Abstract
Northern red oak (Quercus rubra L.) is one of the most important foreign tree species in Germany and considered as a major candidate for prospective sustainable forestry in the face of climate change. Therefore, Q. rubra was subject of many previous studies on its growth traits and attempts to infer the origin of various populations of this species using nuclear and chloroplast DNA markers. However, the exact geographic origin of German red oak stands has still not been identified. Its native range widely extends over North America, and the species can tolerate a broad range of environmental conditions. We genotyped individual trees in 85 populations distributed in Germany and North America using five chloroplast microsatellite and three novel chloroplast CAPS markers, resulting in the identification of 29 haplotypes. The new marker set enabled the identification of several new red oak haplotypes with restricted geographic origin. Some very rare haplotypes helped us narrow down the origin of Q. rubra stands in Germany, especially some stands from North Rhine-Westphalia, to the northern part of the species’ natural distribution area including the Peninsula of Nova Scotia, where the most similar haplotype composition was observed, compared to distinct German stands.
1. Introduction
Northern red oak (Quercus rubra L., section Lobatae) is a forest tree species native to North America, where it occurs from the east coast of Nova Scotia, Canada, and Virginia in the East to Minnesota and Oklahoma in the West. Southern offshoots reach Georgia and South Carolina and almost completely include Alabama [1]. The species was introduced to Europe in the late 17th century [2] and has become the most important foreign deciduous tree species in Germany considering the large forest area [3,4]. Q. rubra has a short rotation period in forestry compared to the native white oaks Q. robur L. and Q. petraea (Matt.) Liebl. and, therefore, has a higher productivity than European oak species. Harvested wood meets good quality standards. Its firewood or fiber properties are similar to that of white oak wood [5]. While its productivity is higher than that of European oak species, its lumber quality is inferior compared to native oaks. For instance, Q. rubra wood is missing thyloses [6], which slow down rotting. Therefore, it is not used for outdoor structures or barrel production. The species is considered as invasive in some European countries [7,8,9] because of its abundant spreading, ability to grow in less fertile soil [8], and in a wide climatic range [4]. However, this is unlikely the case in Germany, because shade tolerance is one of the major factors of successful invasion in German forests [10] and Q. rubra cannot compete with the local predominant tree species Fagus sylvatica L. [11,12]. Moreover, northern red oak from section Lobatae does not hybridize with native German white oaks representing section Quercus [4].
Despite the increasing importance of northern red oak cultivation in Europe, little is known about its origin. Additional northern red oak seed orchards have to be established to meet the growing demand for high quality reproductive material [13]. Therefore, it would be beneficial to identify possible sources of genetic material in the native range of the species. In this study, we aim to infer the origin of Q. rubra stands in Germany by using chloroplast (cp) haplotypes based on five chloroplast simple sequence repeat (cpSSR) and three chloroplast cleaved amplified polymorphic sequence (cpCAPS) markers. Since chloroplasts are maternally inherited in oaks [14] and thus do not undergo recombination, chloroplast haplotypes are highly suitable to draw conclusions about the geographic origin of populations and recolonization routes after the last glacial maximum [15,16,17,18,19,20]. Pettenkofer et al. [21] used five cpSSR markers to study the origin of Q. rubra and concluded that German stands originated from a limited geographic area, possibly located in the northern part of the species’ native distribution range. Using additional haplotypes in red oak stands in Germany and comparing them to reference populations from the native distribution area, we aim to narrow down the geographic origin of German populations. For this purpose, we expanded the sample set by 22 new populations and added three new informative cpCAPS markers. These novel cpCAPS markers were designed for Q. rubra based on comparative plastome genome sequencing [22], which enabled identification of additional haplotypes.
We hypothesize that (1) introduction of Q. rubra to Europe from a restricted native geographic range caused a founder effect reflected in decreased haplotype diversity in German stands, and (2) identification of additional haplotypes using cpCAPS markers would help us narrow down the geographic origin of most German stands to the northeastern part of the natural distribution range.
2. Materials and Methods
2.1. Samples
In total, 1143 different trees were studied, including 592 samples from 44 planted stands in Germany (8–50 per stand, Table 1) and 551 samples from 37 reference naturally grown populations in North America (2–52 per population, Table 1). The sample set included 40 German stands and three North American populations studied in Pettenkofer et al. [21,23], and 16 natural populations from North America—six studied in Lind & Gailing [24], two studied in Lind-Riehl et al. [25], and eight studied in Liesebach & Schneck [3], which all have been genotyped at five cpSSR markers previously [21]. Additionally, four German planted red oak stands, including two stands, Diergardt 1 and 2, which originated from Virginia, and eight Canadian natural populations, were sampled. Furthermore, samples were collected from a provenance trial located in Schleswig-Holstein that was established in 1990 using acorns collected from single trees in different locations in North America (Table 1). Leaves were collected from randomly selected trees within each stand, maintaining a distance of at least 10 m between single trees. In provenance trials, a minimum distance of 10 m was kept between sampled trees and the border of the respective stand. Quality parameters, such as trunk curvature or branching were not taken into account. The experimental plots in the provenance trial consist of descendants of single mother trees each. From each plot, beginning with the Northwestern corner, only the tree in the third column/third row was sampled to exclude border effects, provide reproducibility and avoid the sampling of half-siblings from the same seed parent.

Table 1.
Name, location, and sample size (N) of red oak populations.
2.2. DNA Isolation
The DNeasy™ 96 Plant Kit (Qiagen; Hilden, Germany) was used to extract DNA from a 1 cm2 large piece cut from each fresh or dried leaf sample according to the manufacturer’s manual.
