Abstract
Osmanthus serrulatus Rehder (Oleaceae) is an endemic spring-flowering species in China. It is narrowly distributed in the southwestern Sichuan Basin, and is facing the unprecedented threat of extinction due to problems associated with natural regeneration, habitat fragmentation and persistent and serious human interference. Here, the genetic diversity and population structure of 262 individuals from ten natural populations were analyzed using 18 microsatellites (SSR) markers. In total, 465 alleles were detected across 262 individuals, with a high polymorphic information content (PIC = 0.893). A high level of genetic diversity was inferred from the genetic diversity parameters (He = 0.694, I = 1.492 and PPL = 98.33%). AMOVA showed that a 21.55% genetic variation existed among populations and the mean pairwise Fst (0.215) indicated moderate genetic population differentiation. The ten populations were basically divided into three groups, including two obviously independent groups. Our results indicate that multiple factors were responsible for the complicated genetic relationship and endangered status of O. serrulatus. The concentrated distribution seems to be the key factor causing endangerment, and poor regeneration, human-induced habitat loss and fragmentation seem to be the primary factors in the population decline and further genetic diversity loss. These findings will assist in future conservation management and the scientific breeding of O. serrulatus.
1. Introduction
Osmanthus serrulatus Rehder is an evergreen shrub or small tree with long and slender pedicels and weeping flowers that bloom in spring, which distinguishes it from other species [1,2]. Endemic to China, O. serrulatus is commonly known as Baoxing Osmanthus, and it is sporadically distributed among the high-altitude mountains around the southwest Sichuan Basin [3]. Our previous work on O. serrulatus indicated that persistent human interference and increasing habitat fragmentation were the major external factors posing a critical threat to its wild populations [3], while its characteristic functional androdioecy [4], seed germination every other year [5], and unique habitat requirements [6,7] might be the internal factors leading to a dramatic decline in the population and slow regeneration of the natural habitat. These findings play a crucial role in new Osmanthus cultivar breeding and highlight subtropical forest biodiversity patterns and mechanisms in China. However, research on O. serrulatus, especially genetic studies [8], remains limited. Despite research efforts in regard to the phylogeny and evolution [9,10], genetic diversity and population structure [11,12,13,14] of O. fragrans (Thunb.) Lour., there is an urgent need to explore the genetic relationships of O. serrulatus with neutral markers, e.g., microsatellites, to obtain a better understanding of its genetic diversity and population structure.
As the basis of diversity in species and ecosystems, genetic diversity is an inherent form of biodiversity [15], reflecting the ability of organisms to adapt to the changing environment through natural selection [16,17]. Generally, populations with little genetic variation are vulnerable to the arrival of climate change and habitat destruction, while those of high genetic diversity can withstand the rigorous natural selective pressures [17,18]. It is difficult for rare and endangered species to expand their distribution range and explore new areas for survival because the cumulative loss of genetic variation leads to a continuous reduction in population size [17,19,20]. As an expression vector of genetic traits, the protection of genetic resources is actually to protect the evolutionary potential and environmental adaptability of different species and individuals [21,22]. Exploring genetic diversity and population structure enables an evaluation of the evolutionary potential and adaptability and the prediction of future changes in genetic composition [23]. Genetic conservation is a fundamental task for species preservation and plant germplasm innovation [24,25].
To date, microsatellites (simple sequence repeats, SSRs) are some of the most frequently used molecular markers to assess genetic diversity and population structure, due to their co-dominance inheritance, high polymorphism, high stability and repeatability, rich content distribution, and low cost of detection [19,26]. In this study, we used 18 biparentally inherited polymorphic SSR markers to evaluate the genetic diversity and structure of ten natural populations throughout the current main distribution range. The results will provide molecular evidence for the conservation and utilization of O. serrulatus germplasm.
2. Materials and Methods
2.1. Plant Material and DNA Extraction
Based on the historical data in the literature and for specimens of O. serrulatus, the geographic distribution and location information for this species were synthesized (Figure 1A). Through extensive field investigation over the past few years, ten natural populations from southwestern Sichuan, China, were located and identified as from a wild provenance (Figure 1B). Fresh leaf samples were collected from 262 individuals of the ten populations (Table 1) and immediately dried in silica gel. Total genomic DNA was extracted from dried leaves using the Plant Genomic DNA Kit (Tiangen, Shanghai, China) according to the manufacturer’s instructions. The quality and concentration of DNA were evaluated using a NanoDropTM 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Extracted DNA samples were adjusted to 50 ng/μL and subsequently stored at −20 °C.
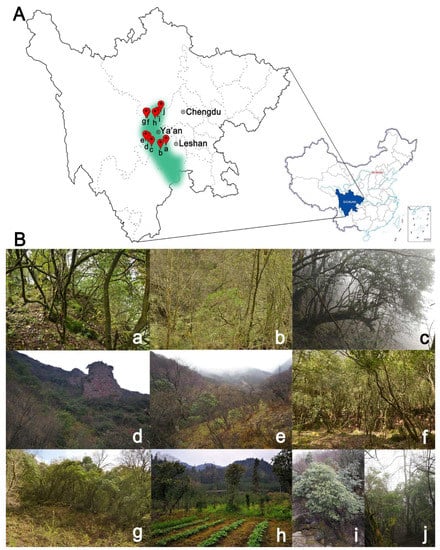
Figure 1.
The geographical distribution and habitats of O. serrulatus in this study. (A) The main distribution range (green) and sampling locations (red dots); (B) the habitat and physiognomy of some O. serrulatus populations. (a) EMS; (b) BCP; (c) XYG; (d) YCP; (e) HZP; (f) DLS1; (g) DLS2; (h) MPZ; (i) WZX; (j) XLXS; the abbreviations are the same as in Table 1.

