Abstract
The argonaute (AGO) protein, as an important member of the small RNA (sRNA) regulatory pathway gene-silencing complex (RNA-induced silencing complex, RISC), is a key protein that mediates gene silencing and plays a key role in the recruitment of sRNAs. In this study, bioinformatics was used to identify the AGO gene family in poplar and study its expression in various tissues and in response to abiotic stress treatments. A total of 15 PtAGO genes were identified in poplar, which were unevenly distributed in 9 chromosomes. Most proteins were predicted to be located in the nucleus and chloroplast. The PtAGOs had similar motif structures and conserved motifs, except for PtAGO3. All the PtAGO genes could be clustered into 3 groups, and Group II, including PtAGO2/3/7, had the smallest number of exons, while the others had more than 20 exons. Cis-regulatory elements involved in light response, growth and development, abiotic stress and hormone-induced responses were found in the promoters of PtAGO members. Further expression analysis found that the PtAGO genes had tissue-specific expression patterns. For example, PtAGO7 and PtAGO10b were mainly expressed in the xylem and might be involved in secondary xylem development. Furthermore, abiotic stress tests, including heat, ABA and PEG treatments, showed that most PtAGO genes could respond quickly to ABA treatment, and multiple PtAGO genes were constantly regulated under heat-shock stress. These results provide a basis for the elucidation mechanism of PtAGO genes and further molecular breeding in poplar.
1. Introduction
Small RNA (sRNA) mediates gene silencing at multiple levels of epigenetic control, including chromatin, transcriptional and post-transcriptional levels, and plays essential roles in plant growth and development, as well as environmental stimuli responses. During this process, sRNA combined with argonaute (AGO) protein forms an RNA-induced silencing complex (RISC). Through binding sRNA and inducing mRNA cleavage, translation inhibition or chromatin modification, RISC mediates the regulation of target gene expression through complementary sequences in plants [1]. The first AGO protein was identified in Arabidopsis thaliana (L.) Heynh. The leaves of AGO1 loss-of-function mutants appear straw-shaped, and the whole plant seems to exhibit the tentacles of a marine organism such as squid; hence, this protein was named AGO [2]. The AGO protein was found to contain PAZ, MID and PIWI domains and a variable N-terminus, in which PAZ and PIWI are conserved domains [3]. The PAZ domain was used to recognize and specifically bind to the 3′ end of the sRNA convex, while the variable N-terminus was used to identify and separate sRNA and further combine the targeted genes [4]. The “bag” structure of the MID domain, however, specifically binds to the 5′end of sRNA, thereby immobilizing the single-stranded sRNA on the AGO protein [5]. Meanwhile, the MID domain may interact with specific proteins [6] and further affect their functions. The PIWI domain has catalytic activity, which is similar to endogenous restriction nucleases [7].
Recently, AGO protein family members were identified in A. thaliana, rice (Oryza sativa L.) [8], tomato (Solanum lycopersicum Miller) [9], soybean (Glycine max (Linn.) Merr.) [10], Capsicum Annuum L. [11] and other plant species. A total of 10 AtAGOs were mainly clustered into three groups, including Group I (AtAGO1/5/10), Group II (AtAGO2/3/7) and Group III (AtAGO4/6/8/9) [12]. The AGO-sRNA regulation pathway has been widely researched regarding Arabidopsis and other herbaceous plants. AGO proteins bind to various sRNA and then play important roles in plant gamete formation, growth and development and stress response. As one of the most widely cultivated trees worldwide, poplar is a key source of raw wood owing to its fast growth and high-quality wood. However, only a few AGO members have been identified in woody plant poplar [13,14].
In this study, we comprehensively identified the members of AGO genes in Populus trichocarpa. Phylogeny, gene structure and chromosomal distribution of the AGOs were determined, and their expression patterns in various tissues and responses to various stress treatments were analyzed. These results will be helpful for the further functional characterization and genetic modification of AGO genes in woody plants.
2. Materials and Methods
2.1. Identification of AGO genes in Poplar
The amino acid (AA) sequences of AGO in Arabidopsis were downloaded from the NCBI database, and the conserved domains were searched for using the Pfam protein family database (Pfam 35.0). Next, BLAST with the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 1 April 2022) and TBtools [15] were used to identify and align AGO genes in 13 selected species, including Volvox carteri, Chlamydomonas reinhardtii, Marchantia polymorpha, Selaginella moellendorffii, Amborella trichopoda, Brachypodium distachyon, O. sativa, Zea mays, Eucalyptus grandis, Populus trichocarpa, A. thaliana, Vitis vinifera and Manihot esculenta. Then, the conserved motifs of these candidate AGO genes were searched for and annotated using the Pfam 35.0 program and SMART. Furthermore, invalid and redundant sequences were removed, and high-fidelity AGO genes were obtained.