2.3. Chloroplast Microsatellites
The newly collected populations from Canada and Germany were genotyped at five cpSSR markers. Two angiosperm cpSSR markers (ccmp2 and ccmp4, [26]), as well as three oak-specific ones (ucd4, udt1, and, udt4) developed by Deguilloux et al. [27] were used. A multiplex PCR was run for ccmp2 and ccmp4 with 1 µL genomic DNA sample, 1.5 µL PCR buffer containing 0.8 M TRIS-HCl and 0.2 M (NH4)2SO4, 4.8 µL ddH2O, 1.5 µL (25 mM) MgCl2, 1µL dNTPs (2.5 mM of each dNTP), 1 µL of each primer (forward and reverse for each, ccmp2 and ccmp4; 5 picomole/µL) and, 0.2 µL (5 U/µL) HOT FIREPol® Taq polymerase (Solis BioDyne; Tartu, Estonia). After initial denaturation for 15 min at 95 °C, 37 cycles with 1 min denaturation (94 °C), 1 min annealing (53 °C), and 1 min of elongation (72 °C) were conducted, which were finally followed by 20 min of elongation terminating the procedure.
Markers ucd4, udt1, and udt4 were amplified using a touchdown PCR with the following parameters: 15 min denaturation at 95 °C, followed by eight cycles of 1 min denaturation (94 °C), 1 min annealing (53 °C, reduced by 1 °C per cycle) and 1 min of elongation (72 °C). Subsequently, 25 cycles with 1 min denaturation (94 °C), 1 min annealing (45 °C) and 1 min elongation (72 °C) were initiated and finally followed by 20 min of elongation. For each primer pair, 1 µL per primer (forward and reverse; 5 picomole/µL), 1 µL genomic DNA sample, 1.5 µL PCR buffer containing 0.8 M TRIS-HCl and 0.2 M (NH4)2SO4, 6.8 µL ddH2O, 1.5 µL (25 mM) MgCl2, 1µL dNTPs (2.5 mM of each dNTP), and, 0.2 µL (5 U/µL) HOT FIREPol® Taq polymerase (Solis BioDyne; Tartu, Estonia) were mixed before undergoing PCR.
Forward primers were labelled with the fluorescent dye 6-FAM™ (Sigma Aldrich; St. Louis, MO, USA). All PCRs were run in a Biometra Tprofessional thermocycler (Jena, Germany). For fragment separation, we used an ABI Genetic Analyzer 3130xl (Applied Biosystems, Foster City, CA, USA). PCR products were diluted 1:650 for ccmp and 1:600 for ucd and udt markers. GeneMapper software v3.7 (Applied Biosystems, Foster City, CA, USA) was used to score the fragment sizes.
2.4. Chloroplast CAPS Markers
In addition to the five cpSSR markers, three cpCAPS markers (4.1, 5.2, and 17.1) were used to define different haplotypes. Each sample was amplified with the specific cpCAPS primer pairs (Table S1) and digested as follows: 4.1 with HaeIII, 5.2 with AciI, and 17.1 with ApaI (all endonucleases were from Thermo Scientific™; Waltham, MA, USA), as described in Pettenkofer et al. [22]. PCRs were done for each primer pair individually with similar settings as for the ccmp cpSSR primers, except for a longer elongation phase in all 37 cycles for 4.1 and 17.1 (90 s instead of 60 s). The vials contained a mixture as described for ucd4, udt1, and udt4. After PCR, digestion was done by mixing 8 µL of PCR product with 3 U of enzyme and 2 µL of the associated buffer. All reactions were incubated at 37 °C overnight.
Fragment separation was done by agarose gel electrophoresis for about 60 min. For 4.1/HaeIII and 5.2/AciI we used 2.5% agarose gels at 3 V/cm and a 100 bp GeneRuler DNA Ladder (Thermo Scientific™; Waltham, MA, USA), for 17.1/ApaI 2% agarose gels, 4 V/cm and a 1kb GeneRuler as DNA Ladder (Thermo Scientific™; Waltham, MA, USA). 4.1/HaeIII had three alleles: a single fragment (uncut), two fragments (cut) and three fragments (cut twice), while 5.2/AciI and 17.1/ApaI had two alleles each (cut and uncut, respectively). Scoring was performed by visual observation (Figure S1). The well-defined DNA fragments were accompanied by faint bands, which exhibited consistent patterns. Since the summarized size of the strong bands added up to the size of the uncut PCR product, the faint bands were not considered for the analysis.
2.5. Chloroplast Haplotypes, Haplotype Diversity and Inter-Population Differentiation
Haplotypes were identified by combining all eight markers (Table 2). For detecting differences between markers and managing the dataset, we used Microsoft Excel 2016 (Redmond, WA, USA).

Table 2.
Marker composition of chloroplast simple sequence repeat (cpSSR) and chloroplast cleaved amplified polymorphic sequences (cpCACPS), and occurrence of Q. rubra chloroplast haplotypes.
Haplotype diversity (HT), average expected within-population haplotypic diversity (HS) and genetic differentiation among populations (GST & RST) were calculated using the software PermutCpSSR_1.2.1 [28,29] and 1000 permutations.
