Abstract
Phytophthora cinnamomi is a devastating pathogen causing root and crown rot and dieback diseases of nearly 5000 plant species. Accurate and rapid detection of P. cinnamomi plays a fundamental role within the current disease prevention and management programs. In this study, a novel effector gene PHYCI_587572 was found as unique to P. cinnamomi based on a comparative genomic analysis of 12 Phytophthora species. Its avirulence homolog protein 87 (Avh87) is characterized by the Arg-Xaa-Leu-Arg (RxLR) motif. Avh87 suppressed the pro-apoptotic protein BAX- and elicitin protein INF1-mediated cell death of Nicotiana benthamiana. Furthermore, a recombinase polymerase amplification-lateral flow dipstick detection assay targeting this P. cinnamomi-specific biomarker was developed. While successfully detected 19 P. cinnamomi isolates of a global distribution, this assay lacked detection of 37 other oomycete and fungal species, including P. parvispora, a sister taxon of P. cinnamomi. In addition, it detected P. cinnamomi from artificially inoculated leaves of Cedrus deodara. Moreover, the RPA-LFD assay was found to be more sensitive than a conventional PCR assay, by detecting as low as 2 pg of genomic DNA in a 50-µL reaction. It detected P. cinnamomi in 13 infested soil samples, while the detection rate was 46.2% using PCR. Results in this study indicated that PHYCI_587572 is a unique biomarker for detecting P. cinnamomi. Although PHYCI_587572 was identified as an effector gene based on the RxLR motif of Avh87 and the avirulence activity on Nicotiana, its exact genetic background and biological function on the natural hosts of P. cinnamomi warrant further investigations.
1. Introduction
Phytophthora cinnamomi is a destructive pathogen of nearly 5000 plant species including many economically important crops [1,2,3]. For example, root rot caused by P. cinnamomi is a widely spread and most severe disease of avocado with an estimated annual loss of $40 million in California, USA alone [4]. Phytophthora cinnamomi also causes root and crown rot, canker and dieback diseases on a wide range of woody ornamental crops such as azalea, boxwood, camellia, cedar, and rhododendron [5]. In addition to its impact on agriculture and horticulture, P. cinnamomi is a notorious invasive pathogen in natural plantations worldwide. In concert with chestnut blight, P. cinnamomi was responsible for the near extinction of wild American chestnut trees in the eastern United States [6,7]. In the forests of south-west Western Australia, P. cinnamomi has permanently altered the composition of plant communities and caused drastic decline of biodiversity by killing over 3500 susceptible endemic plant species and threatening macrofungal and bird species [3,8,9,10]. In China, P. cinnamomi has been found in many provinces and municipalities such as Fujian [11,12,13], Hainan [14], Jiangsu [13], Shaanxi [15], Shanghai [16], and Zhejiang [13], threatening the production of avocado, blueberry, chestnut, kiwifruit, ornamental trees, and the health of natural forests.
Phytophthora species secrete a wide range of effector proteins that overcome host immunity and facilitate the infection of plants [3,17] such as elicitins [18], small cysteine-rich toxins, transglutaminases, and RxLR effectors [3]. RxLR effectors, characterized by an Arg-Xaa-Leu-Arg (RxLR) motif, N-terminal signal peptide, and often Glu-Glu-Arg (EER) motif, are responsible for the translocation of effector proteins from plant apoplast into cytoplasm [19,20,21]. Although the exact number and pathogenesis roles of P. cinnamomi RxLR effectors remain to be determined, a comparative genomic analysis of P. infestans and P. cinnamomi has identified the presence of at least 171 RxLR effector genes in the P. cinnamomi genome [3]. Furthermore, only three out of 10 RxLR effector homologues that have known functional characters are present in the P. cinnamomi genome [3], implying the presence of P. cinnamomi-unique RxLR effector proteins and genes.
Early detection is the first and arguably most effective step within the hierarchy of plant disease control strategies. Although chemical application and soil sanitation could reduce disease severity and inoculum level [5], regular treatments could be costly and ineffective once P. cinnamomi has already widely spread in agricultural settings, and conceivably even more challenging in natural forests due to the scale of the problem. A different approach of disease management is early detection followed by the eradication of the pathogen. It is relatively inexpensive, sustainable, and effective if the pathogen is removed at an early stage. Nevertheless, its success relies on the accuracy and speed of the detection assay.
Detection assays of P. cinnamomi have advanced with molecular techniques, while their accuracy has been challenged by emerging new pathogens. Previous molecular detection assays of P. cinnamomi were developed based on conventional PCR [22,23,24,25,26], real-time PCR [27,28,29], and isothermal amplification assays [28,30]. However, many of these assays have failed in differentiating P. cinnamomi from P. parvispora [31], the species most closely related to P. cinnamomi within the genus Phytophthora [32,33,34].