Table 1.
Geographical location and genetic diversity for ten O. serrulatus populations in southwestern Sichuan Basin, China.
2.2. Primer Screening and PCR Amplification
A total of 100 SSR primers from the developed primers based on high-throughput transcriptome sequencing data [8,27] were randomly selected and 15 primers with stable polymorphic bands were screened by the DNA mixed pooling method [28]. Then, the 15 specific primers with three universal primers (OSM005, OSM010, OSM038) [29] were used to estimate the genetic diversity and structure of the natural O. serrulatus populations (Table S1). The SSR-PCR amplification was performed on the Applied BiosystemsTM Automated Thermal Cycler (Thermo Scientific, Wilmington, DE, USA) as follows: a pre-denaturation at 94 °C for 5 min; followed by 30 cycles of 4 s denaturation at 94 °C, 30 s annealing at 55 °C and 30 s extension at 72 °C; a final extension at 72 °C for 5 min; and then preservation at 4 °C [28]. The reactions were conducted in a 25 μL volume containing 50 ng of template DNA, 1.0 μL of 100 μmol/L of each forward and reverse primer, 12.5 μL of 2× Taq PCR Master Mix, and 9.5 μL ddH2O. The PCR products were analyzed on 8% polyacrylamide gel and silver-stained to detect SSR bands, and the length of the bands were compared with a 50bp DNA ladder (Tiangen, Shanghai, China) to estimate allele size.
2.3. Data Analysis
The genetic diversity parameters were calculated by GenAlEx 6.502 [30] and POPGENE 1.3.2 [31], including the number of observed alleles (Na), number of effective alleles (Ne), Shannon’s information index (I), the mean number of private alleles (Np), observed heterozygosity (Ho), expected heterozygosity (He), percentage of polymorphic loci (PPL), Wright’s fixation index (F), inbreeding coefficient among individuals (Fis), genetic differentiation coefficient among populations (Fst), gene flow (Nm), and the deviation from Hardy–Weinberg equilibrium (HWE) with the chi-squared test for each population. The polymorphic information content (PIC) and null alleles frequency (Fn) were computed by Cervus 3.0.7 [32]. Bottleneck 1.2.02 [33] was employed to test for the recent bottleneck events using the one-tailed Wilcoxon sign-rank test with the infinite alleles model (IAM, 30%), the stepwise mutation model (SMM, 70%) and two-phase model (TPM), and a further mode-shift test was also performed. GenAlEx 6.502 was also used to calculate the pairwise Fst, analysis of molecular variance (AMOVA), principal coordinate analysis (PCoA), and Mantel test based on the pairwise matrix of geographic distance and Nei’s genetic distance in terms of Fst/(1-Fst) with 1000 permutations. The unweighted pair group method with arithmetic mean (UPGMA) clustering based on Nei’s genetic distance among the populations and among the individuals were constructed using MEGA X [34] and visualized by iTOL v6.3 (Interactive Tree of Life) [35]. The population structure was determined with a Bayesian clustering analysis using STRUCTURE v2.3.4 [36]. A total of ten independent simulations for each K ranging from 2 to 10 were performed with a burn-in period of 100,000 steps followed by 100,000 Markov Chain Monte Carlo (MCMC) iterations using the Admixture Model. The most probable number of population groups (K) was determined with delta K (ΔK), following Evanno et al. [37] by web-based STRUCTURE HARVESTER [38]. Repeated sampling analysis and genetic structural plots were analyzed by CLUMPP 1.1.2 [39] and visualized by DISTRUCT 1.1 [40]. The associated membership probability value (Q value) of different genetic clusters in each individual was represented by vertical colored column.
3. Results
3.1. SSR Loci Polymorphism
Across the 18 SSR loci, a total of 465 alleles were detected among 262 individuals from ten populations, and the Na detected per locus ranged from 16 to 41, with an average of 25.83 (Table S2). The mean Ne was 10.748, and ranged from 8.651 (Os48) to 16.837 (Os87). All the values of Ho of 18 SSR loci were much smaller than He, except Os35. The mean Ho was 0.279, and ranged from 0.013 (Os74) to 0.907 (Os35), while the mean He was 0.694, and ranged from 0.602 (Os74) to 0.831 (Os87). The values of PIC and I averaged 0.893 and 1.492, respectively, indicating a high polymorphism and suitability for the genetic analysis. The mean F of 18 SSR loci was 0.623, ranging from 0.480 (Os72) to 0.887 (Os03). The values of Fis and Fst were 0.602 and 0.228 on average, respectively, and the loci Os35 had the minimum values, including a negative inbreeding coefficient (Fis = −0.111). In addition, the mean values of Nm and Fn were 0.969 and 0.556, respectively, and the HWE test across the 262 individuals showed significant deviations for all loci.
3.2. Population Genetic Diversity and Mutation-Drift Equilibrium
At the population level, the genetic diversity index was estimated across the ten populations (Table 1). Overall, the average percentage of polymorphic loci (PPL) was 98.33%, with almost all populations at 100% except EMS (94.44%) and BCP (88.89%). The Na per population ranged from 4.167 (MPZ) to 8.500 (YCP), with an average of 6.894, while the mean Ne was 4.381, ranging from 3.062 (MPZ) to 5.532 (DLS2). The values of Ho and He ranged from 0.136 (MPZ) to 0.373 (DLS1) and from 0.621 (WZX) to 0.774 (DLS2), respectively. The mean I was 1.492 over a range of 1.184 (MPZ) to 1.779 (DLS2). The mean Np was 1.133, and ranged from 0.333 (BCP) to 3.056 (YCP). In addition, the average fixation index (F) was 0.623, ranging from 0.469 (YCP) to 0.813 (MPZ) at the population level, indicating a severe inbreeding and heterozygote deficiency among the natural populations.
