Abstract
Research Highlights: This study identified the cell cycle genes in birch that likely play important roles during the plant’s growth and development. This analysis provides a basis for understanding the regulatory mechanism of various cell cycles in Betula pendula Roth. Background and Objectives: The cell cycle factors not only influence cell cycles progression together, but also regulate accretion, division, and differentiation of cells, and then regulate growth and development of the plant. In this study, we identified the putative cell cycle genes in the B. pendula genome, based on the annotated cell cycle genes in Arabidopsis thaliana (L.) Heynh. It can be used as a basis for further functional research. Materials and Methods: RNA-seq technology was used to determine the transcription abundance of all cell cycle genes in xylem, roots, leaves, and floral tissues. Results: We identified 59 cell cycle gene models in the genome of B. pendula, with 17 highly expression genes among them. These genes were BpCDKA.1, BpCDKB1.1, BpCDKB2.1, BpCKS1.2, BpCYCB1.1, BpCYCB1.2, BpCYCB2.1, BpCYCD3.1, BpCYCD3.5, BpDEL1, BpDpa2, BpE2Fa, BpE2Fb, BpKRP1, BpKRP2, BpRb1, and BpWEE1. Conclusions: By combining phylogenetic analysis and tissue-specific expression data, we identified 17 core cell cycle genes in the Betula pendula genome.
1. Introduction
Many important life processes are closely related to mitosis in higher organisms. The regulation mechanism of eukaryotic cell division cycle is one of the hot topics in cell biology and molecular biology. Research on the regulation of the plant cell cycle started later than that of mammals and yeast. Great progress has been made in the research of cell cycles in higher plants in recent years [1,2,3,4]. The progression of cell cycles is the result of interaction between the gene expression and the external factors. The cell cycle in higher plants is strictly regulated in the course of its growth and development.
The concept of cell cycle was brought forward by Howard and Pelcin 1953 [5], which was divided into the intermitotic phase (G1, S, and G2) and mitotic phase (M). The growth and development of plants depend on accretion, division and differentiation of cells, while cell cycle is involved in these processes. Recent studies have shown that, during the regulation of hormone, nutriment substance, and other growth signals, Cyclin D (CYCD) was expressed first and binds to cyclin dependent kinase A (CDKA) to form a complex. The complex is activated by the action of CDK activating kinase (CAK) and cyclin-dependent kinase inhibitor (CKI) or KIP-related proteins (KRPs). The activated complex attenuates the inhibitory effect of retino-blastoma protein-related (RBR) and E2F (E2 factor) a-b/DP through phosphorylation, and release transcript factor E2Fa-b/DP [6]. While E2F/DPs could promote the expression of genes required for G1 conversion to S phase (DNA synthesis phase). After entering the S phase, CYCA binds to CDKA, and it was combined with CDK subgroup cyclin-dependent kinase subunit (CKS) and CYCB synthesized during the development to the G2 phase. To remove the inhibitory phosphate group from the tyrosine phosphatase, the CDKB must be activated before entering the M phase. At the end of the M phase, cyclin proteins are hydrolyzed through the anaphase promoting complex (APC) protein pathway and exit the mitosis. A whole cell cycle is completed [7,8].
Since the cell cyclins have been found in sea urchins by Hunt in the 1980s [9], tremendous advances have been made in the molecular mechanisms of the cell cycle. This provides a positive direction for the study of tumors and other physiological diseases caused by cell cycle regulation [10]. The most significant molecular structure feature of cyclin is its conserved domain sequence, known as a cyclin box, which consists of about 100 amino acid residues. The cyclin framework is the core structure of cyclin. During the cell cycle, specific cyclins rely on their own unique cyclin frames to recognize specific cyclin-dependent kinase (CDK), and form a complex with it, thus showing specific CDK kinase activity [11]. Many different cyclins have been found, which have different expression patterns in different organs, tissues, and cell types of various organisms [12].
Betula pendula is a pioneer boreal tree that can be induced to flower within one year [13,14]. It is one of the tree species with important application value and development potential in northeast of China. As an important timber tree, it can help us understand how cell cycle genes regulate the growth and development of birch, which will greatly contribute to the application of B. pendula in industrial production and ornamental aspects. Fortunately, the genome sequence of birch [13] has been published in recent years, which has effectively helped us to conduct a series of analysis of Betula platyphylla Suk. cell cycle genes. As the large amount of data we have gathered shows, we identified cell cycle genes that likely play a very important role during plants growth and development. This provides a basis for understanding the expression processes and regulatory mechanism of various cell cycles in Betula platyphylla, and it can be used as a basis for further research on the functions of birch genes.