Recombinase polymerase amplification (RPA) is an emerging technique that has been applied in detection assays of Phytophthora species [28,35,36,37,38]. The RPA process employs three core factors, including recombinase, DNA-binding proteins and polymerase. Recombinases perform to pair oligonucleotide primers with homologous sequence in duplex DNA. Simultaneously, DNA-binding proteins bind to displaced strands of DNA and polymerase begins to synthesize complete double-stranded DNA where the primer has bound to the target DNA. The enzyme mixture conducts exponential amplification of the target region within the template. Compared to PCR and loop-mediated isothermal amplification (LAMP) assays, RPA assays require shorter time span and less complex procedures and equipment for amplifying DNA templates of Phytophthora species [36,37,38,39,40]. In addition, RPA amplified products can be detected by lateral flow strips and the results can be easily read without any specialized equipment, suggesting that combination of RPA with lateral flow dipsticks (RPA-LFD) is suitable for developing a simple, rapid, and specific method for the detection of plant pathogens in resource-limited settings. The aims of this study were to identify a RxLR effector gene PHYCI_587572 using a comparative genomic approach and develop an RPA-LFD assay targeting this P. cinnamomi-specific biomarker.
2. Materials and Methods
2.1. Isolate Selection and DNA Extraction
Phytophthora cinnamomi isolates were obtained from diseased roots of Pinus sp., Rhododendron simsii, and Camellia oleifera from various provinces in China. The roots were surface disinfected, dried, and placed on Phytophthora selective agar PARP-V8 agar [5]. Surface disinfestation of the diseased roots were accomplished incubation in 70% ethanol. When using 70% ethanol, infected roots were given single dips and then dipped in three vials containing autoclaved distilled water for 5–10 min per vial. Following surface disinfestation, roots were surface dried by placing them on filter paper. A total of 76 isolates were used in this study including 19 of P. cinnamomi, 40 samples representing 21 Phytophthora species, one of Globisporangium ultimum, and 16 other fungal species (Table 1). For DNA extraction, each isolate was cultured in 10% clarified V8 juice (oomycetes) or potato dextrose broth (fungi) at 20–25 °C in the dark for 4–5 days. Genomic DNA (gDNA) was extracted from the harvested mycelia using a E.Z.N.A.® HP Fungal DNA Kit (Omega Bio-tek, Inc., Norcross, GA, USA) following the manufacturer’s instructions. Concentrations of gDNA extractions were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA) then adjusted as needed. Extractions were stored at −20 °C until use.

Table 1.
List of oomycete and fungal isolates used in this study and their detection results in the recombinase polymerase amplification-lateral flow dipstick (RPA-LFD) assay targeting the PHYCI_587572 biomarker.
2.2. Genomic Sequences
Genomic sequences and gene models of P. capsici, P. cinnamomi, P. infestans, P. ramorum, and P. sojae were retrieved from the MycoCosm portal [41]. Those of P. asiatica, P. cactorum, P. cambivora, P. hibernalis, P. parvispora, P. syringae, and P. vignae were also included in the subsequent comparative genomic analysis.
2.3. Identification of A P. cinnamomi-Unique RxLR Effector Gene
To identify RxLR effectors, the genome sequence of P. cinnamomi was translated to open reading frames (ORFs). ORFs encoding < 50 amino acids were excluded. SignalP v3.0 was used to identify the N-terminal signal peptide [42] within remaining ORFs. Proteins with Hidden Markov Model (HMM) scores > 0.9 and without predicted transmembrane domain were predicted as secreted proteins. A regular expressions (REGEX) method was performed by script for identifying the (^\w{1,40}\w{1,96}R\wLR\w{1,40} [ED][ED][KR]) motif [43,44] within the predicted secreted proteins. The RxLR-EER domains of the candidates were aligned and then used to construct a HMM profile to search within the predicted secreted proteins using HMMER. Matches with a bit score of >0 were manually validated.
2.4. Construction of Binary Potato Virus X (PVX) Vectors
As no introns were predicted within PHYCI_587572, this gene was directly amplified using the gDNA of isolate Pci1 (Table 1). Each 25-µL PCR reaction include 1 µL of gDNA (100 ng per µL), 1 μL (10 μM) of each of primers PHYCI_587572-PVX-HA(infusion)-F and PHYCI_587572--PVX-HA(infusion)-R (Table 2), 1 µL of PrimeSTAR HS DNA Polymerase (Takara Bio, Kusatsu, Shiga, Japan), 4 µL of dNTP Mixure (2.5 mM), 10 µL of 5× PrimeSTAR Buffer, and 32 µL of nuclease-free water (nfH2O). PCR was carried out using a Veriti 96-Well Thermal Cycler (Applied Biosystems, Singapore) following the program: an initial denaturation step at 95 °C for 3 min, 35 cycles of 95 °C for 15 s, 56 °C for 15 s, and 72 °C for 15 s, plus a final extension at 72 °C for 5 min. Each set of PCR reactions included a NTC. PCR products were examined in 1% agarose gel electrophoresis at 120 V for approximately 25 min. Agarose gel was stained by ethidium bromide and visualized on a transilluminator. The PCR assay was carried out three times. PCR products were digested with the SmaI restriction enzyme (NEB, R0141S) and cloned into the PVX vector pGR107 [45] to form pGR107:PHYCI_587572. The constructs were confirmed by sequencing at Genscript (Nanjing, China) using the primers LBA and LBB (Table 2). Recombinant binary plasmids were maintained and propagated in Escherichia coli strain JM109 grown in Luria-Bertani (LB) media amended with kanamycin (50 mg per mL) and tetracycline (12.5 mg per mL).