2.2. Phylogenetic and Characteristic Analysis of the AGO gene Family
Multiple sequence alignment of the AGO proteins was performed using ClustalW of MEGA 7.0. Additionally, the phylogenetic tree was constructed using the neighbor-joining method with 1000 bootstrap replicates to test its stability. Then, the number of AA substitutions (pairwise distance) between sequences was calculated using MEGA 7.0. Candidate AGO genes were named according to the nomenclature of previously identified genes and their phylogenetic correlations. Then, Evolview (https://www.evolgenius.info/evolview, accessed on 30 March 2023) was further used to adjust the phylogenetic tree for visualization.
The sequence length, CDS information, gene structure and translated protein sequence of the identified PtAGO genes were summarized. The molecular weight, isoelectric point and stability analysis of the PtAGO proteins was conducted through Expasy (https://web.expasy.org/protparam, accessed on 30 March 2023). The subcellular localization of the PtAGO proteins was predicted using WoLF PSORT II (https://www.genscript.com/wolf-psort.html, accessed on 30 March 2023).
2.3. Conserved Motif, Exon, Intron and Chromosome Location of AGO Members in Poplar
The online website MEME (http://meme-suite.org/tools/meme, accessed on 30 March 2023) was used to analyze the conserved motifs, with a maximum number of 10. Gene structure analysis and visualization were performed using TBtools. Meanwhile, the chromosomal mapping of the PtAGO genes was also extracted using TBtools.
2.4. Cis-Regulatory Element Prediction of AGO genes in Poplar
The upstream 2000 bp of the PtAGOs was extracted and submitted to the online website PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 30 March 2023) for the prediction of cis-elements in the gene promoter region. Visualization was performed with TBtools software.
2.5. Plant Materials and Treatments
Poplar 84 K (Populus alba × P. glandulosa) was used throughout the whole experiment. The seedlings were cultured in vitro for 4 months and then transferred to soil with 16 h light/8 h dark at 23 ± 2 °C. After 1 month of culture, the apical terminal, young leaves, mature leaves, lateral roots and main roots were collected. The developing xylem was scratched from the surface of the peeled stem and the xylem was cut from the scratched xylem. The phloem was scratched from the internal surface of peeled bark, including phloem and cambium cells.
Heat treatment was carried out as previously described [16]. The plants were placed in a light incubator of 23 ± 2 °C and then transferred to another incubator of 37 °C (pretreatment) for 3 h, returned to 23 °C for 2 h, transferred to 42 °C (treatment) for 3 h and then returned to the 23 °C incubator for 2 h. The leaves were successively harvested at 7 series time-points, including controls.
For the PEG6000 and ABA treatments, 1-month-old plants were transferred to 1/2 MS liquid medium for 3 days to adapt to the culture conditions. Subsequently, the plants were cultured in 10% (w/v) polyethylene glycol (PEG6000) with 1/2 Hoagland nutrient solution, and leaves grown on the 2–4th internodes were collected at 0, 3, 6, 12, 24, 48 and 96 h after treatment. For the ABA treatment, 100 μM ABA was used, and leaves from the 2–4th internodes were collected at 0, 6, 12, 24, 48 and 96 h after treatment. The collected leaves were snap-frozen using liquid nitrogen and then stored at −80 °C for RNA extraction. Three biological replicates were performed for each treatment.
2.6. RNA Isolation and qRT-PCR
RNA isolation was performed using an RNA extraction kit (TIANGEN), and the quality and concentration were analyzed by gel electrophoresis and an ultramicrophotometer (NanoDrop 2000, Thermo Fisher Scientific, Waltham, MA, USA). cDNA was produced from 1 µg total RNA using the M5 SupperFast qPCR RT kit. qRT-PCR was performed using 2 × M5 HiPer SYBR Premix EsTaq (Mei5bio, Beijing, China) in a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). PagACTIN1 (Potri. 001G309500) and PagUBQ (Potri.001G418500) were used as reference genes to normalize cDNAs of the different treatments. Each reaction was performed in three replicates, and the relative expression value was calculated using the 2−ΔΔCT method and plotted using TBtools. Primer sequences of the PtAGOs and reference gene used for qRT-PCR are listed in Table S1.