2. Materials and Methods
2.1. Identification and Characterization of Cell Cycle Genes in B. pendula
The B. pendula genome was used for the identification of the cell cycle genes according to the previous publication [15]. We downloaded the genomic information and protein sequences of B. pendula form the Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html, accessed on 1 November 2021) and the protein sequence of A. thaliana cell cycle gene family members from the TAIR (https://www.arabidopsis.org/, accessed on 1 November 2021) database. The identification of the cell cycle genes of B. pendula was performed using the BLASTP [16] program with E value of 10−5). In addition, all candidate genes were manually checked through the Pfam database and NCBI Conserved Domain (CD)-search tool [17] to confirm that their conserved domains were correctly annotated. Then, according to the classification in A. thaliana, they are divided into eight subgroups according to their functional types. ExPASy-ProtParam Tool (http://web.expasy.org/protparam/, accessed on 1 November 2021) was used to determine the physical and chemical parameters of the cell cycle genes, including the number of amino acids, molecular weight and isoelectric point (pI).
2.2. Chromosome Distribution of the B. pendula Cell Cycle Genes
According to the locations of these cell cycle genes on the birch chromosomes, the chromosomal distribution of the 59 cell cycle genes was analyzed, and the chromosome position image of them was generated using the TBtools (Toolbox for Biologists, v1.098684) software.
2.3. Phylogenetic, Gene Structure and Conserved Sequence and Specific Motif Analyses of B. pendula Cell Cycle Genes
Based on the previously published results [18], we constructed a phylogenetic tree for each subgroup to facilitate the study of the evolutionary relationship of the birch cell cycle genes. First, we performed a multiple sequence alignment. Then, we used MEGA 5.05 software and 500 bootstrap experiments to construct a phylogenetic tree. Representative trees were selected using the Neighbor-Joining method.
In order to understand the structural diversity of B. pendula cell cycle genes, we performed exon/intron analysis. In order to understand the functional regions of the birch cell cycle proteins and analyze the structural differences of birch cell cycle genes. We used the online software MEME (Multiple Em for Motif Elicitation, Version 5.4.1, http://meme-suite.org/tools/meme, accessed on 1 November 2021) to analyze the conserved amino acid motifs of B. pendula cell cycle genes. The CDS sequence of Betula pendula was extracted from the genomic structure information of the genome, and its intron and exon structure were visualized with TBtools (Toolbox for Biologists, v1.098684).
2.4. RNA-Seq Expression Analysis of Betula pendula Cell Cycle Genes
To investigate the expression patterns of Betula pendula cell cycle genes in different tissues, transcriptome data (PRJNA535361) was downloaded from [15] the public database of NCBI SRA. The clean reads of each sample were obtained by filtering out reads of low quality using fastp. All of the clean reads were aligned to the B. pendula reference genome using bowtie2. The RNA-seq (RNA-sequencing) data were then analyzed using the RSEM (RNA-seq by Expectation-Maximization) pipeline [19] and the data were processed using a paired-end sequencing mode. The number of RNA-seq fragments corresponding to each gene were estimated and normalized to TPM (transcripts per kilobase million) value. The expression profiles of the cell cycle genes were shown as Log2 (TPM + 1) conversion value, and the heat map was constructed by TBtools.
3. Results
3.1. Identification of Betula pendula Cell Cycle Genes and Physical and Chemical Properties Analysis
According to the identified cell cycle genes in Arabidopsis thaliana, a total of 59 cell cycle genes were obtained in birch through homology comparison (Table 1). Of them, there are 15 cyclin-dependent kinases (CDKs), 2 cyclin-dependent kinase subunit (CKSs), 27 Cyclins (CYCs), 3 E2 factor (E2Fs), 2DPs, 2 DP-E2F-like (DELs), 4 KIP-related proteins (KRPs), 2 Rbs, and 2 WEEs, respectively. CYC is the largest family that contains 27 members, while CKS, DEL, Rb and WEE are all of the smallest families containing only two members. Rb and WEE are also the smallest families in A. thaliana containing only one member. Analysis of protein characteristics showed that the lengths of the amino acid sequences encoded by these cell cycle genes range from 69 amino acids (Bpev01.c0457.g0045) to 1316 amino acids (Bpev01.c1113.g0001), and the relative molecular mass ranges from 7 kDa to 14 kDa. The predicted isoelectric point also varies greatly from 4.42 (Bpev01.c0579.g0010) to 9.69 (Bpev01.c1061.g0010), which indicates that different cyclins may work in different microenvironments. The detailed information of the protein molecular weights, isoelectric points and amino acid numbers of the gene family are shown in Table 1.