Table 2.
Primers used in the construction of binary potato virus X (PVX) vectors, infiltration of Agrobacterium tumefaciens, and the recombinase polymerase amplification-lateral flow dipstick (RPA-LFD) assay targeting the PHYCI_587572 biomarker.
2.5. Agrobacterium Tumefaciens Infiltration
Constructs were introduced into A. tumefaciens strain GV3101 by electroporation [46,47]. Recombinants (pGR107:PHYCI_587572) were selected with tetracycline (12.5 mg per mL) and kanamycin (50 mg per mL). Individual colonies were verified in PCR using the primers LBa and LBb (Table 2).
Nicotiana benthamiana plants were grown in a greenhouse at a day/night temperature of 25/16 °C and a 16 h photoperiod for 4–6 weeks. Recombinant GV3101 cells were grown in LB media amended with kanamycin (50 mg per mL) for 48 h, harvested, washed with 10 mM MgCl2 three times, resuspended in 10 mM MgCl2 to a final OD600 of 0.4, then incubated at room temperature for 1 to 3 h prior to infiltration. For pressure infiltration, 100 mL of recombinant GV3101 cell suspension was infiltrated though a small nick into each of three infiltration sites per adaxial side of a N. benthamiana leaf using a syringe. After 0, 12, or 24 h, Agrobacterium tumefaciens cells carrying the Bax gene (pGR107:Bax) or inf1 gene (pGR107:inf1) were infiltrated into each of the three filtration sites. Agrobacterium tumefaciens cells carrying the enhanced green fluorescent protein (eGFP) gene in place of pGR107:Bax or pGR107:inf1 were infiltrated into three parallelly designated filtration sites of the same leaf after 0, 12, or 24 h as controls. Symptom development was monitored daily. Photographs were taken on 5 and 6 days after filtration of pGR107:Bax and pGR107:inf1, respectively. This experiment was conducted twice with three replicate leaves in each repeat.
2.6. RPA-LFD Assay
A set of RPA primers and probe targeting the P. cinnamomi-specific biomarker PHYCI_587572, including a forward primer PciRL587572F, a 5’-biotin-labeled reverse primer PciRL587572R, and a probe PciRL587572P labeled by fluorescein amidite (FAM), tetrahydrofuran (THF), and C3 spacer (Table 2), was designed according to the instruction for the nfo kit in the TwistAmp® DNA Amplification Kits Assay Design Manual. Primers and probe were synthesized by Sangon Biotech (Shanghai, China).
The RPA-LFD assay was performed following the procedures in the TwistAmp® nfo Kit Quick Guide (TwistDx Ltd., Cambridge, UK). Each RPA reaction in a total volume of 50 μL contained 1 μL of DNA template, 29.5 μL of rehydration buffer, 2.1 μL of each of primers PciRL587572F and PciRL587572R (10 μM), 0.6 μL of probe PciRL587572P (10 μM), 12.2 μL of nfH2O, and 2.5 μL of magnesium acetate (280 mM). These reagents were added into reaction microtubes and mixed using a pipette. Freeze-dried pellet magnesium acetate was pre-added to the cap of the microtubes. After brief vortex and centrifugation, the reaction microtubes were incubated in a heating block set at 39 °C for 20 min. To visualize amplicons, 10 μL of RPA product was mixed with 90 μL of phosphate buffered saline with Tween 20 (PBST) running buffer. Then 5 μL of the RPA product per each reaction was added to the sample pad of a Milenia Genline HybriDetect 1 LFD (Milenia Biotec GmbH, Giessen, Germany). The LFD was then vertically immersed into 100 μL of PBST (Milenia Biotec GmbH, Giessen, Germany) in a sterile centrifuge tube for up to 5 min at room temperature (aver. 22 °C) until a clear control line was observed. The dipsticks yielded visible control lines, indicating valid tests. Test lines were visible on all dipsticks correlating to positive RPA reactions.A negative detection or NTC in contrast should not result in any signal or color at the position of the test line.
2.7. Evaluation of RPA Specificity and Sensitivity
The specificity of the RPA-LFD assay was evaluated by testing against all isolates listed in Table 1. One µL of gDNA (10 ng per µL) of each isolate was included in each 50 µL reactions. Each isolate was tested in triplicate.
To evaluate the sensitivity of the RPA-LFD assay, 10-fold serial dilution of gDNA of P. cinnamomi isolate Pci1 ranging from 100 ng to 100 fg (0.0001 ng) per µL were used as templates. Each concentration was evaluated in triplicate. Each set of reactions in the specificity and sensitivity evaluations included an NTC (nfH2O in place of DNA template).