In bottleneck analysis, heterozygosity excess based on the Wilcoxon sign-rank test was highly significant (p < 0.01) for the IAM model in almost all populations except WZX and for the TPM model in populations EMS, BCP, HZP, DLS2, MPZ, and XLXS, while not it was significant in most populations except BCP (p < 0.01) and MPZ (p < 0.05) with the SMM model (Table S3). Meanwhile, the mode-shift test did not detect any significant distortion in the allele frequency with a normal L-shaped distribution indicating the absence of bottleneck in O. serrulatus populations (Figure S1).
3.3. Genetic Differentiation and Gene Flow between Populations
The genetic variation in populations was determined both by AMOVA (Table 2) and pairwise Fst (Table S4). AMOVA demonstrated that only 22.15% of the total molecular variance occurred among populations, while the remainder occurred within populations with 46.84% and 31.61% existing among and within individuals, respectively (p < 0.001). This was confirmed by small Fst and large Nm overall among the populations, representing 0.215 and 0.910 on average, respectively. In total, the pairwise Fst ranged from 0.123 to 0.303, of which most values were between 0.15 to 0.25, and the pairwise Nm ranged from 0.575 to 1.799, with most values around 1.0. Particularly, the smallest Nm and biggest Fst appeared between EMS and BCP, while the biggest Nm and smallest Fst appeared between XLXS and DLS2.

Table 2.
Analysis of molecular variance (AMOVA) of 262 individuals in ten O. serrulatus populations.
3.4. Population Clustering and Genetic Structure
Based on Nei’s genetic distance, UPGMA and PCoA were used to access the population clustering. UPGMA clustering analysis intuitively divided the ten populations into three main groups (Figure 2A) based on a genetic distance threshold of 0.70. Specifically, the population EMS alone formed Group I (red), and population DLS1, DLS2 and XLXS clustered into Group Ⅱ (green), while Group Ⅲ (blue) contained the remaining six populations (YCP, HZP, XYG, WZX, MPZ and BCP). Overall, the result of PCoA (Figure 3) was similar to UPGMA, which approximately divided the individuals of ten populations into three groups. A further UPGMA-based cluster analysis among the 262 individuals was performed to reveal the specific relationship and genetic structure (Figure 2B). The resulting tree showed that the individuals could be divided into five groups. Group I (red cluster) and Ⅱ (green cluster) were consistent with the UPGMA result among populations. The other individuals were separated into three groups. A set of individuals (14) from population HZP was gathered into a small Group Ⅲ (brown cluster), individuals of MPZ and BCP were gathered into another Group Ⅳ (purple cluster), while the remaining individuals were gathered into a complex Group Ⅴ (yellow cluster).
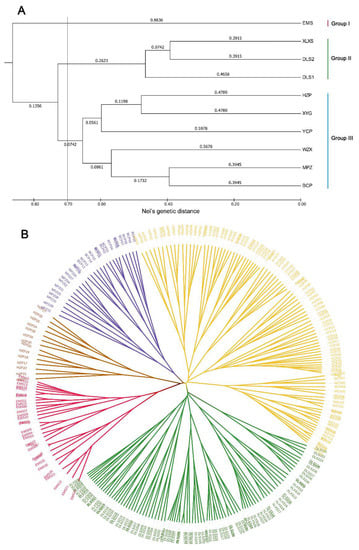
Figure 2.
Genetic divergence of O. serrulatus based on UPGMA cluster analysis. (A) Clustering among ten populations; (B) clustering among 262 individuals.
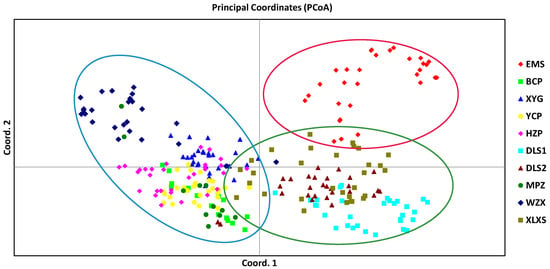
Figure 3.
Principal coordinate analysis (PCoA) of all individuals from ten O. serrulatus populations.
In the population structure analysis, there were two peaks of ΔK (the maximum for 4.853 and the second for 4.120) (Figure S2A), indicating that the entire population could be divided into five or seven clusters (Figure S2B). The genetic composition may be from one or several sub-populations, and the cluster membership probability of each individual represents the genetic relationship among individuals. Every component in an individual takes a certain proportion, which represents the membership probability value (Q value). When the Q value was over 75%, the individual was usually considered as pure, otherwise they are admixed [41]. There were 145 pure individuals and 117 admixed individuals when K = 5, and 199 pure individuals and 63 admixed individuals when K = 7. Nearly all populations except XYG were composed of mostly pure individuals when K = 7, while only half of the populations (EMS, YCP, HZP, DLS2 and XLXS) were pure when K = 5. Among all, the shared genetic origins of sub-populations indicated a frequent gene exchange among the populations, such as BCP, MPZ and WZX, and DLS1, DLS2 and XLXS. The population EMS (red), HZP (orange) and WZX (brown, when K = 7) were isolated as a unique genetic cluster with pure individuals, indicating a certain genetic specificity. Meanwhile, the cluster membership proportion of each population at K = 5 and K = 7 was mapped based on Q values (Figure 4). The resulting chart showed a relatively clear geographical distribution among populations. Additionally, the Mantel test revealed no significant correlation between Nei’s genetic distance and geographical distance (R2 = 0.0001, p ≤ 0.490; Figure S3) and also confirmed that genetic differentiation among populations was not caused by geographic isolation. More broadly, the clustering structures were not in conflict with the results of UPGMA and PCoA, because the population EMS, and the populations DLS1, DLS2 and XLXS seemed to have the same genetic origins in order to gather into independent groups.