Table 1.
Putative cell cycle genes in Betula pendula.
3.2. Chromosome Distribution of Cell Cycle Genes in B. pendula
Based on the genomic information of B. pendula, the chromosomal distribution of the 59 B. pendula cell cycle genes was analyzed. According to chromosome location analysis, these cell cycle genes are unevenly distributed on the 14 chromosomes of B. pendula (Figure 1). Chromosome 11 contains the most cell cycle genes (9), followed by chromosome 6 (8). There are six cell cycle genes on chromosomes 1 and 3, and only one cell cycle gene on chromosomes 2, 8 and 12.
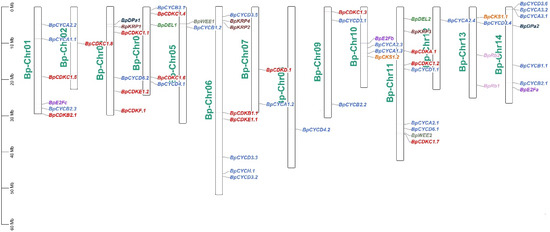
Figure 1.
Chromosome distribution of B. pendula cell cycle genes members in birch.
3.3. Identification and Analysis of Cyclin Dependent Kinases (CDK) Gene Family Members of Betula platyphylla
There are many regulators of the cell cycle in plants; most of them have special serine/threonine protein kinase activity, since they bind to cyclins to function, and are named as cyclin dependent kinases (CDKs). According to their structural and functional similarities with animal and yeast CDKs and their conserved PSTAIRE domains that bind to cyclins, plants CDKs were divided into eight groups: CDKA, CDKB, CDKC, CDKD, CDKE, CDKF, CDKG, and CDKLIKE [4,20]. In this study, we identified five groups of CDKs: BpCDKA, BpCDKB, BpCDKC, BpCDKD, BpCDKE, and BpCDKF. CDKA.1 plays a key role in the process of leaf cell division and differentiation and the development of leaf [21]. CDKB1.1 can prolong hypocotyl cells, promote cotyledon cell development, and regulate stomatal development of Arabidopsis thaliana [2,22]. The mutation of CDKB2 has been shown to impact meristem seriously [23].
We identified 15 BpCDKs in the B. pendula genome. A phylogenetic tree was constructed for the BpCDKs (Figure 2a). We evaluated the evolutionary relationship of each gene in the species. Seven different conserved domains and special motifs of BpCDKs protein were identified using the MEME tool (Figure 2c). All of the BpCDKs proteins contain at least one conserved amino acid motif. For example, BpCDKE1.1 only contains motif 2, while the rest of BpCDKs proteins contain one, two, and three conserved amino acid motifs. The conserved motifs of each BpCDKs protein branch are similar in composition, indicating that these members have a close evolutionary relationship [24]. In addition, most members of the BpCDKs protein contain motif 1, motif 2, motif 3, and motif6. These conservative motifs may have an important influence on the function of BpCDKs protein. The gene structure helps to further understand the gene family. In the BpCDK family, there are at most 13 introns (BpCDKC1.1 and BpCDKE1.2), and at least one intron (BpCDKC1.8 and BpCDKE1.1). Most genes in the BpCDKs family contain seven to eight introns (Figure 2b), and the fact that most members of the same subfamily share a similar exon/intron structure strengthens the observed phylogenetic distribution.
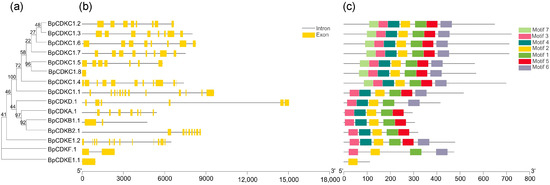
Figure 2.
Phylogenetic analysis; exon/Intron genomic structure and protein motif organization of CDK in B. pendula. An unrooted phylogenetic tree was constructed using MEGA5.05 by the neighbor-joining method. Gene structure of the corresponding BpCDKs genes, TBtools software was used to visualize the gene structure. The yellow boxes represent exons and grey lines represent introns. Use MEME web server to analyze the distribution of conserved motifs in BpCDKs protein. The protein motif figure of BpCDKs was constructed by TBtools software. (a) Phylogenetic analysis of BpCDKs; (b) exon/intron genomic structure of BpCDKs; (c) protein motif organization of BpCDKs.