2.8. Detecting P. cinnamomi in Artificially Inoculated Pine Needles Using RPA-LFD
Pine needles were collected from a healthy deodar cedar tree (Cedrus deodara) at a landscape site of Nanjing Forestry University (Nanjing, China). They were artificially inoculated by P. cinnamomi. A 2 × 2 mm mycelial plug of P. cinnamomi isolate Pci1 was mixed with approximately 100 g of pine needles and incubated in each of triplicate sterile 250 mL flasks at 22 °C, 100% relative humidity, and a 12-h photoperiod for 5 days. Sterile agar plugs were mixed with pine needles to produce three non-inoculated control samples. Total DNAs were extracted from P. cinnamomi-colonized and non-inoculated samples using an NaOH lysis method. Briefly, 20 mg of plant tissues collected from the wound site of each Pine needle were placed into a 1.5-mL microtube containing 200 µL of NaOH (0.5 N). They were grinded for approximately 1 min until no large pieces of plant tissues were visible using a sterile tissue grinder pestle. Then, 5 µL of grinded tissues in NaOH were transferred to a new microtube containing 495 µL of Tris buffer (100 mM, pH 8.0). Two µL of the mixture were used as used as templates in the RPA-LFD assay. Purified gDNA (10 ng per µL) of isolate Pci1 and nfH2O were used as a positive control and NTC, respectively, in two repeats of the experiment.
2.9. Detecting P. cinnamomi in Soil Samples Using the RPA-LFD Assay
A total of 13 soil samples were collected from five provinces in China (Table 3). A previous study found the presence of P. cinnamomi in all 13 samples [30]. Each soil sample was mixed well and divided into two equal subsamples. DNAs were directly extracted from the first set of 13 soil subsamples using a Plant DNA Mini Kit (Omega Biotek, Norcross, GA, USA). For the second set, a baiting method [5] using C. deodara leaves was deployed. After 5 days, DNA was extracted from leaf baits of each subsample using the NaOH lysis method. Both RPA-LFD and conventional PCR assays were performed using DNA extractions of both soil samples and leaf baits. Each DNA extraction was tested three times using each assay. Purified gDNA (10 ng) of isolate Pci1 was used in positive control reactions.

Table 3.
Detection results of Phytophthora cinnamomi in 13 infested soil samples using the recombinase polymerase amplification-lateral flow dipstick (RPA-LFD) and PCR assays.
3. Results
3.1. Identification of A P. cinnamomi-Unique RxLR Effector Gene PHYCI_587572
A protein (Avh87), translated from the PHYCI_587572 gene located at scaffold_77:29462-29848, was found containing a signal peptide cleavage site at residue 20, an RLLR motif at 44–47, followed by an SEER motif at 63–66 (Figure 1). Sequences of the PHYCI_587572 gene and Avh87 were used to blast against the genome sequences and predicted ORFs of P. asiatica, P. cactorum, P. cambivora, P. capsici, P. hibernalis, P. infestans, P. parvispora, P. ramorum, P. sojae, P. syringae, and P. vignae, respectively. No homologs of PHYCI_587572 gene or Avh87 were found within these non-target species.

Figure 1.
Amino acid sequences of avirulence homolog protein 87 (Avh87) derived from the PHYCI_587572 gene open reading frame of Phytophthora cinnamomi. Boxed residues 1–20: predicted signal peptide; 44–47: an RxLR (RLLR) motif; 63–66: a dEER (SEER) motif.
3.2. Suppression of Programmed Cell Death by Avh87
Avh87 encoded by the PHYCI_587572 gene consistently suppressed BAX- and INF1-mediated cell death among three replications between two repeats of the A. tumefaciens infiltration experiment. BAX (pGR107:Bax) could triggered apoptosis of N. benthamiana leaves when simultaneously infiltrated along with Avh87 (Figure 2A). However, infiltration leaves with pGR107:PHYCI_587572 12 or 24 h prior to that with pGR107:Bax inhibited the BAX-mediated cell death (Figure 2A). The inhibition of Avh87 appeared as more effective against INF1. As shown in Figure 2B, INF1-mediated cell death was suppressed when pGR107:Inf1 was infiltrated promptly following that with pGR107:PHYCI_587572. The eGFP protein in place of BAX or INF1 expressed as negative controls consistently induced minimum cell death (Figure 2) in both sets of experiments.
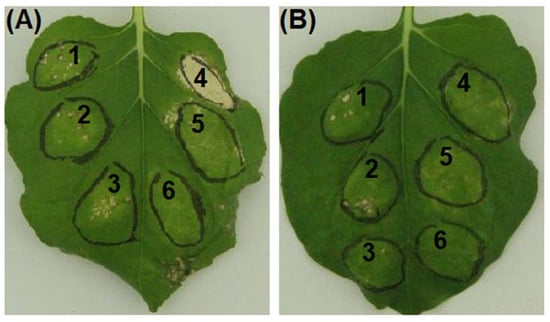
Figure 2.