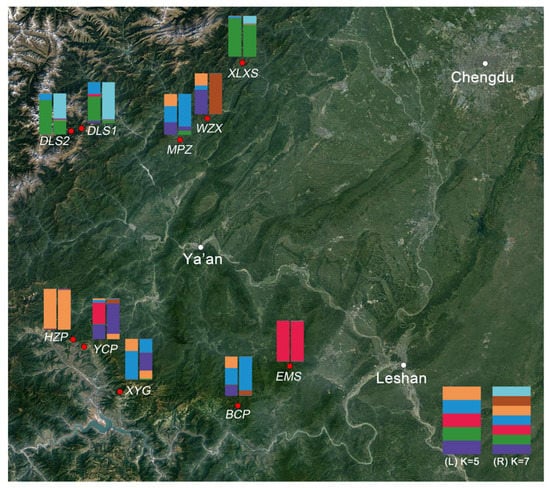
Figure 4.
Geographical distribution of the plant materials. Red dots indicate the populations collected from southwestern Sichuan, China, and the white dots indicate the main cities. Colored bars show the mean proportions of cluster memberships of analyzed individuals in each of the ten O. serrulatus populations based on structure at K = 5 (left) and K = 7 (right).
4. Discussion
4.1. Genetic Diversity of Osmanthus serrulatus
Generally, the PIC value is used as an indicator for the genetic informativeness of markers among the accessions [42]. As shown in this study, the highly polymorphic markers with a mean PIC value of 0.893 were very suitable for revealing the genetic diversity of the ten natural populations of O. serrulatus from southwestern Sichuan, especially the primer Os87. There was a high level of genetic diversity shown by the high values of expected heterozygosity (He > 0.6) and Shannon’s information index (I > 1.0) of O. serrulatus (Table S2). Population size does indeed affect genetic diversity [43]. Among the populations, DLS2 with a larger local population size had the highest level of genetic diversity (He = 0.774, I = 1.779, Table 1), and its remote habitat with very little human interference might result in greater genetic diversity and lead to more intraspecific genetic variation in recent history. At the same time, the ex situ population MPZ with less samples had the lowest level of genetic diversity, and habitat changes and severe human disturbance including damage during transplanting might be other factors affecting the genetic diversity.
The mating system, that is, functional androdioecy, and the contrasting levels of gene flow might explain the high genetic diversity of this narrowly distributed species in some way [44,45]. The presence of null and dropout alleles will lead to a high deviation between Ho and He, and a high frequency of null alleles will cause the deviation from HWE in all loci [42,46]. In this study, all values of Ho were much smaller than He among the ten populations as well as among the 18 loci (except Os35), which may be caused by the high frequency of null alleles at all loci (0.017–0.966, mean Fn = 0.556), and further lead to an underestimation of population genetic diversity [46]. Meanwhile, the significant HWE deviation (p < 0.001) might present signs of inbreeding, which suggests high heterozygosity deficiency and nonrandom mating among individuals [47].
In general, the genetic diversity of plants is positively correlated to the geographical range and population size, and the population decline leads to a loss of genetic diversity [43,45,48]. Endangered, endemic and narrowly distributed species usually have low genetic diversity, but many studies have shown that even endangered plants can have a moderate or high level of genetic diversity, such as Michelia coriacea Hung T. Chang & B. L. Chen (He = 0.505) [49], Rhododendron jingangshanicum P. C. Tam (He = 0.642) [50], Houpoea officinalis (Rehder & E. H. Wilson) N. H. Xia & C. Y. Wu (He = 0.600, I = 1.158) [51], Populus wulianensis S. B. Liang & X. W. Li (He = 0.611, I = 1.114, PPL = 98.6%) [26], Pteroceltis tatarinowii Maxim. (He = 0.696, I = 1.452, PPL = 95.19%) [52], and Emmenopterys henryi Oliv. (He = 0.755, I = 1.717) [42], etc. As an endemic species, a high genetic diversity (He = 0.694, I = 1.492, PPL = 98.33%) of O. serrulatus was demonstrated based on the microsatellite analysis, much higher than the average value of He (0.42) in endemic plants [53]. In addition, in contrast with the previous study on wild Osmanthus species using SSR markers, the observed genetic diversity of O. serrulatus was close to the high-mountain-endemic species, O. delavayi Franch. (He = 0.723, I = 1.657) [54] and widespread species, O. fragrans (He = 0.673) [13], but much higher than O. cooperi Hemsl. (He = 0.1419, I = 0.5559) [55], a widespread but sporadic species, indicating relatively stronger environmental adaptability and evolutionary potential.
Additionally, the genetic bottleneck effect was tested under the IAM, SMM and TPM models. The transient effect of population decline is usually measured by heterozygosity excess [56]. In bottleneck analysis, the IAM is usually recommended for allozyme data and microsatellites tend to evolve under the SMM and TPM models [33]. However, SMM is generally more appropriate for long-term dynamic analysis of large populations and only a few microsatellite loci strictly conform with it [57]. Consequently, the TPM is apparently even more appropriate than IAM and SMM in this study, and the populations EMS, BCP, HZP, DLS2, MPZ and XLXS might actually have experienced bottleneck events in recent history (p < 0.01, Table S3).