2.6. Minimum Spanning Tree
A minimum spanning tree (Figure 1) was created using Microsoft Office 2016 (Redmond, WA, USA) by comparing different haplotypes at each marker with Microsoft Excel and sorting them according to their similarity. Consequently, the haplotypes were linked with the most similar one in the dataset and visualized using Microsoft Publisher. Linkages were not always additive. Thus, if a haplotype 1 (H1) differed from a haplotype 2 (H2) at two positions, and H2 differed from haplotype 3 (H3) at three positions, H1 and H3 do not necessarily differ at five positions. The length of the links between two haplotypes was sometimes adjusted to fit the graph. Furthermore, base pair differences between cpSSR marker alleles were not considered (Table 2). For better visualization, three haplotype groups were assembled by grouping similar haplotypes, which differ from each other in less than three markers. Haplotypes thus compiled, were highlighted in different colors (blue, purple, and orange).
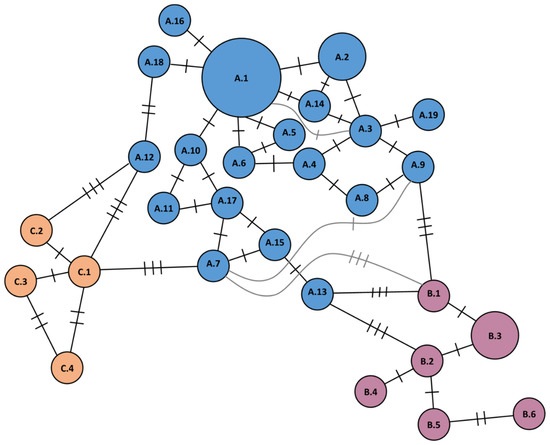
Figure 1.
Minimum spanning tree of Q. rubra haplotypes. Each haplotype is linked to the others by the minimum number of marker differences necessary. Each crossbar represents one mutation separating haplotypes. The area of the circle reflects sample sizes, where the largest circle represents the most common haplotype A.1 found in 690 trees, and the second largest circle represents two haplotypes A.2 and B.3 found in 126 and 167 trees, respectively (see also Table 2).
3. Results
3.1. Minimum Spanning Tree
Related haplotypes within each cluster are usually separated by differences in only one marker, with the exception of three haplotypes that show two differences instead. The majority of the samples was assigned to haplotypes A.1, A.2, and B.3 with 690, 126, and 167 samples, respectively.
3.2. Haplotype Diversity
Sixteen of the 29 haplotypes (Figure 1, Table 2) (A.4, A.6–A.9, A.12–A.16, A.19, B.2, B.6, C.1, C.2, and C.4) occurred only in North America and six exclusively in Germany (A.10, A.11, A.17, A.18, B.4, and C.3).
Seven haplotypes (A.1, A.2, A.3, A.5, B.1, B.3, and B.5) occurred in both North America and Germany. Haplotypes A.6, A.8–A.15, A.18, A.19, B.2, B.4, B.6, and C.3 were solely observed in single trees, while haplotypes A.4, A.7, A.16, A.17, B.1, B.5, C.1, C.2, and C4 were observed in four and more (up to ten) trees. A.1–A.3, A.5, and B.3 were observed in more than 35 trees, with A.1 being the most common haplotype observed in 690 trees (Table 2, Figure 2 and Figure 3).
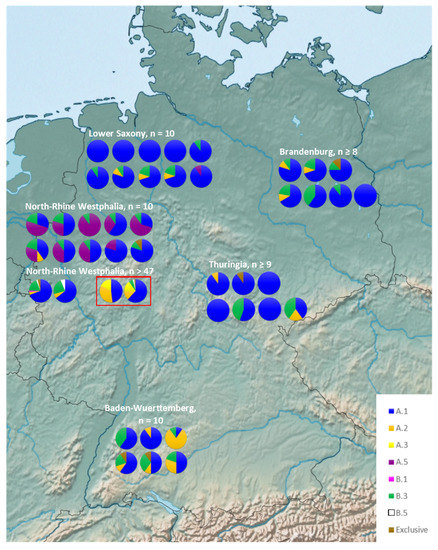
Figure 2.
Haplotype distribution of Q. rubra in Germany. For better visualization, haplotypes that were exclusive to Germany and did not occur in North America (brown) are pooled together (see Table 2). Those provenances in North Rhine-Westphalia that originated from Virginia (“Sonderherkunft Bornheim”) are depicted by a red frame. “Exclusive” haplotype in those provenances is haplotype C.3.
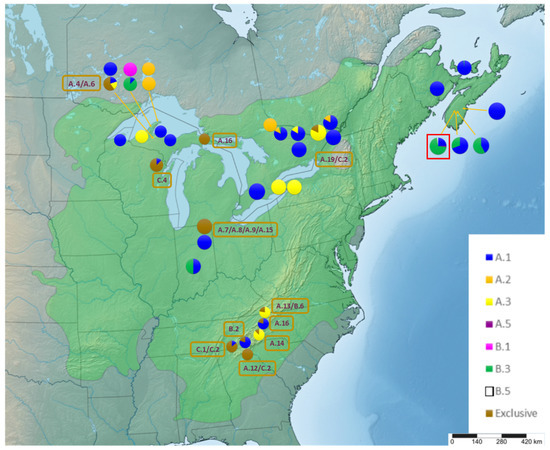
Figure 3.
Distribution of Q. rubra chloroplast haplotypes in North America. The native red oak range is highlighted in green. “Exclusive” are haplotypes that were found only in North America, but not in Germany. For better visualization, they were highlighted by brown color and pooled together in pie charts, but their individual names are listed within the brown frames placed next to the respective population. A5 and B.5 were represented by a single tree each in one population of Nova Scotia highlighted by the red frame.