3. Results
3.1. Identification and Evolution Analysis of AGO genes in Poplar
The conserved domain of the AGO family was first analyzed using the Pfam protein family database. Then, the conserved domains were used to blast the candidate AGO genes in 13 plant species. The genes with incomplete domains and low reliability values were deleted. Finally, 145 AGO genes with high reliability were identified in the 13 plant species. Although AGO genes were found in all the selected plant species, the number of members varies among different species. The smallest AGO gene family was found in algae, and only 2 members were identified, while the largest AGO family was found in monocotyledon, with 19 in rice and 18 in maize. Hence, these results indicate that the members of the AGO family gradually increased with the evolution from algae, bryophyte, fern and early angiosperm to dicotyledon and monocotyledon. To discover the relationships of all the AGO genes in these 13 species, a phylogenetic tree was constructed (Figure 1). All the AGOs were clustered into three groups, including Group I (AGO1/5/10), Group II (AGO2/3/7) and Group III (AGO4/6/8/9), as observed with the groups in Arabidopsis. The detailed gene information is listed in Table 1. The number of genes in Group I (73/152) was greater than that in Group II (31/152) and Group III (41/152). Meanwhile, the AGOs in algae including V. carter and C. reinhardtii, were grouped in Group III, while the AGOs in M. polymorpha and S. moellendorffii were mainly grouped in Group I and Group III. Interestingly, all AGOs in Group II originated from angiosperm, which suggests that these genes were newly developed. Furthermore, distinct numbers of AGO genes were found in different groups among various species. A total of 15 PtAGO genes were identified in P. trichocarpa, with 8, 3 and 4 genes in Group I, Group II and Group III, respectively (Table 1).
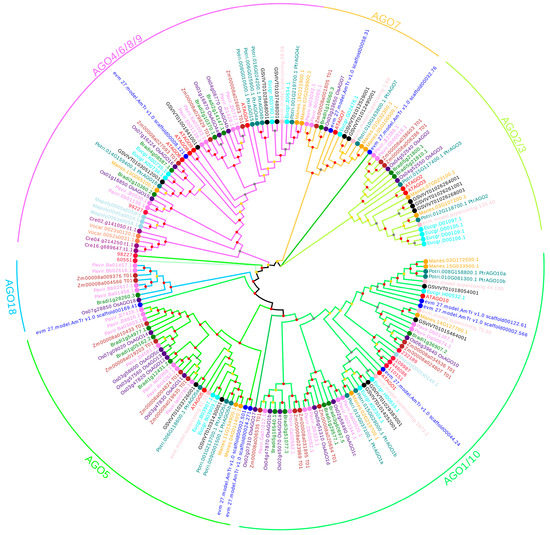
Figure 1.
Phylogenetic analysis of plant AGO proteins. Note: the phylogenetic tree was constructed using full-length amino acid sequences of PtAGO proteins in selected species via the neighbor-joining (NJ) method with 1000 bootstrap replicates. Bootstrap support values are indicated on each node.

Table 1.
Number of AGO family genes in different species.
3.2. Characteristic Analysis of PtAGOs
To further understand the PtAGO genes, the characteristics of the PtAGOs were analyzed. The gene lengths of the PtAGOs showed obvious differences. The longest gene, 2PtAGO4c, was 9978 bp, while the shortest gene, PtAGO3, was only 3795 bp. The CDS length varied from PtAGO1b with 2628 bp to PtAGO1a with 3189 bp. Interestingly, the exon number was more than 20 for most PtAGOs, except for PtAGO2, PtAGO3 and PtAGO7, which contained 3–4 exons. This difference in gene structure may correlate with various PtAGO gene functions. Meanwhile, the physical and chemical properties of the PtAGOs were also determined (Table 2). The isoelectric point of the PtAGOs ranged from 8.91 to 9.47, carrying a negative charge. The molecular weight of the PtAGO proteins ranged from 98,054.72 to 117,666.56 Da. The protein stability index of the PtAGOs, except for PtAGO2 and PtAGO3, was over 40, indicating that most PtAGOs were unstable. The prediction of the subcellular localization of the PtAGO proteins showed most PtAGOs were localized in the cell nucleus, cell membrane and chloroplast, which may correspond with their functions (Table 2).

Table 2.
Information on the AGO genes from P. trichocarpa.