3.4. Identification and Analysis of Cyclins (CYC) Gene Family Members of in Birch
Monomeric CDKs have no kinase activity and must associate with regulatory proteins called cyclins to be activated. There is common molecular structure among various cyclins, which contain a rather conservative amino acid sequence called cyclin frame to mediate the binding to CDK and regulate the activity of CDK. In plants, cyclins can be grouped into M-cyclin (containing A-and B-type cyclins) and G1-specific cyclins (designated D-type cyclins). C-cyclin and H-cyclin have been confirmed, and only CYCH.1 could activate CDK [25].
All four types of cyclins known in plants were identified. A total of 27 BpCYCs genes were detected in the B. pendula genome, including nine A-type, six B-type, eleven D-type, and one H-type. An evolutionary tree was built for BpCYCs. The MEME tool was used to identify five different conserved amino acid motifs of the CYC protein (Figure 3c). All BpCYCs proteins contain at least one conserved amino acid motif. For example, BpCYCD3.4, BpCYCD3.2, and BpCYCD3.3 only contain motif 2, BpCYCA1.2 only contains motif 3, and most of the other BpCYCs proteins contains one, two, three, and four conservative amino acid motifs, indicating that these motifs may have an important influence on the function of BpCYCs protein. It can be seen from Figure 3b that the BpCYCs family has a similar intron structure (Figure 3b). The intron-exon organization of the BpCYCs family is similar to that of Arabidopsis, this indicates that CYC is highly conserved in plants in an evolutionary manner.
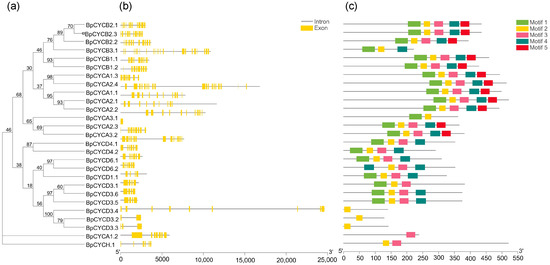
Figure 3.
Phylogenetic analysis; exon/intron genomic structure and protein motif organization of CYC in B. pendula. An unrooted phylogenetic tree was constructed using MEGA5.05 by the neighbor-joining method. Gene structure of the corresponding BpCYCs genes, TBtools software was used to visualize gene structure. The yellow boxes represent exons and grey lines represent introns. Use MEME web server to analyze the distribution of conserved motifs in BpCYCs protein. The protein motif figure of BpCYCs was constructed by TBtools software. (a) Phylogenetic analysis of BpCYCs; (b) exon/intron genomic structure of BpCYCs; (c) protein motif organization of BpCYCs.
3.5. Identification and Analysis of Cyclin Dependent Kinases Subunit (CKS) Gene Family Members in Birch
CDK subunit (CKS) proteins act as docking factors that mediate the interaction of CDKs with putative substrates and regulatory proteins. There are two CDK subunit genes in Arabidopsis described previously [4]. In a large amount of experimental data, we found two BpCKSs in the B. pendula genome (Figure 4). It can be seen that these two genes have the same motif, but their gene structures are quite different.
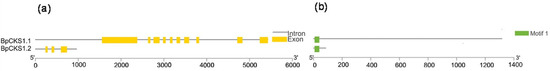
Figure 4.
Exon/Intron genomic structure and protein motif organization of CKS in B. pendula. Gene structure of the corresponding BpCKSs genes, TBtools software was used to visualize gene structure. The yellow boxes represent exons and grey lines represent introns. Use MEME web server to analyze the distribution of conserved motifs in BpCKSs protein. The protein motif figure of BpCKSs was constructed by TBtools software. (a) Exon/intron genomic structure of BpCKSs; (b) protein motif organization of BpCKSs.