A.1 was observed in every single stand within Germany, but only in ~65% of the North American populations (Figure 2 and Figure 3). Haplotype B.3 occurs in 30 German stands (~68% of German stands), while in North America it is only present in seven populations (~19% of American populations), mostly observed at the northern edge of the distribution range (Nova Scotia, Ontario and northern Michigan) (Figure 2 and Figure 3). The distribution of A.2 shows a similar pattern. It was found in 41% of the German stands, distributed over all federal states but occurs only in five (~13.5%) of the North American populations in the Great Lakes region of Michigan and Canada (Figure 2 and Figure 3).
Haplotype A.5 is widespread in North Rhine-Westphalia (64% of NRW stands) and present once in Lower Saxony (Germany). Thus, overall, it is present in 23% of German stands (Figure 2). Nevertheless, it could only be found in a single individual in the Nine Mile Woods population from Nova Scotia in North America (Figure 3). One of the rare haplotypes, B.5, was also observed in the same Nine Miles Woods population from Nova Scotia (Table 1) and in three of the German stands in North Rhine-Westphalia (Figure 2 and Figure 3). Haplotype A.3 was detected in the Appalachian Mountains and the Great Lakes region around Michigan but does not occur in northeastern populations of New Brunswick, Prince Edward Island, and Nova Scotia (Figure 3). In Germany, A.3 was only observed in two stands Diergardt 1 and 2 (“Bornheimer Roteiche”), with documented origin from Virginia, USA.
3.3. Haplotype Structure and Distribution
The 29 defined haplotypes (Figure 1) are assumed to be closely related within their assigned major clusters or haplogroups: A (blue), B (purple), and C (orange). Most of them are related to another haplotype differing at only one marker. Nevertheless, haplotypes A.12, B.6, and C.4 each differed from other haplotypes within their clusters by two mutations instead of one. Therefore, it is not certain whether they belong to their haplogroups.
The majority of defined haplotypes, including the most common haplotype A.1 observed in 690 trees, formed cluster or haplogroup A. Haplotype A.1 is followed by A.2 observed in 126 trees. In total, there were 940 trees in cluster A (~82.2% of all trees). Most of the remaining trees belonged to cluster or haplogroup B (~16.2% of all trees) with B.3 being the most frequent haplotype observed in 185 trees. Cluster C shows minor occurrences (~1.7% of all trees) with less than 10 trees per each haplotype and a total of 19 trees. All three clusters are present in Germany (Figure 2) and the native range (Figure 3). However, haplotypes in cluster C are mostly located near the Great Lakes region of Michigan and the southern parts of the Appalachian Mountains (Figure S2). Additionally, one population in eastern Ontario holds a small number of haplotypes from cluster C, but it was not observed at the northeastern edge of the distribution area in Canada. Haplotype C.3 occurred only in a single tree in German stand Diergardt 2, which originated presumably from Virginia, USA (“Bornheimer Roteiche”). Haplogroup A is characterized by an allele with fragment size of 228 bp at ccmp2, while typical for cluster B was an allele with fragment size of 115 bp at ccmp4, and cluster C was characterized by an allele with usually a single cut at 5.2/AciI and by an allele with fragment size of 226 bp at ccmp2 and an allele with fragment sizes of 117 or 118 bp at ccmp4 (see Table 2). The haplotypes from clusters A and B were widespread over the native range and Germany. Cluster C contained only haplotypes exclusive to one of the continents. Haplotypes A.6, A.8-A.15, A.18, A.19, B.2, B.4, B.6, and C.3 occurred only in single trees. A.7 was found in multiple trees, but only in a northern population of Indiana.
Haplotype A.1 was predominant in Germany and was present in all sampled stands (Figure 2). Haplotype B.3 was also common and present in 68% of all German stands, followed by A.2 present in 41% of stands. The distribution of haplotype A.5 was remarkable; it was present in 86% of NRW stands, but was observed only once in Lower Saxony. Haplotypes that were exclusive to Germany were distributed evenly over the federal states. B.5 can only be observed in the stands of NRW with larger sample size of 49–50 trees. These were also stands with the largest diversity containing up to six different haplotypes, probably due to their sample size. By contrast, the smaller stands with 8–10 trees contained a maximum of four different haplotypes (Figure 2). Haplotype A.3 was observed only in the two German stands Diergardt 1 and 2 (“Bornheimer Roteiche”) in the provenances that originated presumably from Virginia, USA. Furthermore, one of these stands contained haplotype C.3, which is the only haplotype of the C cluster observed in Germany (Figure S3) and was not observed in North America.
A.1 was the most common haplotype and observed in 65% of the North American populations (Figure 3), followed by A.3, B.3, and A.2 observed in 21.6%, 18.9%, and 13.5% of the North American populations, respectively. The 16 different populations that have haplotypes exclusive to North America are spread mainly from the Northwest to the South of the natural range, with the exception of one population near Ottawa. Six populations with large sample sizes of 30-50 trees located in Nova Scotia and on Prince Edward Island contained four different haplotypes in total, while six smaller populations in the Appalachian Mountains with sample sizes of 4-9 trees contained together 10 haplotypes.
