspectives on feline coronavirus evolution. Feline Pract. 1995, 23, 34–39. [Google Scholar]
- Hickman, M.A.; Morris, J.G.; Rogers, Q.R.; Pedersen, N.C. Elimination of Feline Coronavirus Infection from a Large Experimental Specific Pathogen-Free Cat Breeding Colony by Serologic Testing and Isolation. Feline Pract. 1995, 23, 96–102. [Google Scholar]
- Poland, A.M.; Vennema, H.; Foley, J.E.; Pedersen, N.C. Two related strains of feline infectious peritonitis virus isolated from immunocompromised cats infected with a feline enteric coronavirus. J. Clin. Microbiol. 1996, 34, 3180–3184. [Google Scholar] [PubMed]
- Vennema, H.; Poland, A.; Foley, J.; Pedersen, N.C. Feline infectious peritonitis viruses arise by mutation from endemic feline enteric coronaviruses. Virol. 1998, 243, 150–157. [Google Scholar] [CrossRef]
- Rottier, P. J.; Nakamura, K. Acquisition of macrophage tropism during the pathogenesis of feline infectious peritonitis is determined by mutations in the feline coronavirus spike protein. J. Virol. 2005, 79, 14122–14130. [Google Scholar] [CrossRef] [PubMed]
- Dye, C.; Siddell, S. G. Genomic RNA sequence of feline coronavirus strain FCoV C1Je. J Feline Med. Surg. 2007, 9, 202–213. [Google Scholar] [CrossRef]
- Herrewegh, A.A.; Mähler , M.; Hedrich, H.J.; Haagmans, B.L.; Egberink, H.F.; Horzinek, M.C.; Rottier, P.J.; de Groot, R.J. Persistence and evolution of feline coronavirus in a closed cat-breeding colony . Virol. 1997, 4 , 349–363. [Google Scholar]
- Lin, C-N.; Su, B-L.; Huang, H-P.; Lee, J-J.; Hsieh, M-W.; Chueh, L-L. Field strain feline coronaviruses with small deletions in ORF7b associated with both enteric infection and feline infectious peritonitis. J. Feline Med. Surg. 2009, 11, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.; Boedeker, N.; Gibbs, P.; Kania, S. Deletions in the 7a ORF of feline coronavirus associated with an epidemic of feline infectious peritonitis. Vet. Microbiol. 2001, 8, 227–234. [Google Scholar] [CrossRef]
- Kiss, I.; Kecskeméti, S.; Tanyi, J.; Klingeborn, B.; Belák S. Preliminary studies on feline coronavirus distribution in naturally and experimentally infected cats. Res. Vet. Sci. 2000, 68, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Addie, D. D.; Kennedy, L. J.; Nart, P ; Radford, A. D. Feline leucocyte antigen class II polymorphism and susceptibility to feline infectious peritonitis. J. Feline Med. Surg. 2004, 6, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Kipar, A.; Baptiste, K.; Barth, A.; Reinacher, M. Natural FCoV infection: cats with FIP exhibit significantly higher viral loads than healthy infected cats. J. Feline Med. Surg. 2006, 8, 69–72. [Google Scholar] [CrossRef]
- Kipar , A.; Meli, M. L.; Failing, K.; Euler, T.; Gomes-Keller, M. A.; Schwartz, D.; Lutz, H.; Reinacher, M. Natural feline coronavirus infection: differences in cytokine patterns in association with the outcome of infection. Vet. Immunol. Immunopathol. 2006, 112, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Meli, M.; Kipar, C.; Müller, A.; Jenal, K.; Gönczi, E.; Borel, N.; Gunn-Moore, D.; Chalmers, S.; Linn, F.; Rinacher, M.; Lutz, H. High viral loads despite absence of clinical and pathological findings in cats experimentally infected with feline coronavirus (FCoV) type I and in naturally FCoV-infected cats. J. Feline Med. Surg. 2004, 6, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Paltrinieri, S.; Gelain, M.E.; Ceciliani, F.; Ribera, A.M.; Battilani, M. Association between faecal shedding of feline coronavirus and serum alpha1-acid glycoprotein sialylation. J. Feline Med. Surg. 2008, 10, 514–518. [Google Scholar] [CrossRef] [PubMed]