3.6. Identification and Analysis of Rb and Ubiquitin-Conjugating Enzyme Factor and DP (E2F/DP) Gene Family Members in Birch
Rb regulates the expression of many essential genes in the cell cycle progression by regulating the activity of E2F transcription factor. Only one Rb could be identified in the Arabidopsis genome [4]. We identified two BpRbs in the B. pendula genome. E2F transcription factors, composed of E2F and DP, play a decisive role in plants cell size control [26]. We identified three BpE2Fs and two BpDPs in the Betula pendula genome. Two DP-E2F-like (DEL) were identified in the B. pendula genome, since they form a distinct class. A phylogenetic tree was generated for these genes, which contains for groups (Figure 5a). Through the analysis of conservative motifs, it can be seen that both E2F and DP families contain conservative motif 1 (Figure 5c), indicating that conservative motif 1 is highly conserved during evolution. Except for BpRb2 and BpDPa2, both intron and exon structures contain highly similar and numerous introns (Figure 5b).
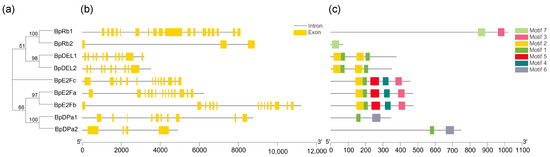
Figure 5.
Phylogenetic analysis; exon/Intron genomic structure and protein motif organization of E2F, DP, DEL, and Rb in B. pendula. An unrooted phylogenetic tree was constructed using MEGA5.05 by the neighbor-joining method. Gene structure of the corresponding BpE2Fs, BpDPs, BpDELs, and BpRbs genes, TBtools software was used to visualize gene structure. The yellow boxes represent exons and grey lines represent introns. Use MEME web server to analyze the distribution of conserved motifs in BpE2Fs, BpDPs, BpDELs, and BpRbs protein. The protein motif figure of BpE2Fs, BpDPs, BpDELs, and BpRbs was constructed by TBtools software. (a) Phylogenetic analysis of BpE2Fs, BpDPs, BpDELs and BpRbs; (b) exon/intron genomic structure of BpE2Fs, BpDPs, BpDELs, and BpRbs; (c) protein motif organization of BpE2Fs, BpDPs, BpDELs, and BpRbs.
3.7. Identification and Analysis of KIP-Related Proteins (KRP) and WEE Gene Family Members of Betula pendula
The activity of CYC-CDK is also regulated by an inhibitory protein CKI (also known as KRP). Seven CKI genes belonging to the group of Kip/Cip CKIs have been described previously for Arabidopsis, designated KRP1 to KRP7 [27]. In this study, we have identified four BpKRPs in the B. pendula genome. These four genes all have motif 1 (Figure 6c). BpKRP1 and BpKRP2 also contain the same motif 2, and both contain three to four introns (Figure 6b), and have similar structures.
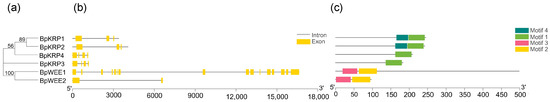
Figure 6.
Phylogenetic analysis; exon/Intron genomic structure and protein motif organization of KRP and WEE in B. pendula. An unrooted phylogenetic tree was constructed using MEGA5.05 by the neighbor-joining method. Gene structure of the corresponding BpKRPs, BpWEEs genes, TBtools software was used to visualize gene structure. The yellow boxes represent exons and grey lines represent introns. Use MEME web server to analyze the distribution of conserved motifs in BpKRPs, BpWEEs protein. The protein motif figure of BpKRPs, BpWEEs was constructed by TBtools software. (a) Phylogenetic analysis of BpKRPs, BpWEEs; (b) exon/intron genomic structure of BpKRPs, BpWEEs; (c) protein motif organization of BpKRPs, BpWEEs.
CDK/cyclin activity is regulated negatively by phosphorylation of the CDK subunit by the WEE1 kinase and positively when the inhibitory phosphate groups are removed by the CDC25 phosphatase. Two BpWEEs were identified in the B. pendula genome, their conserved motifs are similar in structure, while there are only two introns in BpWEE2.
3.8. RNA-Seq Expression Analysis of Betula pendula Cell Cycle Genes
According to the abundance and specificity of transcripts, the specific expression data of the four tissue parts of birch xylem, roots, leaves and flowers are used to analyze cell cycle genes. We count the specific expression data of all cell cycle genes. The results showed that there are 17 cell cycle genes that are highly expressed in leaves, xylem or flowers (Figure 7). The 17 cell cycle genes were BpCDKA.1, BpCDKB1.1, BpCDKB2.1, BpCKS1.2, BpCYCB1.1, BpCYCB1.2, BpCYCB2.1, BpCYCD3.1, BpCYCD3.5, BpDEL1, BpDpa2, BpE2Fa, BpE2Fb, BpKRP1, BpKRP2, BpRb1, and BpWEE1.