3.3. Location of the PtAGOs in Chromosomes
The location of the PtAGOs in the chromosomes was investigated (Figure 2), and the results showed that 15 AGO genes were distributed unevenly among 9 chromosomes of poplar. Chromosomes 1, 6, 8, 10, 12 and 15 each had two AGO genes, while chromosomes 9, 14 and 16 only had one. Furthermore, the locations of the AGOs in duplicated chromosomal blocks were analyzed, and it was found that segmental duplication is the major contributor to the expansion of the AGO gene family.
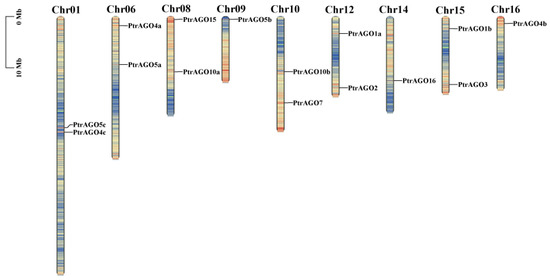
Figure 2.
Chromosomal localization of PtAGO genes in poplar.
3.4. Conserved Motifs and Gene Structure Analysis of the PtAGO Family in Poplar
To investigate the PtAGO genes’ structural diversity, the conserved motifs and positions of introns/exrons were analyzed. As shown in Figure 3, 10 conserved motifs were identified in the PtAGOs. Interestingly, except for PtAGO3, which was without motif 6, the other PtAGOs contained all 10 motifs. As for the introns/exrons, most of the PtAGOs contained more than 20 introns, except for PtAGO2, PtAGO3 and PtAGO7, with only one or two introns. As expected, PtAGOs with similar gene structures showed closer genetic distance in the phylogenetic tree, suggesting that PtAGOs maintained high conservation and consistency in the process of evolution.
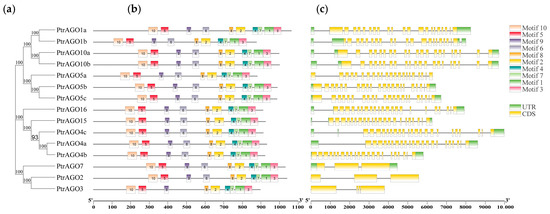
Figure 3.
Motifs and gene structure analysis of AGO genes in poplar: (a) phylogenetic tree constructed based on the full-length sequence of PtAGO protein, (b) conserved motif and (c) exon–intron structure of AGO genes.
3.5. Cis-Regulatory Element Analysis of PtAGO Promoters
In order to investigate the possible biological processes that the PtAGOs are involved in, cis-regulatory element analysis was performed in the promoters of PtAGO genes (Figure 4). Cis-regulatory elements related to light response, growth and development, abiotic stress and hormone response were identified, including GA motif, Box 4, Box II, G-box, ACE, ACA motif, MRE, AE-box, Sp1, I-box and chs-CMA1a elements. Among them, the most widely distributed element was that related to light response, with a number of 137 in all 15 PtAGO genes.
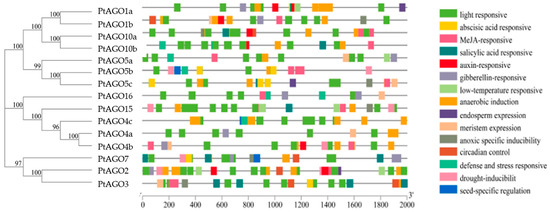
Figure 4.
Distribution of cis-acting elements in the promoter region of the PtAGO genes.
Cis-regulatory elements involved in stress response include those related to hypoxic, drought and cryogenic responses, anaerobic induction and defense and the response to adversity stress. Two to eight ART elements related to hypoxic stress were distributed in almost all of the PtAGOs, except PtAGO16. The LTR cis-acting elements involved in the low-temperature reaction, however, were only distributed in some specific PtAGO genes with two LTR in PtAGO15 and PtAGO2 and one LTR in PtAGO1a and PtAGO5a. Meanwhile, two TC-rich repeat cis-regulatory elements involved in defense and adversity stress responses were found in PtAGO16 and PtAGO10b, and one was found in PtAGO4c, PtAGO5a, PtAGO5b and PtAGO5c. Moreover, it was found that cis-regulatory elements related to gibberellin, abscisic acid, jasmonic acid, salicylic acid and auxin response were in the promoters of the PtAGO genes. ABRE cis-regulatory elements were widely distributed in all promoters of the PtAGO genes except for PtAGO16 and PtAGO4c. TGACG motif and CGTCA motif cis-regulatory elements associated with the jasmonic acid response were also found in 12 PtAGO gene promoters with 2 to 6 elements. The P-box and GARE-motif cis-regulatory elements involved in gibberellin response were distributed in the promoters of PtAGO16, PtAGO1a, PtAGO1b, PtAGO2, PtAGO4a, PtAGO4b, PtAGO5a and PtAGO7.