Suppression of BAX- and elicitin protein INF1-mediated cell death of Nicotiana benthamiana leaves infiltrated with Agrobacterium tumefaciens strains containing a PVX vector carry the PHYCI_587572 gene (Avh87) or a control gene (eGFP). (A): 1, Avh87→eGFP; 2, Avh87→12h→eGFP; 3, Avh87→24h→eGFP; 4, Avh87→BAX; 5, Avh87→12h→BAX; 6, Avh87→24h→BAX. (B): 1, Avh87→eGFP; 2, Avh87→12h→eGFP; 3, Avh87→24h→eGFP; 4, Avh87→INF1; 5, Avh87→12h→INF1; 6, Avh87→24h→INF1.
3.3. Specificity and Sensitivity of the RPA-LFD Assay
In the evaluation of specificity, identical results were obtained among three repeats of all isolates. All LFDs had a visible control line, indicating valid tests. Test lines were visible only on LFDs with genomic DNAs (gDNAs) of 19 P. cinnamomi isolates (Table 1). No test lines were observed on those of 22 other oomycete and 15 fungal species (Figure 3).
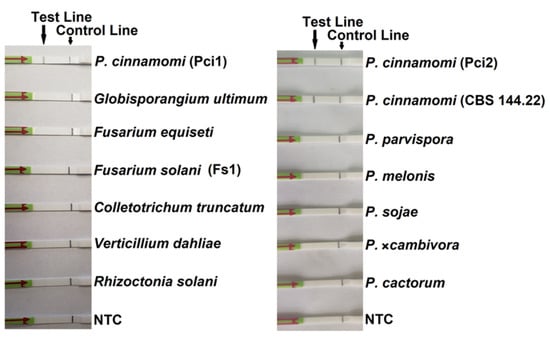
Figure 3.
Detection results of the recombinase polymerase amplification-lateral flow dipstick assay targeting the PHYCI_587572 biomarker. Genomic DNAs (10 ng per 50-µL reaction) of Phytophthora cinnamomi isolates and other oomycete and fungal species. Nuclease-free water was used in place of DNA templates in no template controls (NTC).
In the sensitivity evaluation, all LFDs had a visible control line. Solid test lines were observed on LFDs of RPA reactions containing 0.1–100 ng of P. cinnamomi gDNA (Figure 4). They were also visible on LFDs of 0.01 to 0.001 ng of gDNA (Figure 3). No test lines were observed on those of 0.0001 ng (100 fg) of gDNA or no template control (NTC). These results were consistent across three repeats of the evaluation. In the comparison PCR assay, ≥1 ng of gDNA was required for consistent positive results using primers PciRL587572F and PciRL587572F (Supplementary Figure S1). The result indicates that RPA-LFD assay was more sensitive than PCR by using LFD strips.
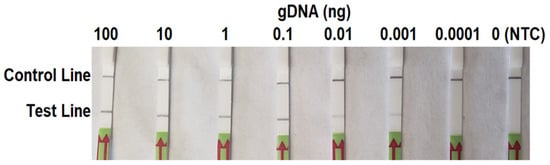
Figure 4.
Evaluation results of sensitivity of the recombinase polymerase amplification-lateral flow dipstick assay targeting the PHYCI_587572 biomarker using a 10-fold serial dilutions of genomic DNA (gDNA) of Phytophthora cinnamomi isolate Pci1. Nuclease-free water was used in place of DNA templates in no template controls (NTC).
3.4. Detecting P. cinnamomi in Artificial Inoculated Pine Needles
All LFDs had a visible control line indicating valid tests. Phytophthora cinnamomi was detected in the total DNAs extracted from three P. cinnamomi-infested pine needle samples and the positive control, whereas not detectable in three non-inoculated samples or NTC (Figure 5). Identical results were recorded in both repeats of the experiment.
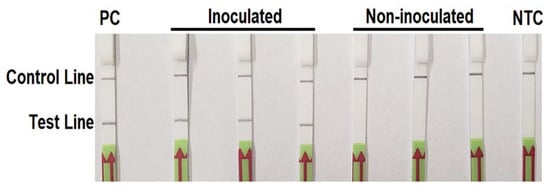
Figure 5.
Detection of Phytophthora cinnamomi in artificially inoculated pine needles of Cedrus deodara using the recombinase polymerase amplification-lateral flow dipstick assay. Genomic DNA (10 ng per µL) of P. cinnamomi isolate Pci1 was used as the positive control (PC). Nuclease-free water was used in place of DNA templates in the no template control (NTC). Dipsticks of the first repeat are shown. Identical detection results were observed in the second repeat of the experiment.
3.5. Detection of P. cinnamomi in Infested Soil Samples
The RPA-LFD assay was applied to 13 P. cinnamomi-infested soil samples. Detection results were consistent among three repeats of the experiment. Using DNA samples directly extracted from soil samples, P. cinnamomi was detected in all 13 samples using the RPA-LFD assay, while it was detected in only six samples using PCR (Table 3). Using DNAs extracted from C. deodara leaf baits, both RPA-LFD and PCR assays detected P. cinnamomi in all 13 samples.