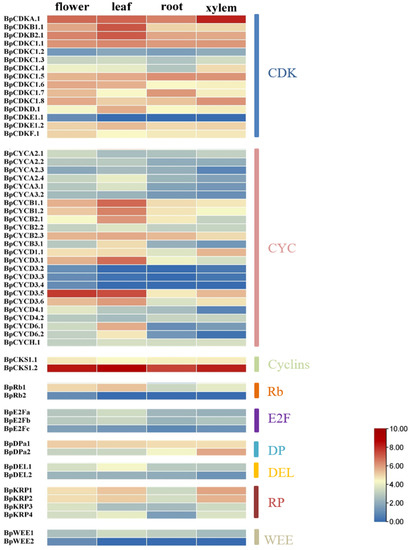
Figure 7.
The heat map shows the expression of cell cycle genes in different parts of the birch tissue. Highly or lowly expressed genes are colored red or blue, respectively.
In the BpCDK family, BpCDKA.1 is abundant in xylem. In addition, BpCDKA.1, BpCDKB1.1 and BpCDKB2.1 were highly expressed in leaves. BpCDKA.1 is highly expressed in all of the four investigated tissues, which indicated BpCDKA.1 may play multiple roles in different tissues. The most similar genes to BpCDKA.1, BpCDKB1.1 and BpCDKB2.1 in A. thaliana are CDKA; 1 (AT3G48750), CDKB1.1 (AT3G54180) and CDKB2.1 (AT1G76540).
A total of 27 BpCYCs were detected in the B. pendula genome, of which BpCYCD3.5 is abundant in flower and leaves. The gene most similar to BpCYCD3.5 in A. thaliana is AT3G50070. In addition to this, BpCYCB1.1, BpCYCB1.2, BpCYCB2.1 and BpCYCD3.1 are highly expressed in leaves.
In the CKS family of birch, BpCKS1.2 was most abundant in the leaf and expressed at moderate levels in the other three tissues. The gene most similar to BpCKS1.2 in A. thaliana is AT2G27960.
BpRb1 is abundant in leaves BpRb1 is most similar to AT3G12280. ZmRb1 binds to D- type cyclins in plants, is highly expressed in differentiated cells, and regulates leaf development at temporal and spatial level [28]. BpE2Fa and BpE2Fb are abundant in leaves. BpE2Fa and BpE2Fb are most similar to AT2G36010 and AT5G22220, respectively. Two BpDPs were identified in the Betula pendula genome, of which BpDP2 is abundant in xylem, and this gene is similar to AT5G02470. BpDEL1 is abundant in leaves. This gene is similar to AT3G48160 in A. thaliana.
In the KRP family of Betula pendula, BpKRP1 was most abundant in the xylem and BpKRP2 also was expressed at a high level in the xylem. These two genes are most similar to AT2G23430 in A. thaliana. Moreover, BpKRP1 and BpKRP2 are also highly expressed in flower and leaves. BpWEE1 is abundant in leaves. This gene is similar to AT1G02970.
4. Discussion
According to the previous reports, many cell cycle genes have been identified [29], but it is still necessary to screen and identify birch cell cycle genes in order to better understand their functions in birch. Using bioinformatics methods, a total of 59 cell cycle genes of B. pendula have been identified.
Plant cell cycles could be regulated by altered expression of some G1-S and G2-M checkpoints genes in cells [3]. G1-S phase was one of the most important checkpoints among all of the cell cycles, and CycD genes have been indicated as a sensor of extracellular growth condition [1]. Over expression of CycD3;1 in Arabidopsis thaliana could induce B-type cyclin expression, resulting in not only an increase in endoreduplication but also in mitosis [30]. A further study revealed that CYCLIN B1; 2 was the mitosis promoting factor [31]. CYCLIN B1;2 expression can promote nuclear and cellular division, which is sufficient to trigger endoreduplication to mitosis, but not sufficient enough to increase cell cycle rounds [31]. In contrast with our results, BpCYCB1.1, BpCYCB1.2, BpCYCB2.1, and BpCYCD3.1 are highly expressed in leaves, and BpCYCD3.5 is abundant in flower and leaves (Figure 7). These genes with high expression levels in birch tissues contain CYCD3.1 and CYCB1.2, indicating that these two genes in birch may also play a very important role in cell division. Gene structure analysis found that the gene sequence structure of BpCYCs family members is similar (Figure 3b), indicating that their gene structure is highly conserved during evolution. Both the pistil cell death and stamen cell arrest are involved in cell cycle regulation in maize sex determination. CYCA, CYCB, and CDK were highly expressed in the developing pistil and stamen, while WEE1 and CKI were only expressed in the arresting stamen [32]. In our study, part of genes was highly expressed in flower, such as BpCYCD3.5, BpCKS1.2, and BpCDKA.1 (Figure 7). However, birch has unisexual flowers on separate male and female inflorescences (catkins) [12,33,34]. The question concerning how the cell cycle genes regulate the flower development process of birch requires further research.