4.2. Genetic Differentiation and Population Structure of Osmanthus serrulatus
Population structure was manifested mainly as genetic differentiation among populations [58]. Nybom [53] proposed that the genetic variability of species with long-lived or outcrossing is mostly retained within populations, while in annual or selfing species, variability is mainly allocated among populations. In the present study, AMOVA results showed that only 21.55% genetic variations existed among O. serrulatus populations, suggesting that genetic variation mostly occurred within populations. According to Wright [59,60], Fst has usually been used for evaluating the genetic differentiation among populations with the following rules: Fst < 0.15, low; 0.15 ≤ Fst < 0.25, medium; Fst ≥ 0.25, high. The larger Fst and smaller Nm indicated greater differences among populations. Similar mean values of moderate genetic differentiation and medium gene flow were detected for all populations across the 18 SSR loci (Fst = 0.228, Nm = 0.969, Table S2) and between the populations (Fst = 0.215, Nm = 0.910, Table 2). Particularly, the pairwise Fst of EMS-associated populations was mostly more than 0.25, highlighting a high differentiation between these populations. Moreover, Nm was one of the main factors for homogenization, given that gene exchange between populations could counteract the genetic differentiation caused by genetic drift and geographical isolation when Nm > 1.0 [61]. Most pairwise Nm around 1.0 indicated a relatively frequent gene flow among O. serrulatus populations, suggesting the Nm partly replaced the effect of genetic drift or geographical isolation between some populations.
For population structure classification, the analysis of PCoA, UPGMA and STRUCTURE complement and validate each other, because PCoA and UPGMA only expound intuitive relationships without fully categorizing them, while STRUCTURE can categorize the populations objectively and determine the probability and degree of genetic exchange among different populations [62]. In the present study, we obtained five or seven sub-populations from the ten O. serrulatus populations using STRUCTURE (Figure S2). Overall, the EMS population, and the populations, DLS1, DLS2 and XLXS could definitely be separated into two independent groups. They had a relatively distinct genetic composition, whereas the remainder showed marginal differences based on the three methods, indicating complicated genetic relationships among different populations and different locations. Especially, population EMS and HZP were isolated as a unique genetic cluster with pure individuals whether K = 5 or K = 7, indicating a certain genetic specificity and limited gene exchange with others. Furthermore, some populations were geographically adjacent but quite different in genetic structure, such as EMS and BCP, HZP and YCP (Figure 4), suggesting no correlation between genetic structure and geographical distribution. This was also confirmed by the Mantle test (R2 = 0.0001, p ≤ 0.490, Figure S3). As for the individuals, most individuals were pure in theory, whereas most populations had moderate gene flow. This may be explained by the patchy distribution and the mating system of O. serrulatus [4], i.e., the geographic barriers of altitude and distance restricted the gene exchange between different populations to some extent, and the functional androdioecy strengthened mating fitness within populations. According to the current results and the biological characteristics [4,5] and the demands of special habitats [6], we believe it is very likely that there is no population differentiation of O. serrulatus across the local area, and that the concentrated distribution might be the key factor in the endangerment of O. serrulatus.
4.3. Conservation Proposal of Osmanthus serrulatus
Sweet Osmanthus is one of the top ten traditional famous flowers in China. As a rare spring-flowering endemic Osmanthus species, O. serrulatus occupies an important phylogenetic node in genus Osmanthus, and is a crucial germplasm resource for genetic breeding of Sweet Osmanthus. Like rare and endangered species, endemic species preservation is challenged by habitat destruction and fragmentation [63]. Previous field studies have showed that the existing wild habitats of O. serrulatus populations were isolated in some fragmented, independent and narrow regions in high-altitude mountain areas in Sichuan. At the same time, a lot of historical sampling sites have disappeared due to land clearing, grazing and destructive exploitation. Some initial characteristics of Plant Species with Extremely Small Populations (PSESP) are associated with O. serrulatus, such as a decline in population size, natural regeneration is difficult, habitat fragmentation, and persistent and serious human interference [3,4]. Therefore, effective and efficient conservation practices should be put in place as soon as possible to reduce the population decline. Considering the high genetic diversity and moderate genetic differentiation of the wild O. serrulatus populations, human-induced habitat loss and poor regeneration are mainly responsible for the decline in the population size, while the accompanying genetic erosion and genetic drift might influence the genetic diversity in subsequent generations.
Maintenance of genetic diversity is a primary objective in the management of wild populations of threatened species [58]. Based on the results of present and previous work, both in situ and ex situ efforts on the original habitat and genetic germplasm are needed in order to formulate a conservation strategy for O. serrulatus [48]. Since genetic variation mainly occurs within populations, it is vital that preferential in situ conservation is implemented for populations with high genetic diversity and individuals with unique genes, such as populations DLS2, XLXS, YCP, and HZP. Simultaneously, it is necessary to introduce individuals from different gene pools into other gene pools to improve the reproductive fitness and maintain genetic diversity of different populations [58], especially the pure individuals of EMS and HZP. Additionally, ex situ conservation of as many individuals as possible via seeds or branches could be used as an auxiliary measure to establish a germplasm resources library to preserve the genetic diversity, by providing sufficient materials for germplasm innovation and possible reintroduction of this endemic species in the future.