However, only a few cis-acting elements related to growth and development regulation, including endosperm expression, seed-specific regulation, meristem expression regulation and circadian rhythm regulation, were identified. The cis-regulatory elements associated with meristem development were widely distributed in the promoter regions of PtAGO genes, including PtAGO16, PtAGO1b, PtAGO3, PtAGO4a, PtAGO4b, PtAGO5a and PtAGO5c. The GCN4-motif, which is mainly involved in endosperm development, was distributed in the promoter regions of PtAGO1a, PtAGO2 and PtAGO5c. Meanwhile, the RY-element, involved in seed-specific regulation, was observed in the promoter regions of the genes PtAGO4c, PtAGO5b and PtAGO7, and cis-regulatory elements involved in circadian rhythm control were identified in PtAGO10a, PtAGO1b, PtAGO2 and PtAGO7, respectively. These results suggested that different PtAGO genes may be induced by abiotic stress or a variety of hormones, and they may play important roles in poplar growth and development and environmental adaptation.
3.6. Expression Pattern of PtAGOs in Various Tissues
The expression levels of the PtAGOs were detected in various tissues, including the shoot tips, lateral roots, young leaves, mature leaves, main roots, immature xylem, xylem and phloem (Figure 5). Most of the PtAGOs showed relatively high expression in young leaves and low expression in the xylem, suggesting that these PtAGOs may play distinct roles in leaf development and wood formation. Meanwhile, it was also found that several PtAGOs were mainly expressed in the lateral roots, such as PtAGO1b, PtAGO2 and PtAGO4a, suggesting that these genes may be involved in lateral root formation, growth and elongation. Moreover, PtAGO4b, PtAGO4c, PtAGO5a and PtAGO5b showed relatively high expression in the phloem, while PtAGO5b and PtAGO1b showed relatively high expression in the developing xylem, indicating that these AGOs are possibly involved in xylem development.
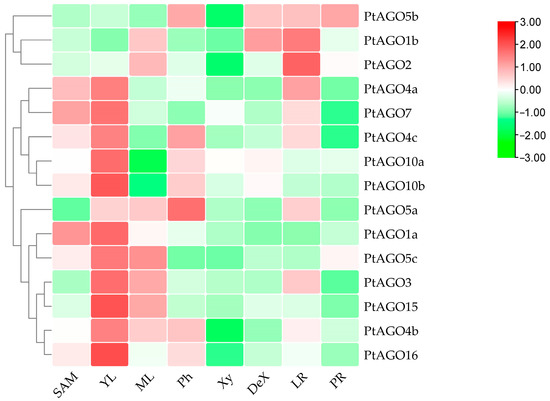
Figure 5.
Relative expression of PtAGO genes in different tissues of poplar. SAM, shoot tips; YL, young leaf; ML, mature leaf; Ph, phloem; Xy, xylem; DeX, developing xylem; LR, lateral root; PR, primary root.
3.7. Expression Pattern of PtAGOs in Response to PEG6000, ABA and Heat Treatments
To further identify the functions of PtAGOs under various abiotic stresses, the expression pattern of the PtAGOs under PEG6000, ABA and heat treatments was also detected. Under the 10% PEG6000 treatment, most PtAGOs showed similar expression trends: their expression increased and reached the highest level at 12 h and then decreased gradually (Figure 6). However, high expression points of PtAGO2, PtAGO3 and PtAGO5a were also observed after 96 h of treatment. This indicated that different PtAGOs might have different response patterns to drought.
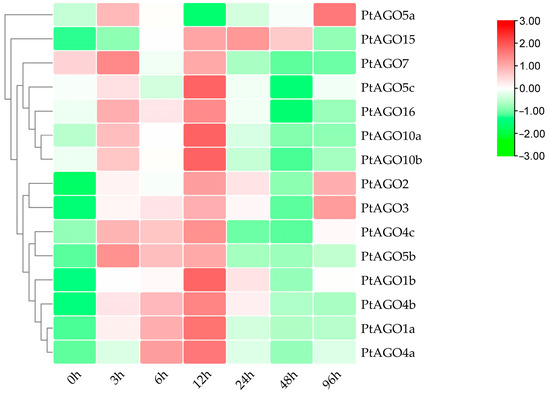
Figure 6.