4. Discussion
Phytophthora cinnamomi is one of the most invasive plant pathogens with a global impact [3,6]. Early and accurate detection of P. cinnamomi is paramount for implementing disease management promptly and avoiding unnecessary costs due to misdiagnosis. The present study describes PHYCI_587572, a potentially P. cinnamomi-specific RxLR effector gene identified using a comparative genomics approach, and a rapid and equipment-free RPA-LFD detection assay for P. cinnamomi that targets this novel biomarker. The findings in this study have important implications on the P. cinnamomi-hosts interactions and the management of this aggressive pathogen.
Despite the potential importance of RxLR proteins in the infection of plants by P. cinnamomi, to date, the molecular function of only a few RxLR effectors and genes has been determined [3]. Identification of oomycete pathogenicity effector genes has been facilitated by the increasing availability of genome sequences and the recognition of the RxLR-dEER motif. In this study, the PHYCI_587572 effector gene encoding Avh87 was identified from the genome sequence of P. cinnamomi. During this process, we also identified 265 other RxLR candidate genes. Efforts to identify their molecular functions are underway. Additionally, a comparative analysis of 12 Phytophthora genomes indicated the specificity of PHYCI_587572 to P. cinnamomi genome. Furthermore, the inhibition of Avh87 on BAX- and INF1-mediated cell death indicated the potentially fundamental role of Avh87 in the pathogenicity of P. cinnamomi. Further investigations are warranted on the biological function of PHYCI_587572 (Avh87) on the natural hosts of P. cinnamomi.
Given the important role of effector genes in oomycete pathogenicity, they are unique candidates for developing novel biomarkers. The RxLR effector gene PHYCI_587572 identified in this study was found lack of homolog in other reference Phytophthora species. This implies its unique function in the pathogenicity of P. cinnamomi. However, because many effector genes have been found rapidly evolving [21,48], thus have shown a higher degree of intra-specific polymorphism than universal housekeeping genes [49], utilizing a novel effector gene as a diagnostic biomarker may bear the risk of false-negative detections against mutated isolates of the target Phytophthora species. In this study, the novel RPA-LFD assay targeting the RxLR effector gene PHYCI_587572 accurately detected all 19 P. cinnamomi isolates recovered from at least eight host species in three provinces of China, two U.S. states, Puerto Rico, and Indonesia, including the ex-type isolate CBS 144.22 (Table 1). This finding indicates that the novel RPA-LFD assay has a high degree of inclusivity within P. cinnamomi isolates. Nevertheless, additional isolates should be tested in the future to confirm its inclusivity.
Rapid pathogen detection is extremely important to the management of aggressive diseases. The key advantage of RPA-based assays is their short reaction time span and requirement for less complex instrumentation. The RPA-LFD assay in this study could be finished within 30 min, which is only half of the time taken by a LAMP assay [30] and a quarter of PCR reactions. Also, it does not require a thermal cycler or gel electrophoresis and imaging system, as its results could be visualized on LFDs. Compared to the LAMP assay for P. cinnamomi [30], the RPA-LFD assay also needed fewer primers and lower incubation temperature. All these advantages make the RPA-LFD assay an ideal method for detecting P. cinnamomi under field conditions and in diagnostic laboratories.
The development of this novel RPA-LFD assay has further improved the sensitivity of isothermal detection assays for P. cinnamomi. As determined in this study, this RPA-LFD assay detected 0.001 ng of P. cinnamomi gDNA (~12 copies of nuclei) in a 50-µL reaction system. This is at least 200 times more sensitive than our previously developed LAMP assay and 2000 times more sensitive than a PCR assay using primers F3 and B3 [30]. However, this sensitivity appears lower than a previously reported qPCR-based assay [29]. Although the RPA-LFD assay has the advantages of short time span and less instrumentation requirement against qPCR, future studies are warranted to further improve the sensitivity of isothermal assays for P. cinnamomi.
5. Conclusions
A novel RxLR effector PHYCI_587572 was bioinformatically identified from the genome sequence of P. cinnamomi. A comparative analysis of genome sequences of 12 Phytophthora species indicated that this effector gene is unique to P. cinnamomi, thus an ideal biomarker for developing diagnostic assays. This assay was specific to P. cinnamomi, while it lacked detection against 37 other oomycete and fungal species, including P. parvispora, a sister taxon of P. cinnamomi. It was also found to be 200 times more sensitive than a previously developed LAMP assay. In addition, it detected P. cinnamomi in artificially inoculated leaves of Cedrus deodara. Results in this study indicated that the RPA-LFD assay is an ideal method for detecting P. cinnamomi under field conditions and in diagnostic laboratories, while understanding the precise molecular and biological functions of PHYCI_587572 and its avirulence homolog protein Avh87 requires further investigation.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4907/11/3/306/s1.