5. Conclusions
This is the first study to perform an analysis of the genetic diversity and population structure of O. serrulatus in southwestern Sichuan using 18 SSR markers. Our results show high adaptability and evolutionary potential in the natural O. serrulatus populations. There was a high level of genetic diversity within populations, and moderate genetic differentiation among populations. The ten populations were mainly divided into three groups, with two distinct, independent groups and a complex mixed group. There was no correlation between genetic structure and geographical distribution. The geographic barriers of altitude and distance restricted the gene exchange between different populations to some extent, but only a moderate genetic differentiation occurred between populations. Multiple factors attributed to the complicated genetic relationship and endangered status O. serrulatus. Poor regeneration and human-induced habitat loss and fragmentation are mainly responsible for the decline in population size and genetic diversity, and the concentrated distribution might be the key cause of endangerment. Our study sheds light on the genetic diversity, population structure and genetic relationships among populations, and provides important information on the conservation strategy development, genetic improvement and scientific breeding of O. serrulatus.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/f12101365/s1, Figure S1: Mode shift graph showing L-shaped distribution and indicating the absence of bottleneck in O. serrulatus populations, Figure S2: Population genetic structure. (A) The appropriate K value determined by estimated value ΔK. (B) estimate population structure for ten O. serrulatus populations when K=5 and K=7, Figure S3: Mantel test for matrix correlation between Nei’s genetic distance and geographical distance of ten O. serrulatus populations. Table S1: Characteristics of 18 polymorphic SSR loci in O. serrulatus, Table S2. Polymorphsim parameters of 18 SSR loci in O. serrulatus, Table S3. Summary of population bottleneck tests with four models (IAM, TPM, SMM, and Model-shift) in ten O. serrulatus populations, Table S4. Genetic differentiation coefficient Fst (below diagonal) and gene flow Nm (above diagonal) values between O. serrulatus populations across 18 loci.
Author Contributions
Conceptualization, L.C. and X.W.; methodology, L.C. and H.Q.; software, L.C., T.P. and H.Q.; validation, L.C., M.Z. and T.P.; investigation, L.C., G.Y., H.Q. and X.W.; resources, L.C.; writing—original draft preparation, L.C., T.P. and H.Q.; writing—review and editing, L.C. and M.Z.; project administration, L.C.; funding acquisition, L.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 31300558, 32071782; the Natural Science Foundation of Jiangsu Province, grant number BK20130972; and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analysed during this study are included in this published article and its Supplementary Materials.
Acknowledgments
We appreciate Meng Xu (Nanjing Forestry University) and Longna Li (Nanjing Agricultural University) for their constructive advice in revising the manuscript, and appreciate Li Guo (Xi’an Jiaotong University) for his linguistic assistance.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Chang, M.; Qiu, L.; Wei, Z.; Green, P.S. Osmanthus. In Flora of China; Wu, Z., Raven, P., Hong, D., Eds.; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 1996; Volume 15, pp. 286–292. [Google Scholar]
- Xiang, Q.B.; Liu, Y.L. An Illustrated Monograph of the Sweet Osmanthus Cultivars in China; Zhejiang Science & Technology Press: Hangzhou, China, 2008; pp. 2–12. [Google Scholar]
- Chen, L.; Yang, G.D.; Qian, H.R.; Li, L.N.; Cheng, Y. Analyses on characteristics and species diversity of Osmanthus serrulatus community in Dongla Mountain of Sichuan. J. Plant. Resour. Environ. 2017, 26, 74–83. [Google Scholar] [CrossRef]
- Yang, G.D.; Qian, H.R.; Chen, L.; Wang, X.R. Reproduction system of Osmanthus serrulatus, an endemic plant to China. Sci. Silvae Sin. 2018, 54, 17–29. [Google Scholar] [CrossRef]
- Yang, G.D.; Qian, H.R.; Chen, L.; Wang, X.R. Ultrastructure observation in the development of Osmanthus serrulatus seed mature. J. Cent. South. Univ. For. Technol. 2018, 38, 19–25, 34. [Google Scholar] [CrossRef]
- Yang, G.D.; Qian, H.R.; Chen, L.; Wang, X.R. Analysis of community structure of Osmanthus serrulatus based on TWINSPAN classification and DCCA sequencing. Acta Ecol. Sin. 2018, 38, 3059–3068. [Google Scholar] [CrossRef]
- Yang, G.D.; Qian, H.R.; Chen, L.; Wang, X.R. Effect of soil nutrients on spatial distribution of Osmanthus serrulatus community in Dongla Mountains. Acta Bot. Boreali-Occident. Sin. 2018, 38, 343–352. [Google Scholar] [CrossRef]
- Chen, L.; Li, L.; Dai, Y.; Wang, X.; Duan, Y.; Yang, G. De novo transcriptome analysis of Osmanthus serrulatus Rehd. flowers and leaves by Illumina sequencing. Biochem. Syst. Ecol. 2015, 61, 531–540. [Google Scholar] [CrossRef]