Relative expression levels of PtAGO genes in poplar treated with PEG6000 at different times.
Under the ABA treatment, the expression levels of most PtAGOs significantly increased after 12 to 24 h of treatment (Figure 7). Then, the expression of these genes gradually decreased. Accordingly, most PtAGOs may respond to ABA treatment in the early stage. However, PtAGO5c PtAGO7 and PtAGO10b showed the highest expression after 96 h of ABA treatment. These results indicate that different PtAGOs might play different roles in the response to ABA treatment.
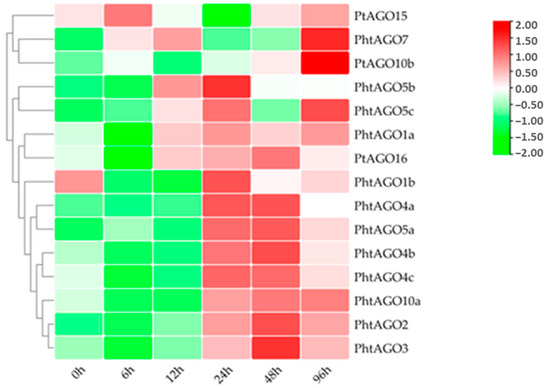
Figure 7.
Relative expression levels of PtAGO genes in poplar treated with 100 μM ABA at different times.
The response of the AGOs to different temperature treatments was also investigated. As shown in Figure 8, for almost all the PtAGO genes, expression was induced by heat shock, but the response patterns differed between different members. Most of the PtAGO genes responded quickly to the temperature increase and maintained a high expression level during the treatment. Meanwhile, the expression of PtAGO5a, PtAGO5c and PtAGO7 was up-regulated till the high-temperature treatment. Interestingly, the expression levels of all PtAGO genes quickly restored to the normal level when the temperature was recovered. These results indicate that different PtAGOs may play distinct roles in the response to heat stress.
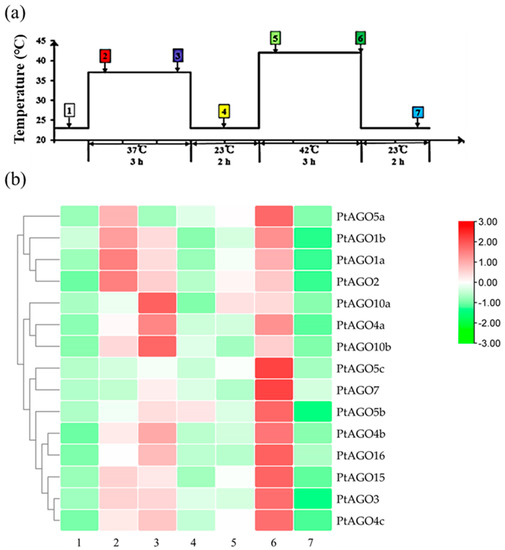
Figure 8.
Relative expression of PtAGO genes in poplar subjected to heat shock: (a) heat-stress treatment. Seedlings were cultured at 37 °C for 3 h, returned to 23 °C for 2 h, cultured at 42 °C for 3 h and then allowed to recover at 23 °C for 2 h. Here, 1–7 represents the control, 30 min after pretreatment at 37 °C, 2 h after pretreatment at 37 °C, 1 h after recovery at 23 °C, 30 min after treatment at 42 °C, 2 h after treatment at 42 °C and 2 h after recovery at 23 °C, respectively; (b) relative expression of PtAGO genes subjected to heat-shock treatment.
4. Discussion
AGOs are essential proteins of the RISC that regulate gene expression through post-transcriptional modification. The AGO gene family existed in all the plant species investigated, indicating that the AGO gene family was conserved during the plants’ evolution. In this study, we surveyed AGO genes on the whole-genome scale, and a total of 15 members were found in poplar. Phylogenetic analysis showed that the PtAGO genes were classified into three groups, which were similar to the groups observed in Arabidopsis. The AGO genes in Platycophytes and C. reinthine were clustered in Group III, while the AGO genes in the bryophytes and ferns were mainly distributed in Group I and Group III. The AGO genes in angiosperms were distributed in all three groups. Notably, Group II only contained AGO genes in angiosperms and none from other low-grade plants. In the early angiosperms, true dicots and monocots had different numbers of AGO genes. The number of AGO genes gradually increased from algae, C. reinthine and the early angiosperms to true dicots and monocots, which correspond with the evolution from simple to complex species. Therefore, it can be speculated that the emergence and amplification of AGO genes may be correlated with plant evolution. Meanwhile, the increase in the number of AGO genes may be crucial for the adaptability of plants to terrestrial environments.