Author Contributions
T.D., A.W., X.Y., X.Y., and W.T. conceived and designed the experiments. T.D. and A.W. contributed the reagents and materials. T.D., A.W., X.Y., Y.X., and T.H. performed the experiments. T.D., A.W., X.Y., and W.T. analyzed the data. T.D., A.W., and X.Y. contributed the analysis tools and wrote the first draft of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Natural Science Foundation of Jiangsu Province, China (BK 20191389), Jiangsu University Natural Science Research Surface Project (19KJB220003), National Natural Science Foundation of China (31500526), China Postdoctoral Science Foundation (2016T790467), Overseas Research and Study Project of Excellent Young and Middle-aged Teachers and Principals in Colleges and Universities of Jiangsu Province of 2018, and the Priority Academic Program Development of Jiangsu Higher Education Institutions(PAPD).
Acknowledgments
The authors would like to thank Brett Tyler at Oregon State University (Corvallis, OR, USA) and Jin Huo Peng at Dalian Animal and Plant Quarantine Bureau (Dalian, China) for providing isolates, DNAs, and sequences of Phytophthora species used in this study. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cahill, D.M.; Rookes, J.E.; Wilson, B.A.; Gibson, L.; McDougall, K.L. Phytophthora cinnamomi and Australia’s biodiversity: Impacts, predictions and progress towards control. Aust. J. Bot. 2008, 56, 279–310. [Google Scholar] [CrossRef]
- Jung, T.; Colquhoun, I.J.; Hardy, G.E.S. New insights into the survival strategy of the invasive soilborne pathogen Phytophthora cinnamomi in different natural ecosystems in Western Australia. For. Pathol. 2013, 43, 266–288. [Google Scholar] [CrossRef]
- Hardham, A.R.; Blackman, L.M. Phytophthora cinnamomi. Mol. Plant Pathol. 2018, 19, 260–285. [Google Scholar] [CrossRef] [PubMed]
- Coffey, M.D. Phytophthora root rot of avocado. In Plant Diseases of International Importance. Volume III. Diseases of Fruit Crops; Kumar, J., Chaub, H.S., Singh, U.S., Mukhopadhyay, A.N., Eds.; Prentice Hall: Englewood Cliffs, NJ, USA, 1992. [Google Scholar]
- Erwin, D.C.; Ribeiro, O.K. Phytophthora Diseases Worldwide; APS Press: St. Paul, MN, USA, 1996. [Google Scholar]
- Zentmyer, G.A. Phytophthora cinnamomi and the Diseases It Causes; American Phytopathological Society: St. Paul, MN, USA, 1980. [Google Scholar]
- Freinkel, S. American Chestnut: The Life, Death, and Rebirth of a Perfect Tree; University of California Press: Berkeley, CA, USA, 2007. [Google Scholar]
- Shearer, B.L.; Crane, C.E.; Cochrane, A. Quantification of the susceptibility of the native flora of the South-West Botanical Province, Western Australia, to Phytophthora cinnamomi. Aust. J. Bot. 2004, 52, 435–443. [Google Scholar] [CrossRef]
- Anderson, P.; Brundrett, M.; Grierson, P.; Robinson, R. Impact of severe forest dieback caused by Phytophthora cinnamomi on macrofungal diversity in the northern jarrah forest of Western Australia. For. Ecol. Manag. 2010, 259, 1033–1040. [Google Scholar] [CrossRef]
- Davis, R.A.; Valentine, L.E.; Craig, M.D.; Wilson, B.; Bancroft, W.J.; Mallie, M. Impact of Phytophthora-dieback on birds in Banksia woodlands in south west Western Australia. Biol. Conserv. 2014, 171, 136–144. [Google Scholar] [CrossRef]
- Lan, C.Z.; Ruan, H.C.; Yao, J.A. First report of Phytophthora cinnamomi causing root and stem rot of blueberry (Vaccinium corymbosum) in China. Plant Dis. 2016, 100, 2537. [Google Scholar] [CrossRef]
- Lan, C.Z.; Ruan, H.C.; Yao, J.A. First report of Phytophthora cinnamomi causing root rot of Castanea mollissima (Chinese chestnut) in China. Plant Dis. 2016, 100, 1248. [Google Scholar] [CrossRef]
- Zheng, X.B.; Lu, J.Y. Studies on Phytophthora species in Fujian, Zhejiang, Jiangsu Provinces and Shanghai, China. Acta Mycol. Sin. 1989, 8, 161–168. [Google Scholar]
- Zeng, H.C.; Ho, H.H.; Zheng, F.C. A survey of Phytophthora species on Hainan Island of South China. J. Phytopathol. 2009, 157, 33–39. [Google Scholar] [CrossRef]
- Bi, X.Q.; Hieno, A.; Otsubo, K.; Kageyama, K.; Liu, G.; Li, M.Z. A multiplex PCR assay for three pathogenic Phytophthora species related to kiwifruit diseases in China. J. Gen. Plant Pathol. 2019, 85, 12–22. [Google Scholar] [CrossRef]