3.4. Haplotype Diversity and Inter-Population Differentiation
Total haplotype diversity in Germany (HT = 0.516 ± 0.047) was lower than in North America (HT = 0.754 ± 0.059). However, haplotype diversity within German stands (HS = 0.385 ± 0.037) was higher than in North American populations (HS = 0.218 ± 0.042). Genetic differentiation was higher in North America (GST = 0.710 ± 0.054, RST = 0.717 ± 0.039), compared with German stands (GST = 0.254 ± 0.068, RST = 0.095 ± 0.031). Total haplotype diversity decreased from the South (HT = 0.882 ± 0.040, Figure S4) and the Northwest (HT = 0.860 ± 0.073, Figure S4) towards the Northeast (HT = 0.439 ± 0.124, Figure S4) in the native distribution area. Since some populations had higher sample sizes for North American and German stands, we tested whether there was an influence of the computed parameters by reducing the sample size of German and North American stands with more than eleven sampled trees to an amount of equal-sized randomly taken samples and recalculated the computed values. Resulting values were similar to the values of the entire dataset (Table S3).
4. Discussion
In this study, we successfully detected several new chloroplast haplotypes in German and North American red oaks, which enabled us to validate our hypothesis of a founder effect in German red oak reflected in decreased haplotype diversity compared to the native range. Furthermore, we found evidence for an introduction of the species from various North American populations from the northeastern part of the natural distribution range displayed by the presence of many haplotypes from the northern Great Lakes Region and the northeastern part of the natural range in German stands. This distribution pattern led us to the conclusion that North Rhine-Westphalian stands might originate from the Peninsula of Nova Scotia at the northeastern edge of the native distribution area.
4.1. Haplotype Diversity and Distribution
Q. rubra exhibits comparatively low intraspecific chloroplast variation [17,30], and Pettenkofer et al. [21] detected thirteen different red oak haplotypes in 62 stands based on five chloroplast SSR markers. Our results demonstrate that the resolution of both haplotype diversity and inter-population genetic differentiation increased with the use of additional chloroplast markers (cpCAPS) and the inclusion of more German stands and North American red oak populations. Pettenkofer et al. [21] observed a relatively low overall haplotype diversity (HT = 0.337 ± 0.046) of red oak in Germany and a relatively low genetic differentiation among stands (GST = 0.137 ± 0.067). Our study with additional cpCAPS markers (Table S2) and red oak populations revealed overall higher levels of chloroplast haplotype diversity in German stands as well as overall higher estimates of differentiation among German stands. In particular, stands from North Rhine-Westphalia could now be differentiated from other stands in Germany (Figure 2). Similarly, a finer haplotype resolution and the analysis of additional populations revealed higher levels of the total haplotype diversity in North American populations in comparison to that reported in Pettenkofer et al. [21] (HT = 0.652 ± 0.096). Genetic differentiation among the North American populations reported in Pettenkofer et al. [21] was high (GST = 0.729 ± 0.086) and it was similar to that we observed in this study. While the RST value in North America was not significantly different from the observed GST value, in Germany we observed a significantly (p = 0.014) lower value for RST than for GST. This finding is a result of the German haplotype composition that often differs only by a single nucleotide motif in the chloroplast SSR markers and was already observed in Pettenkofer et al. [21].
Due to the predominance of haplotypes A.1 and B.3, total haplotype diversity and genetic differentiation of northeastern North American populations (Figure S4) were low (HT = 0.439 ± 0.124, GST = 0.465 ± 0.095), compared with the Appalachian region (Figure S4) in the South (HT = 0.882 ± 0.040, GST = 0.503 ± 0.052) or the Great Lakes Region of Michigan and Wisconsin (Figure S4) in the West (HT = 0.860 ± 0.073, GST = 0.864 ± 0.087), where a higher number of haplotypes were observed. The reduced number of haplotypes in the Canadian populations, despite their large sample size, may be a result of postglacial migration from the south or a margin effect, which reduces occurring haplotypes to adapted ones. Haplotype diversity within populations was lowest for the western region (HS = 0.117 ± 0.077, Figure S4) as a result of fixation of many populations on one haplotype. The Northeastern populations had an intermediate level of haplotype diversity (HS = 0.235 ± 0.100), while the southern populations showed the highest haplotype diversity within populations (HS = 0.439 ± 0.056) and a high number of different haplotypes.
These findings are in agreement with previous studies that found a continuous trend of increasing genetic variation from the Northeast towards the Southeast for nuclear DNA markers (SNPs) [31]. Furthermore, Merceron et al. [31] identified three different genetic clusters (G1, G2, and G3) in native red oak based on the examination of 73 native and 38 European populations at 69 SNP markers. The G2 cluster represented the southern region from North Carolina and Alabama to Indiana, while the G1 cluster represented mainly the Northeast of the native distribution area. The trees in the third cluster G3 were more or less equally distributed across the species range [31]. Dispersion of the trees in the northeastern cluster G1 follows well the distribution of haplotypes A.2 and B.3 in our study. The haplotype A.3 is associated well with the G2 cluster in the South. The clusters based on the nuclear DNA markers [31] seem to be in agreement with the clusters based on the cpDNA haplotypes observed in this study.
The lower haplotype diversity in German stands compared to North American populations might indicate a founder effect due to introduction of a relatively small number of trees from a limited range in North America. Since German red oak chloroplast haplotype diversity is still high, we consider multiple introductions, possibly at different times and probably from different stands within the Northeastern parts of the native range. Consequently, German stands showed a higher genetic variation within stands than North American populations, which were often fixed on one haplotype. Haplotype diversity was especially high in North Rhine-Westphalia (HS = 0.532 ± 0.043) and Baden Wuerttemberg (HS = 0.526 ± 0.082) stands compared to stands in other German federal states (HS = 0.296 ± 0.050). These results suggest plausible multiple introductions or mixtures of reproductive material from several separated geographic areas. The low differentiation among stands of Germany compared to the differentiation among North American populations indicates that German stands do not fully represent the haplotype diversity contained in North American populations. This can be explained by a genetic bottleneck during the introduction of the species to Europe. Pettenkofer et al. [21] also mentioned a possible increase in variation within stands due to admixture and multiple introductions of different North American provenances in Southern Germany. Our findings support this conclusion, especially for the federal states North Rhine-Westphalia and Baden-Wuerttemberg.