The cis-regulatory elements existing in the promoters of PtAGO genes were mainly related to light response, abiotic stress, growth and development regulation and hormones. The ABA-related signal transduction of plant stress resistance was divided into PP2C-phosphatase- and ABA-induced stomatal closure [17]. The promoters of all the PtAGO genes, except for PtAGO16 and PtAGO4c, contained multiple ABRE elements, indicating that PtAGOs are likely to participate in the ABA-dependent signaling pathway. Here, the expression of the PtAGOs under ABA treatment was also detected, and most of the PtAGOs could be rapidly up-regulated under ABA treatment. These results further support the notion that most PtAGOs may participate in ABA-mediated stress response and mainly act in the early stage. However, the way in which PtAGO genes operate in ABA-related abiotic stress response needs to be studied further.
In Arabidopsis, AGO specifically binds to the sRNA and further participates in the regulation of cotyledon differentiation, flower organ formation and stem tip meristem differentiation via the RNAi pathway [18]. For example, AGO1 and AGO10 were reported to regulate the expression of AG, LFY and AP1 through the RNAi mechanism and ultimately affect the establishment of meristems and the determination of the flowers, leaves and pollen tube [19]. Meanwhile, previous studies also showed that most AGO genes can cooperate with DCL and DRB to induce RNA silencing and further participate in plant growth and development or abiotic stress response [20]. In addition, AGO1 was reported to play a key role in plant stress response and signaling through co-coordinated regulation with miR168, and this regulatory mechanism is relatively conserved [21]. For example, tomato AGO1 and miR168 were reported to be involved in plant development stage alteration, growth rate and fruit formation [9]. In woody plants, however, only AGO1 and AGO10 were reported to specifically bind to miR165/166 and synergistically affect the differentiation of the stems’ apex meristem and cambium [22]. In this study, we found that PtAGO1 was expressed in various tissues and organs in poplar, similar to the patterns observed in other species. It can be speculated that PtAGO1 may have a similar function to that of AGO1 in other species. PtAGO1b and PtAGO5b were highly expressed in the developing xylem, indicating their functions in wood formation and secondary xylem development. Considering the important roles of AGOs in plant development and abiotic stress response, more studies on AGO genes in woody plants are required.
5. Conclusions
In this study, a total of 15 AGO genes were identified from the poplar genome. The PtAGOs were unevenly distributed on nine chromosomes, and most PtAGO proteins were localized in the nucleus and chloroplasts. Except for PtAGO3, all the PtAGOs had similar motif structures. All the PtAGO genes could be clustered into 3 groups, and Group II, including PtAGO2/3/7, had the smallest number of exons, while others had more than 20 exons. Cis-regulatory elements involved in light response, growth and development, abiotic stress and hormone-induced responses were found in the promoters of the PtAGO genes. The expression analysis of PtAGO genes in various tissues showed that the expression of the PtAGO genes had a certain expression specificity. Most PtAGO genes may be involved in young leaf development, and several PtAGO genes, including PtAGO1b and PtAGO5b, might be involved in secondary xylem development. Furthermore, the expression alteration of the PtAGO genes under abiotic stress showed that most PtAGO genes can quickly respond to ABA treatment, and multiple PtAGO genes successively respond to heat-shock stress.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f14051015/s1. Table S1: List of primers for quantitative real-time PCR.