- Chen, H.; Zeng, X.; Yang, J.; Cai, X.; Shi, Y.; Zheng, R.; Wang, Z.; Liu, J.; Yi, X.; Xiao, S.; et al. Whole-genome resequencing of Osmanthus fragrans provides insights into flower color evolution. Hortic. Res. 2021, 8, 98. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, M.; Wang, X.; Sylvester, S.P.; Xiang, Q.; Li, X.; Li, M.; Zhu, H.; Zhang, C.; Chen, L.; et al. Revisiting the phylogeny and taxonomy of Osmanthus (Oleaceae) including description of the new genus Chengiodendron. Phytotaxa 2020, 436, 283–292. [Google Scholar] [CrossRef]
- Chen, Y.L.; Luo, Y.S.; Tao, X.H. The genetic relationship of two new colorful sweet Osmanthus fragrans Cultivars revealed by AFLP and SSR markers. Agric. Sci. Technol. 2019, 20, 40–48. [Google Scholar] [CrossRef]
- Duan, Y.; Li, M.; Yi, X.G.; Wang, H.; Wang, X.; Xiang, Q. Assessment of genetic diversity among androdioecious ancient Osmanthus fragrans trees by SSR markers. Biochem. Syst. Ecol. 2015, 61, 179–185. [Google Scholar] [CrossRef]
- Hu, W.; Luo, Y.; Yang, Y.; Zhang, Z.Y.; Fan, D.M. Genetic diversity and population genetic structure of wild sweet osmanthus revealed by microsatellite markers. Acta Hortic. Sin. 2014, 41, 1427–1435. [Google Scholar]
- Duan, Y.; Wang, X.; Xiang, Q.; Liang, L.; Li, X.; Liu, Y.; Li, M. Genetic diversity of androdioecious Osmanthus fragrans (Oleaceae) cultivars using microsatellite markers. Appl. Plant. Sci. 2013, 1, 1200092. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Hong, D.Y. Genetic diversity and its detection methods. In Principles and Methodologies of Biodiversity Studies; Qian, Y.Q., Ma, K.P., Eds.; China Science and Technology Press: Beijing, China, 1994; pp. 123–124. [Google Scholar]
- Jump, A.S.; Marchant, R.; Peñuelas, J. Environmental change and the option value of genetic diversity. Trends Plant. Sci. 2009, 14, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Krutovsky, K.; Neale, D.B. Forest genomics and new molecular genetic approaches to measuring and conserving adaptive genetic diversity in forest trees. In Conservation and Management of Forest Genetic Resources in Europe; Geburek, T., Turok, J., Eds.; Arbora Publishers: Zvolen, Slovakia, 2005; pp. 369–390. [Google Scholar]
- Liu, H.Y.; Zang, F.Q.; Wu, Q.C.; Ma, Y.; Zheng, Y.Q.; Zang, D.K. Genetic diversity and population structure of the endangered plant Salix taishanensis based on CDDP markers. Glob. Ecol. Conserv. 2020, 24, e1242. [Google Scholar] [CrossRef]
- Li, S.; Liu, S.; Pei, S.; Ning, M.; Tang, S. Genetic diversity and population structure of Camellia huana (Theaceae), a limestone species with narrow geographic range, based on chloroplast DNA sequence and microsatellite markers. Plant. Divers. 2020, 42, 343–350. [Google Scholar] [CrossRef]
- Soltis, P.S.; Soltis, D.E. Genetic variation in endemic and widespread plant species: Examples from Saxifragaceae and Polystichum. Aliso 1991, 13, 215–223. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Zheng, P.; Ni, B.; Miao, X.; Zhao, Z.; Li, M. Population genetic diversity and structure analysis of wild apricot (Prunus armeniaca L.) revealed by SSR markers in the Tien-Shan mountains of China. Pak. J. Bot. 2018, 50, 609–615. [Google Scholar]
- Kazemeini, F.; Asri, Y.; Mostafavi, G.; Kalvandi, R.; Mehregan, I. Assessment of genetic diversity, population structure and morphological analyses in an Iranian endemic species Rhabdosciadium aucheri Boiss. (Apiaceae) using ISSR markers. Biologia 2021, 76, 441–451. [Google Scholar] [CrossRef]
- Iwaizumi, M.; Tsuda, Y.; Ohtani, M.; Tsumura, Y.; Takahashi, M. Recent distribution changes affect geographic clines in genetic diversity and structure of Pinus densiflora natural populations in Japan. For. Ecol. Manag. 2013, 304, 407–416. [Google Scholar] [CrossRef]
- Da Silva Almeida Leal, G.; Leal, F.A.; Gomes, H.T.; de Souza, A.M.; Ribeiro, S.C.; Scherwinski-Pereira, J.E. Structure and genetic diversity of natural populations of Guadua weberbaueri in the southwestern Amazon, Brazil. J. For. Res. 2021, 32, 755–763. [Google Scholar] [CrossRef]
- Gregorius, H. Gene conservation and the preservation of adaptability. In Species Conservation: A Population-Biological Approach; Seitz, A., Loeschcke, V., Eds.; Birkhäuser Basel: Basel, Switzerland, 1991; pp. 31–47. [Google Scholar]
- Wu, Q.; Zang, F.; Ma, Y.; Zheng, Y.; Zang, D. Analysis of genetic diversity and population structure in endangered Populus wulianensis based on 18 newly developed EST-SSR markers. Glob. Ecol. Conserv. 2020, 24, e1329. [Google Scholar] [CrossRef]
- Chen, L.; Li, L.; Yang, G.; Dai, Y. Microsatellites characteristics of transcriptomic sequences from Osmanthus serrulatus. Mol. Plant. Breed. 2016, 14, 959–965. [Google Scholar] [CrossRef]
- Qian, H.R.; Chen, L.; Yang, G.D.; Pan, T.T.; Wang, X.R. Optimization of SSR-PCR reaction system in Osmanthus serrulatus and rapid primer selection based on DNA mixing pool. Mol. Plant. Breed. 2019, 17, 525–530. [Google Scholar] [CrossRef]
- Zhang, Z.R.; Fan, D.M.; Guo, S.Q.; Li, D.Z.; Zhang, Z.Y. Development of 29 microsatellite markers for Osmanthus fragrans (Oleaceae), a traditional fragrant flowering tree of China. Am. J. Bot. 2011, 98, e356–e359. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]