4.2. Haplotype Distribution and German Red Oak Origin
The number of haplotypes on the North American continent (16 haplotypes) that are completely absent in Germany exceeds by far the number of exclusively German haplotypes (6 haplotypes). Moreover, haplotype A.2 was frequent in many German stands, but rarely present in North American populations, where it is restricted to the Great Lakes region of Upper Michigan and Canada. Finally, the absence in Germany of diverse haplotypes distributed from the Northwestern to Southern areas of the native range points to an origin of most German red oaks from the Northeast of North America (Figure 3, brown frames) [21].
A.3, which is frequent in the center of the native range, is not only rare in German stands, but occurs only in those two stands that supposedly originated from Virginia, USA [32]. One of those two stands from Virginia also contains a single tree with haplotype C.3, which is the only haplotype from the C cluster in Germany. Cluster C is absent in the northeastern edge of the natural range, as well as in other German stands supporting a separate origin of these two stands.
It is unlikely that German private haplotypes are completely novel and cannot be found in North America. It is more likely that rare haplotypes were not found in our sampled trees because of their low frequency and restricted geographic distribution, which caused failure to detect them in the natural range of the species. Thus, additional and more comprehensive collections of populations in North America would be desirable. Rare haplotypes that are exclusive to Germany, such as A.10 or B.4 could then also be of interest to trace the origin of Q. rubra. Their identification in a restricted geographic area in North America could further delineate the origin of German stands. Private German haplotype A.18 was found only in a single tree in the stand “Schutterwald 3”and differed from haplotype A.1 only by one mutation (Table 1) suggesting the possibility of a recent mutation.
4.3. Red Oak Stands in North Rhine-Westphalia Might Have Originated from Nova Scotia
The presence of haplotypes B.5 and A.5 only in several stands in North Rhine-Westphalia, Germany, with their simultaneous absence in nearly all other German stands and in most populations of North America suggests a distinct origin of these stands from a very limited geographic area within the natural distribution range. In particular, B.5 and A.5 were observed with low frequency in a Canadian stand on the Peninsula of Nova Scotia. It is possible that A.5 and B.5 haplotypes were not found in other areas of the natural range due to the smaller sample size of North American populations compared to the northeastern Canadian populations. However, the southern parts of Nova Scotia exhibit good geographical exposition for seaborne trade with Boston Harbor, which was assumed to be one of the main transshipment points for northern red oak introduction to Germany [33]. Our inference is consistent with the settlement history of the Nova Scotia area from where the red oak populations were sampled. According to Library and Archives Canada [34] over 2000 German settler arrived in Nova Scotia in 1750-1752, and in 1753, some of these settlers established the town of Lunenburg. Later, the town of New Germany was established. The sampled Nova Scotia red oak populations are from the New Germany and Lunenburg area. The German settlers may have introduced red oak from Nova Scotia to Germany. Consequently, some North Rhine-Westphalian stands may have originated from the southern parts of Nova Scotia. Since seed traders do not necessarily take genetic variety into account while harvesting acorns, it is plausible that descendants from a limited number of trees resulted in widespread dissemination of single haplotypes that may be rare in the region of origin. This way, anthropogenic influence could create novel haplotype compositions as observed in North Rhine-Westphalian stands.
Haplotype A.5 was identified as a new haplotype based on the analysis of the new cpCAPS markers highlighting the importance of whole plastome sequencing of different provenances for the identification of rare and diagnostic genetic variants. The large sample size of four German stands provided a much better resolution of chloroplast haplotype diversity because of the discovery of several new haplotypes. While the most common haplotype A.1 showed similar shares within the single stands, and therefore was comparable to the chloroplast haplotype based on the cpSSR in Pettenkofer et al. [21], rarer haplotypes with restricted geographic range in North America such as B.1 and B.5 were especially useful.
5. Conclusions
We were able to provide additional evidence that German Q. rubra stands originated from the Northern edge of the native distribution area [21,23]. Similar observations were made for French red oak stands [31]. The low haplotype diversity in Germany as compared to North America supports the introduction of Q. rubra to Germany from a limited geographic range. Furthermore, our data suggest an introduction of Q. rubra from different restricted geographic areas within the natural distribution area and helped us narrow down the putative origin of most German stands in North Rhine-Westphalia to the Peninsula of Nova Scotia in the northeast of the native range. Additional high coverage sampling and cpDNA genotyping in the native range could provide further evidence for the origin of introduced red oak stands.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4907/11/9/1025/s1, Figure S1: Cutting patterns for 4.1/HaeIII (top), 5.2/AciI (middle) and 17.1/ApaI (bottom) cpCAPS markers. Separated by the black bar on the right is the control that shows uncut and cut patterns for 5.2/AciI and 17.1/ApaI and uncut, cut, cut twice patterns for 4.1/HaeIII from left to right, Figure S2: Distribution of the three cpDNA clusters in North America (see also the haplotype network in Figure 1), Figure S3: Distribution of the three cpDNA clusters in Germany, Figure S4: Haplotype distribution in North America. Regions for which GST, HS and HT were calculated individually are shown within red frames, Table S1: PCR primer nucleotide sequences for the 4.1, 5.2, and 17.1 cpCAPS markers, Table S2: Chloroplast haplotypes based on five cpSSR markers studied in Pettenkofer et al. [21] and the new chloroplast haplotypes in this study based on the five cpSSR and three novel cpCAPS markers, Table S3: HS, HT, and GST of the entire datasets for Germany and North America, compared to corrected ones, where populations with enlarged sample sizes were reduced to n = 10 in German stands and n = 8 in North American populations by taking a random sample.