5. Conclusions
Cell cycle genes are closely related to all life activities of plants, we identified 17 core cell cycle genes in the genome of Betula pendula by analyzing phylogenetic analysis, gene structure analysis and tissue specific expression data, provide some help for better application of cell cycle genes and modern molecular breeding.
Author Contributions
Conceptualization, H.H.; software, Y.L. (Yijie Li) and S.C.; validation, H.H.; writing-original draft preparation, Y.L. (Yijie Li) and S.C.; writing-review and editing, Y.L. (Yijie Li); literature search, Y.L. (Yuhang Liu); study design, Y.L. (Yijie Li) and Y.L.; project administration, H.H.; funding acquisition, H.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Fundamental Research Funds for the Central Universities (2572018BW06), the National Natural Science Foundation of China (31800556).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- De Veylder, L. The Discovery of Plant D-Type Cyclins. Plant Cell 2019, 31, 1194–1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boudolf, V.; Lammens, T.; Boruc, J.; Van Leene, J.; Van Den Daele, H.; Maes, S.; Van Isterdael, G.; Russinova, E.; Kondorosi, E.; Witters, E.; et al. CDKB1;1 Forms a Functional Complex with CYCA2;3 to Suppress Endocycle Onset. Plant Physiol. 2009, 150, 1482–1493. [Google Scholar] [CrossRef] [Green Version]
- Boudolf, V.; Vlieghe, K.; Beemster, G.T.S.; Magyar, Z.; Acosta, J.A.T.; Maes, S.; Van Der Schueren, E.; Inze, D.; De Veylder, L. The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 2004, 16, 2683–2692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandepoele, K.; Raes, J.; De Veylder, L.; Rouze, P.; Rombauts, S.; Inze, D. Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 2002, 14, 903–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelc, S.R.; Howard, A. Effect of Irradiation on DNA Synthesis in Vicia as Shown by Autoradiographs. Acta Radiol. 1954, 41, 699. [Google Scholar] [CrossRef]
- Inze, D.; De Veylder, L. Cell cycle regulation in plant development. Annu. Rev. Genet. 2006, 40, 77–105. [Google Scholar] [CrossRef]
- Kosugi, S.; Ohashi, Y. Interaction of the Arabidopsis E2F and DP proteins confers their concomitant nuclear translocation and transactivation. Plant Physiol. 2002, 128, 833–843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boniotti, M.B.; Gutierrez, C. A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J. Cell Mol. Biol. 2001, 28, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Evans, T.; Rosenthal, E.T.; Youngblom, J.; Distel, D.; Hunt, T. Cyclin: A protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell 1983, 33, 389–396. [Google Scholar] [CrossRef]
- Drakare, S.; Lennon, J.J.; Hillebrand, H. The imprint of the geographical, evolutionary and ecological context on species-area relationships. Ecol. Lett. 2006, 9, 215–227. [Google Scholar] [CrossRef]
- Finlay, B.J. Global dispersal of free-living microbial eukaryote species. Science 2002, 296, 1061–1063. [Google Scholar] [CrossRef] [Green Version]
- Fenchel, T.; Finlay, B.J. The ubiquity of small species: Patterns of local and global diversity. Bioscience 2004, 54, 777–784. [Google Scholar] [CrossRef]
- Salojarvi, J.; Smolander, O.P.; Nieminen, K.; Rajaraman, S.; Safronov, O.; Safdari, P.; Lamminmaki, A.; Immanen, J.; Lan, T.Y.; Tanskanen, J.; et al. Genome sequencing and population genomic analyses provide insights into the adaptive landscape of silver birch. Nat. Genet. 2017, 49, 904–912. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.J.; Wang, S.; Jiang, J.; Liu, G.F.; Li, H.Y.; Chen, S.; Xu, H.W. Overexpression of BpAP1 induces early flowering and produces dwarfism in Betula platyphylla x Betula pendula. Physiol. Plant. 2014, 151, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Lin, X.; Zhang, D.W.; Li, Q.; Zhao, X.Y.; Chen, S. Genome-Wide Analysis of NAC Gene Family in Betula pendula. Forests 2019, 10, 741. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST plus: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef] [PubMed]