- Yeh, F.C.; Yang, R.C.; Boyle, T. POPGENE Version 1.32. Microsoft Windows-Based Freeware for Population Genetic Analysis; University of Alberta: Edmonton, AB, Canada, 1999. [Google Scholar]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Piry, S.; Luikart, G.; Cornuet, J. BOTTLENECK: A computer program for detecting recent reductions in the effective size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, N.A. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Zhong, Y.; Yang, A.; Li, Z.; Zhang, H.; Liu, L.; Wu, Z.; Xu, M.; Yu, F. Genetic diversity and population genetic structure of Cinnamomum camphora in South China revealed by EST-SSR markers. Forests 2019, 10, 1019. [Google Scholar] [CrossRef] [Green Version]
- Niu, Y.; Bhatt, A.; Peng, Y.; Chen, W.; Gao, Y.; Zhan, X.; Zhang, Z.; Hu, W.; Song, M.; Yu, Z. Genetic diversity and population structure analysis of Emmenopterys henryi Oliv., an endangered relic species endemic to China. Genet. Resour. Crop. Evol. 2021, 68, 1135–1148. [Google Scholar] [CrossRef]
- Hague, M.T.; Routman, E.J. Does population size affect genetic diversity? A test with sympatric lizard species. Heredity 2016, 116, 92–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamrick, J.L.; Godt, M.J.W.; Sherman-Broyles, S.L. Factors influencing levels of genetic diversity in woody plant species. New For. 1992, 6, 95–124. [Google Scholar] [CrossRef]
- Martin, S.L.; Benedict, L.; Wei, W.; Sauder, C.A.; Beckie, H.J.; Hall, L.M. High gene flow maintains genetic diversity following selection for high EPSPS copy number in the weed kochia (Amaranthaceae). Sci. Rep. 2020, 10, 18864. [Google Scholar] [CrossRef]
- Barros, J.; Winkler, F.M.; Velasco, L.A. Assessing the genetic diversity in Argopecten nucleus (Bivalvia: Pectinidae), a functional hermaphrodite species with extremely low population density and selffertilization: Effect of null alleles. Ecol. Evol. 2020, 10, 3919–3931. [Google Scholar] [CrossRef] [Green Version]
- Stark, A.E. A clarification of the Hardy–Weinberg law. Genetics 2006, 174, 1695–1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lammi, A.; Siikamäki, P.; Mustajärvi, K. Genetic diversity, population size, and fitness in central and peripheral populations of a rare plant Lychnis viscaria. Conserv. Biol. 1999, 13, 1069–1078. [Google Scholar] [CrossRef]
- Zhao, X.; Ma, Y.; Weibang, S.; Wen, X.; Milne, R. High genetic diversity and low differentiation of Michelia coriacea (Magnoliaceae), a critically endangered endemic in Southeast Yunnan, China. Int. J. Mol. Sci. 2012, 13, 4396–4411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.W.; Chen, S.F.; Shi, S.; Zhang, Z.; Liao, W.B.; Wu, W.; Zhou, R.C.; Fan, Q. High genetic diversity and weak population structure of Rhododendron jinggangshanicum, a threatened endemic species in Mount Jinggangshan of China. Biochem. Syst. Ecol. 2015, 58, 178–186. [Google Scholar] [CrossRef]
- Yang, X.; Yang, Z.; Li, H. Genetic diversity, population genetic structure and protection strategies for Houpoëa officinalis (Magnoliaceae), an endangered Chinese medical plant. J. Plant. Biol. 2018, 61, 159–168. [Google Scholar] [CrossRef]
- Fan, J.J.; Zhang, X.P.; Liu, K.; Liu, H.J.; Zhang, L.; Wang, X.P.; Li, X.H. The population genetic diversity and pattern of Pteroceltis tatarinowii, a relic tree endemic to China, inferred from SSR markers. Nord. J. Bot. 2019, 37, e1922. [Google Scholar] [CrossRef]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef]
- Duan, Y.; Li, W.; Zheng, S.; Sylvester, S.P.; Li, Y.; Cai, F.; Zhang, C.; Wang, X. Functional androdioecy in the ornamental shrub Osmanthus delavayi (Oleaceae). PLoS ONE 2019, 14, e221898. [Google Scholar] [CrossRef]
- Zuo, M.Y. Study on the Phenotypic Variation and Genetic Diversity of Osmanthus cooperi Hemsl. Master’s Thesis, Nanjing Forestry University, Nanjing, China, 2013. [Google Scholar]
- Cristescu, R.; Sherwin, W.B.; Handasyde, K.; Cahill, V.; Cooper, D.W. Detecting bottlenecks using BOTTLENECK 1.2.02 in wild populations: The importance of the microsatellite structure. Conserv. Genet. 2010, 11, 1043–1049. [Google Scholar] [CrossRef]
- Cornuet, J.M.; Luikart, G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 1996, 144, 2001–2014. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK, 2002; pp. 227–253. [Google Scholar]
- Wright, S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 1965, 19, 395–420. [Google Scholar] [CrossRef]
- Wright, S. Evolution and the Genetics of Populations. Volume 4. Variability within and among Natural Populations; The University of Chicago Press: Chicago, IL, USA, 1978; pp. 82–89. [Google Scholar]
- Loveless, M.D.; Hamrick, J.L. Ecological determinants of genetic structure in plant populations. Annu. Rev. Ecol. Syst. 1984, 15, 65–95. [Google Scholar] [CrossRef]
- Wang, S.Q. Genetic diversity and population structure of the endangered species Paeonia decomposita endemic to China and implications for its conservation. BMC Plant. Biol. 2020, 20, 510. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Hong, D.; Fu, C.; Cameron, K.M. Genetic variation in the endangered and endemic species Changium smyrnioides (Apiaceae). Biochem. Syst. Ecol. 2004, 32, 583–596. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).