Author Contributions
Conceptualization, O.G., L.L., and K.V.K.; methodology, O.G. and J.G.; validation, O.G., K.V.K., L.L., M.M, and J.G.; field sampling design and sampling in Canada, O.P.R.; formal analysis, J.G.; investigation, J.G.; resources, O.G., L.L. and O.P.R.; data curation, J.G.; writing—original draft preparation, J.G.; writing—review and editing, O.G., K.V.K., J.G., L.L., M.M. and O.P.R.; visualization, J.G.; supervision, O.G.; project administration, O.G.; funding acquisition, O.G., O.P.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the German Federal Ministry of Food and Agriculture, grant number 22023314 and in part by the Natural Sciences and Engineering Research Council of Canada Discovery Grant RGPIN 17-04589 to Om P. Rajora.
Acknowledgments
We thank Alexandra Dolynska, Tim Pettenkofer, and Ying Wang for technical support and Katharina Burkardt, as well as Martin Rogge from the “Landesbetrieb Wald und Holz Nordrhein-Westfalen” for sample collection in the Fander Ville 1 and 2, and Diergardt 1 and 2 (“Bornheimer Roteiche”) stands. We thank Mary Jane Rodger, General Manager and Jennica Hunsinger, Research Intern of Medway Community Forest Co-op, Andree Morneault of Nipissing Forest Resource Management Inc, Steve Chenier, Gary Schneider of MacPhail Woods, and Chandler Colpit of Mcrea Farms for their help in locating and field sampling of red oak populations in Canada. We acknowledge support by the Open Access Publication Funds of the Georg-August University of Göttingen. Finally, we would like to thank anonymous reviewers for providing suggestions for improvement.
Conflicts of Interest
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Sander, I.L. Quercus rubra L. In Silvics of North America: Hardwoods; Handbook, 654; Burns, R.M., Honkala, B.H., Eds.; US Department of Agriculture Forest Service: Washington, DC, USA, 1990; Volume 2, pp. 727–733. [Google Scholar]
- Göhre, K.; Wagenknecht, E. Die Roteiche und ihr Holz; Deutscher Bauernverlag: Berlin, Germany, 1955; pp. 22–45. [Google Scholar]
- Liesebach, M.; Schneck, V. Entwicklung von amerikanischen und europäischen Herkünften der Roteiche in Deutschland. Forstarchiv 2011, 82, 125–133. [Google Scholar]
- Nagel, R.-V. 4.13 Roteiche (Quercus rubra L.). In Potentiale und Risiken Eingeführter Baumarten. Baumartenportraits mit Naturschutzfachlicher Bewertung, 1st ed.; Vor, T., Spellmann, H., Bolte, A., Ammer, C., Eds.; Universitätsverlag Göttingen: Göttingen, Germany, 2015; pp. 166–205. [Google Scholar]
- Woziwoda, B.; Kope, D.; Witkowski, W.J. The negative impact of intentionally introduced Quercus rubra L. on a forest community. Acta Soc. Bot. Pol. 2014, 83, 39–49. [Google Scholar] [CrossRef]
- Zürcher, E.; Kucera, L.; Bosshard, H.H.; Zürich, E.T.H. Bildung und Morphologie der Thyllen: Eine Literaturübersicht. Vierteljahrsschr. Nat. Ges. Zürich 1985, 130, 311–333. [Google Scholar]
- Chmura, D. Penetration and naturalisation of invasive alien plant species [Neophytes] in woodlands of the Silesian Upland [Southern Poland]. Nat. Conserv. 2004, 60, 3–11. [Google Scholar]
- Riepšas, E.; Straigyte, L. Invasiveness and ecological effects of red oak (Quercus rubra L.) in lithuanian forests. Balt. For. 2008, 14, 122–130. [Google Scholar]
- Chmura, D. Impact of alien tree species Quercus rubra L. on understorey environment and flora: A study of the Silesian upland (Southern Poland). Pol. J. Ecol. 2013, 61, 431–442. [Google Scholar]
- Martin, P.H.; Marks, P.L. Intact forests provide only weak resistance to a shade-tolerant invasive Norway maple (Acer platanoides L.). J. Ecol. 2006, 94, 1070–1079. [Google Scholar] [CrossRef]
- Vor, T.; Lüpke, B. Das Wachstum von Roteiche, Traubeneiche und Rotbuche unter verschiedenen Lichtbedingungen in den ersten beiden Jahren nach der Pflanzung. Forstarchiv 2004, 75, 13–19. [Google Scholar]
- Ninements, Ü.; Valladares, F. Tolerance to shade, drought and waterlogging of temperate northern hemisphere trees and shrubs. Ecol. Monogr. 2006, 76, 521–547. [Google Scholar] [CrossRef]
- Steiner, W. Hochwertiges Vermehrungssaatgut durch züchterische Verbesserung: Ein Vergleich verschiedener möglichkeiten am Beispiel der Roteiche (Quercus rubra L.). Forstarchiv 2012, 83, 85–92. [Google Scholar]