Author Contributions
Conceptualization, H.L. and S.Z.; methodology, Z.W. and Y.G.; software, Y.G., M.C. and X.C.; validation, H.L. and Z.W.; formal analysis, Y.G., Z.H. and L.G.; writing—original draft preparation, H.L.; writing—review and editing, C.F. and S.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Scientific Research Project of China (Grant Number: 2021YFD2200201) and the Outstanding Youth Fund Project of the Natural Science Foundation of Henan Province (Grant Number: 202300410119).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef] [PubMed]
- Bohmert, K.; Camus, I.; Bellini, C.; Bouchez, D.; Caboche, M.; Benning, C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. Embo J. 2014, 17, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Tolia, N.H.; Joshua-Tor, L. Slicer and the argonautes. Nat. Chem. Biol. 2007, 3, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Mallory, A.; Vaucheret, H. Form, function, and regulation of ARGONAUTE proteins. Plant Cell 2010, 22, 3879–3889. [Google Scholar] [CrossRef]
- Hutvagner, G.; Simard, M.J. Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008, 9, 22–32. [Google Scholar] [CrossRef]
- Frank, F.; Hauver, J.; Sonenberg, N.; Nagar, B. Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs. EMBO J. 2012, 31, 3588–3595. [Google Scholar] [CrossRef]
- Yuan, Y.; Pei, Y.; Ma, J.; Kuryavyi, V.; Zhadina, M.; Meister, G.; Chen, H.; Dauter, Z.; Tuschl, T.; Patel, D.J. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell 2005, 19, 405–419. [Google Scholar] [CrossRef]
- Kapoor, M.; Arora, R.; Lama, T.; Nijhawan, A.; Khurana, J.P.; Tyagi, A.K.; Kapoor, S. Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA Polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genom. 2008, 9, 451. [Google Scholar] [CrossRef]
- Bai, M.; Yang, G.; Chen, W.; Mao, Z.; Kang, H.; Chen, G.; Yang, Y.; Xie, B. Genome-wide identification of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analyses in response to viral infection and abiotic stresses in Solanum lycopersicum. Gene 2012, 501, 52–62. [Google Scholar] [CrossRef]
- Liu, X.; Lu, T.; Dou, Y.; Yu, B.; Zhang, C. Identification of RNA silencing components in soybean and sorghum. BMC Bioinform. 2014, 15, 4. [Google Scholar] [CrossRef]
- Qin, L.; Mo, N.; Muhammad, T.; Liang, Y. Genome-wide analysis of DCL, AGO, and RDR gene families in pepper (Capsicum Annuum L.). Int. J. Mol. Sci. 2018, 19, 1038. [Google Scholar] [CrossRef] [PubMed]
- Morel, J.-B.; Godon, C.; Mourrain, P.; Béclin, C.; Boutet, S.; Feuerbach, F.; Proux, F.; Vaucheret, H. Fertile hypomorphic ARGONAUTE (AGO1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 2002, 14, 629–639. [Google Scholar] [CrossRef]
- Wang, M.; Qi, Z.; Pei, W.; Cheng, Y.; Mao, K.; Ma, F. The apple Argonaute gene MdAGO1 modulates salt tolerance. Environ. Exp. Bot. 2023, 207, 105202. [Google Scholar] [CrossRef]
- Xu, M.; Xie, W.; Pan, H.; Su, X.; Zhang, S.; Huang, M. Cloning and Characterization of ARGONAUTE Genes in Populus. Sci. Silvae Sin. 2011, 47, 46–51. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Zhang, J.; Li, J.; Liu, B.; Zhang, L.; Chen, J.; Lu, M. Genome-wide analysis of the Populus Hsp90 gene family reveals differential expression patterns, localization, and heat stress responses. BMC Genom. 2013, 14, 532. [Google Scholar] [CrossRef]
- Zhu, J. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Iwakawa, H.-O.; Tomari, Y. Molecular insights into microRNA-mediated translational repression in plants. Mol. Cell 2013, 52, 591–601. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, W.; Yan, D.; Du, B.; Zhang, J.; Liu, Q.; Chen, S. Characterization of a DRE-binding transcription factor from a halophyte Atriplex hortensis. Theor. Appl. Genet. 2003, 107, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Vaucheret, H. Plant argonautes. Trends Plant Sci. 2008, 13, 350–358. [Google Scholar] [CrossRef]
- Dalmadi, Á.; Miloro, F.; Bálint, J.; Várallyay, É.; Havelda, Z. Controlled RISC loading efficiency of miR168 defined by miRNA duplex structure adjusts ARGONAUTE1 homeostasis. Nucleic Acids Res. 2021, 49, 12912–12928. [Google Scholar] [CrossRef] [PubMed]
- Sarkar Das, S.; Majee, M.; Nandi, A.K.; Karmakar, P. Sequestering miR165/166 enhances seed germination in Arabidopsis thaliana under normal condition and ABA treatment. J. Plant Biochem. Biotechnol. 2020, 29, 838–841. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).