- Foley, J. E.; Pedersen, N. C. Inheritance of susceptibility of feline infectious peritonitis in purebred catteries. Feline Pract. 1996, 24, 14–22. [Google Scholar]
- Foley, J. E.; Poland, A.; Carlson, J.; Pedersen, N.C. Risk factors for feline infectious peritonitis among cats in multiple-cat environments with endemic feline enteric coronavirus. J. Am. Vet. Med. Assoc. 1997, 210, 1313–1318. [Google Scholar] [PubMed]
- Battilani, M.; Coradin, T.; Scagliarini, A.; Ciulli, S.; Ostanello, F.; Prosperi, S.; Morganti, L. Quasispecies composition and pylogenetic analysis of feline coronaviruses (FCoVs) in naturally infected cats. FEMS 32. Immunol. Med. Microbiol. 2003, 39, 141–147. [Google Scholar] [CrossRef]
- Mochizuki, M.; Mitsutake, Y.; Miyanohara, Y. Antigenic and plaque variations of serotype II feline infectious peritonitis coronaviruses. J. Vet. Med. Sci. 1997, 59, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Addie, D.D.;Schaap; Nicolson, L; Jarrett, O. Persistence and transmission of natural type I feline coronavirus infection. J. Gen. Virol. 2003, 84, 2735–2744. [Google Scholar] [CrossRef] [PubMed]
- Herrewegh, A.A.; Vennema, H.; Horzinek, M.C.; Rottier, P.J.; de Groot, R.J. The molecular genetics of feline coronaviruses: comparative sequence analysis of the ORF7a/7b transcription unit of different biotypes. Virol. 1995, 212, 622–631. [Google Scholar] [CrossRef]
- Hardy, Jr., W.D.; Hurvitz, L. Feline infectious peritonitis: Experimental studies. J Am. Vet. Med. Assoc. 1971, 158, 994–1002. [Google Scholar] [PubMed]
- Can-Sahna, K.; Soydal Ataseven, V.; Pinar, D.; Oğuzoğlu, T. C. The detection of feline coronaviruses in blood samples from cats by mRNA RT-PCR. J. Feline Med. Surg. 2004, 9, 369–372. [Google Scholar] [CrossRef]
- Oostra, M.; de Haan, C.A.M.; de Groot, R.J.; Rottier P.J.M. Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M. J. Virol. 2006, 80, 2326–2336. [Google Scholar] [CrossRef] [PubMed]
- Ito, N.; Mossel, E. C.; Narayanan, K.; Popov, V. L.; Huang, C.; Inoue, T.; Peters, C. J.; Makino, S. Severe acute respiratory syndrome coroanvirus 3a protein is a viral structural protein. J. Virol. 2005, 79, 3182–3186. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Guo, Z.; Yang, H.; Peng, L.; Xie, Y.; Wong, T. Y.; Lai, S. T.; Guo, Z. Amino terminus of the SARS coronavirus protein 3a elicits strong, potentially protective humoral responses in infected patients. J. Gen. Virol. 2006, 87, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y. J. The Severe Acute Respiratory Syndrome (SARS)-coronavirus 3a protein may function as a modulator of the trafficking properties of the spike protein. Virol. J. 2005, 10, 2–5. [Google Scholar]
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Share and Cite
Pedersen, N.C.; Liu, H.; Dodd, K.A.; Pesavento, P.A. Significance of Coronavirus Mutants in Feces and Diseased Tissues of Cats Suffering from Feline Infectious Peritonitis. Viruses 2009, 1, 166-184. https://doi.org/10.3390/v1020166
Pedersen NC, Liu H, Dodd KA, Pesavento PA. Significance of Coronavirus Mutants in Feces and Diseased Tissues of Cats Suffering from Feline Infectious Peritonitis. Viruses. 2009; 1(2):166-184. https://doi.org/10.3390/v1020166
Chicago/Turabian StylePedersen, Niels C., Hongwei Liu, Kimberly A. Dodd, and Patricia A. Pesavento. 2009. "Significance of Coronavirus Mutants in Feces and Diseased Tissues of Cats Suffering from Feline Infectious Peritonitis" Viruses 1, no. 2: 166-184. https://doi.org/10.3390/v1020166
APA StylePedersen, N. C., Liu, H., Dodd, K. A., & Pesavento, P. A. (2009). Significance of Coronavirus Mutants in Feces and Diseased Tissues of Cats Suffering from Feline Infectious Peritonitis. Viruses, 1(2), 166-184. https://doi.org/10.3390/v1020166