- Dumolin, S.; Demesure, B.; Petit, R.J. Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theor. Appl. Genet. 1995, 91, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Petit, R.J.; Brewer, S.; Bordács, S.; Burg, K.; Cheddadi, R.; Coart, E.; Cottrell, J.; Csaikl, U.M.; Van Dam, B.; Deans, J.D.; et al. Identification of refugia and post-glacial colonisation routes of European white oaks based on chloroplast DNA and fossil pollen evidence. For. Ecol. Manag. 2002, 156, 49–74. [Google Scholar] [CrossRef]
- Heuertz, M.; Fineschi, S.; Anzidei, M.; Pastorelli, R.; Salvini, D.; Paule, L.; Frascaria-Lacoste, N.; Hardy, O.J.; Vekemans, X.; Vendramin, G.G. Chloroplast DNA variation and postglacial recolonization of common ash (Fraxinus excelsior L.) in Europe. Mol. Ecol. 2004, 13, 3437–3452. [Google Scholar] [CrossRef]
- Magni, C.R.; Ducousso, A.; Caron, H.; Petit, R.J.; Kremer, A. Chloroplast DNA variation of Quercus rubra L. in North America and comparison with other Fagaceae. Mol. Ecol. 2005, 14, 513–524. [Google Scholar] [CrossRef]
- Molina-Cano, J.L.; Russell, J.R.; Moralejo, M.A.; Escacena, J.L.; Arias, G.; Powell, W. Chloroplast DNA microsatellite analysis supports a polyphyletic origin for barley. Theor. Appl. Genet. 2005, 110, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Imazio, S.; Labra, M.; Grassi, F.; Scienza, A.; Failla, O. Chloroplast microsatellites to investigate the origin of grapevine. Genet. Resour. Crop Evol. 2006, 53, 1003–1011. [Google Scholar] [CrossRef]
- Birchenko, I.; Feng, Y.; Romero-Severson, J. Biogeographical distribution of chloroplast diversity in northern red oak (Quercus rubra L.). Am. Midl. Nat. 2009, 161, 134–145. [Google Scholar] [CrossRef]
- Pettenkofer, T.; Burkardt, K.; Ammer, C.; Vor, T.; Finkeldey, R.; Müller, M.; Krutovsky, K.; Vornam, B.; Leinemann, L.; Gailing, O. Genetic diversity and differentiation of introduced red oak (Quercus rubra) in Germany in comparison with reference native North American populations. Eur. J. For. Res. 2019, 138, 275–285. [Google Scholar] [CrossRef]
- Pettenkkofer, T.; Finkeldey, R.; Müller, M.; Krutovsky, K.V.; Vornam, B.; Leinemann, L.; Gailing, O. Development of novel Quercus rubra chloroplast genome CAPS markers for haplotype identification. Silvae Genet. 2020, 69, 78–85. [Google Scholar] [CrossRef]
- Pettenkofer, T.; Finkeldey, R.; Müller, M.; Krutovsky, K.V.; Vornam, B.; Leinemann, L.; Gailing, O. Genetic variation of introduced red oak (Quercus rubra) stands in Germany compared to North American populations. Eur. J. For. Res. 2020, 139, 321–331. [Google Scholar] [CrossRef]
- Lind, J.F.; Gailing, O. Genetic structure of Quercus rubra L. and Quercus ellipsoidalis E. J. Hill populations at gene-based EST-SSR and nuclear SSR markers. Tree Genet. Genomes 2013, 9, 707–722. [Google Scholar] [CrossRef]
- Lind-Riehl, J.F.; Sullivan, A.R.; Gailing, O. Evidence for selection on a CONSTANS-like gene between two red oak species. Ann. Bot. 2014, 113, 967–975. [Google Scholar] [CrossRef]
- Weising, K.; Gardener, R.C. A set of conserved PCR Primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome 1999, 42, 9–19. [Google Scholar] [CrossRef]
- Deguilloux, M.F.; Dumolin-Lapègue, S.; Gielly, L.; Grivet, D.; Petit, R.J. A set of primers for the amplification of chloroplast microsatellites in Quercus. Mol. Ecol. Notes 2003, 3, 24–27. [Google Scholar] [CrossRef]
- Burban, C.; Petit, R.J.; Carcreff, E.; Jactel, H. Rangewide variation of the maritime pine bast scale Matsucoccus feytaudi Duc. (Homoptera: Matsucoccidae) in relation to the genetic structure of its host. Mol. Ecol. 1999, 8, 1593–1602. [Google Scholar] [CrossRef]
- Pons, O.; Petit, R.J. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 1996, 144, 1237–1245. [Google Scholar]
- Feng, Y.; Sun, Y.L.; Romero-Severson, J. Heterogenity and spatial autocorrelation for chloroplast haplotypes in three old growth populations of northern red oak. Silvae Genet. 2008, 57, 212–220. [Google Scholar] [CrossRef]
- Merceron, N.R.; Leroy, T.; Chancerel, E.; Romero-Severson, J.; Borkowski, D.S.; Ducousso, A.; Monty, A.; Porté, A.J.; Kremer, A. Back to America: Tracking the origin of European introduced populations of Quercus rubra L. Genome 2017, 60, 778–790. [Google Scholar] [CrossRef]
- Holthausen, R. Die Bornheimer Roteiche. Allg. Forstz. 1987, 42, 44. [Google Scholar]
- Bauer, F. Zur Rassenfrage der Roteiche. Allg. Forstz. 1954, 9, 470–474. [Google Scholar]
- Library and Archives Canada. Available online: https://www.bac-lac.gc.ca/eng/discover/immigration/history-ethnic-cultural/Pages/German.aspx (accessed on 1 June 2020).
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).