- Gang, H.X.; Li, R.H.; Zhao, Y.M.; Liu, G.F.; Chen, S.; Jiang, J. Loss of GLK1 transcription factor function reveals new insights in chlorophyll biosynthesis and chloroplast development. J. Exp. Bot. 2019, 70, 3125–3138. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Song, J.; Wang, F.; Zhang, X.S. Genome-wide identification and expression analysis of rice cell cycle genes. Plant Mol. Biol. 2007, 64, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Adachi, S.; Nobusawa, T.; Umeda, M. Quantitative and cell type-specific transcriptional regulation of A-type cyclin-dependent kinase in Arabidopsis thaliana. Dev. Biol. 2009, 329, 306–314. [Google Scholar] [CrossRef] [Green Version]
- Boudolf, V.; Barroco, R.; Engler, J.D.; Verkest, A.; Beeckman, T.; Naudts, M.; Inze, D.; De Veylder, L. B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 2004, 16, 945–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wang, Q.; Zhao, Y.; Han, G.; Zhu, S. Systematic analysis of maize class III peroxidase gene family reveals a conserved subfamily involved in abiotic stress response. Gene 2015, 566, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Andersen, S.U.; Buechel, S.; Zhao, Z.; Ljung, K.; Novak, O.; Busch, W.; Schuster, C.; Lohmann, J.U. Requirement of B2-type cyclin-dependent kinases for meristem integrity in Arabidopsis thaliana. Plant Cell 2008, 20, 88–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huntley, R.; Healy, S.; Freeman, D.; Lavender, P.; Murray, J.A.H. The maize retinoblastoma protein homologue ZmRb-1 is regulated during leaf development and displays conserved interactions with G1/S regulators and plant cyclin D (CycD) proteins. Plant Mol. Biol. 1998, 37, 155–169. [Google Scholar] [CrossRef]
- Sabelli, P.A.; Larkins, B.A. Regulation and function of retinoblastoma-related plant genes. Plant Sci. 2009, 177, 540–548. [Google Scholar] [CrossRef]
- Shimotohno, A.; Umeda-Hara, C.; Bisova, K.; Uchimiya, H.; Umeda, M. The plant-specific kinase CDKF;1 is involved in activating phosphorylation of cyclin-dependent kinase-activating kinases in Arabidopsis. Plant Cell 2004, 16, 2954–2966. [Google Scholar] [CrossRef]
- De Veylder, L.; Beeckman, T.; Beemster, G.T.; Krols, L.; Terras, F.; Landrieu, I.; van der Schueren, E.; Maes, S.; Naudts, M.; Inze, D. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 2001, 13, 1653–1668. [Google Scholar] [CrossRef] [Green Version]
- Schnittger, A.; Schobinger, U.; Bouyer, D.; Weinl, C.; Stierhof, Y.D.; Hulskamp, M. Ectopic D-type cyclin expression induces not only DNA replication but also cell division in Arabidopsis trichomes. Proc. Natl. Acad. Sci. USA 2002, 99, 6410–6415. [Google Scholar] [CrossRef] [Green Version]
- Schnittger, A.; Schobinger, U.; Stierhof, Y.D.; Hulskamp, M. Ectopic B-type cyclin expression induces mitotic cycles in endoreduplicating Arabidopsis trichomes. Curr. Biol. 2002, 12, 415–420. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.R.; Kim, J.C. Temporal and Spatial Regulation of Cell Cycle Genes during Maize Sex Determination. J. Life Sci. 2006, 16, 828–833. [Google Scholar]
- Wang, S.; Huang, H.J.; Han, R.; Liu, C.Y.; Qiu, Z.N.; Liu, G.F.; Chen, S.; Jiang, J. Negative feedback loop between BpAP1 and BpPI/BpDEF heterodimer in Betula platyphylla x B. pendula. Plant Sci. 2019, 289, 110280. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Huang, H.J.; Han, R.; Chen, J.Y.; Jiang, J.; Li, H.Y.; Liu, G.F.; Chen, S. BpAP1 directly regulates BpDEF to promote male inflorescence formation in Betula platyphylla x B. pendula. Tree Physiol. 2019, 39, 1046–1060. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).