- Zhou, X.G.; Zhu, Z.Y.; Lu, C.P.; Wang, S.J.; Ko, W.H. Phytophthora cinnamomi in Shanghai and it possible origin. Mycopathologia 1992, 120, 29–32. [Google Scholar] [CrossRef]
- Wang, W.J.; Jiao, F.C. Effectors of Phytophthora pathogens are powerful weapons for manipulating host immunity. Planta 2019, 250, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Duclos, J.; Fauconnier, A.; Coelho, A.C.; Bollen, A.; Cravador, A.; Godfroid, E. Identification of an elicitin gene cluster in Phytophthora cinnamomi. DNA Seq. 1998, 9, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Dou, D.L.; Kale, S.D.; Wang, X.; Jiang, R.H.Y.; Bruce, N.A.; Arredondo, F.D.; Zhang, X.M.; Tyler, B.M. RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell 2008, 20, 1930–1947. [Google Scholar] [CrossRef]
- Wawra, S.; Belmonte, R.; Lobach, L.; Saraiva, M.; Willems, A.; van West, P. Secretion, delivery and function of oomycete effector proteins. Curr. Opin. Microbiol. 2012, 15, 685–691. [Google Scholar] [CrossRef]
- Jiang, R.H.Y.; Tripathy, S.; Govers, F.; Tyler, B.M. RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. USA 2008, 105, 4874–4879. [Google Scholar] [CrossRef]
- Kong, P.; Hong, C.X.; Richardson, P.A. Rapid detection of Phytophthora cinnamomi using PCR with primers derived from the Lpv putative storage protein genes. Plant Pathol. 2003, 52, 681–693. [Google Scholar] [CrossRef]
- Schena, L.; Duncan, J.M.; Cooke, D.E.L. Development and application of a PCR-based ‘molecular tool box’ for the identification of Phytophthora species damaging forests and natural ecosystems. Plant Pathol. 2008, 57, 64–75. [Google Scholar] [CrossRef]
- Trzewik, A.; Nowak, K.J.; Orlikowska, T. A simple method for extracting DNA from rhododendron plants infected with Phytophthora spp. for use in PCR. J. Plant Prot. Res. 2016, 56, 104–109. [Google Scholar] [CrossRef]
- Williams, N.; Hardy, G.E.S.J.; O’Brien, P.A. Analysis of the distribution of Phytophthora cinnamomi in soil at a disease site in Western Australia using nested PCR. For. Pathol. 2009, 39, 95–109. [Google Scholar] [CrossRef]
- Langrell, S.R.; Morel, O.; Robin, C. Touchdown nested multiplex PCR detection of Phytophthora cinnamomi and P. cambivora from French and English chestnut grove soils. Fungal Biol. 2011, 115, 672–682. [Google Scholar] [CrossRef] [PubMed]
- Engelbrecht, J.; Duong, T.A.; van den Berg, N. Development of a nested quantitative real-time PCR for detecting Phytophthora cinnamomi in Persea americana rootstocks. Plant Dis. 2013, 97, 1012–1017. [Google Scholar] [CrossRef] [PubMed]
- Miles, T.D.; Martin, F.N.; Coffey, M.D. Development of rapid isothermal amplification assays for detection of Phytophthora spp. in plant tissue. Phytopathology 2015, 105, 265–278. [Google Scholar] [CrossRef]
- Kunadiya, M.B.; Dunstan, W.D.; White, D.; Hardy, G.E.S.J.; Grigg, A.H.; Burgess, T.I. A qPCR assay for the detection of Phytophthora cinnamomi including an mRNA protocol designed to establish propagule viability in environmental samples. Plant Dis. 2019, 103, 2443–2450. [Google Scholar] [CrossRef]
- Dai, T.T.; Yang, X.; Hu, T.; Li, Z.Y.; Xu, Y.; Lu, C.C. A novel LAMP assay for the detection of Phytophthora cinnamomi utilizing a new target gene identified from genome sequences. Plant Dis. 2019, 103, 3101–3107. [Google Scholar] [CrossRef]
- Kunadiya, M.B.; White, D.; Dunstan, W.A.; Hardy, G.E.S.J.; Andjic, V.; Burgess, T.I. Pathways to false-positive diagnoses using molecular genetic detection methods; Phytophthora cinnamomi a case study. FEMS Microbiol. Lett. 2017, 364, fnx009. [Google Scholar] [CrossRef]
- Yang, X.; Tyler, B.M.; Hong, C.X. An expanded phylogeny for the genus Phytophthora. IMA Fungus 2017, 8, 355–384. [Google Scholar] [CrossRef]
- Scanu, B.; Hunter, G.C.; Linaldeddu, B.T.; Franceschini, A.; Maddau, L.; Jung, T. A taxonomic re-evaluation reveals that Phytophthora cinnamomi and P. cinnamomi var. parvispora are separate species. For. Pathol. 2014, 44, 1–20. [Google Scholar]
- Martin, F.N.; Blair, J.E.; Coffey, M.D. A combined mitochondrial and nuclear multilocus phylogeny of the genus Phytophthora. Fungal Genet. Biol. 2014, 66, 19–32. [Google Scholar] [CrossRef]
- Rojas, J.A.; Miles, T.D.; Coffey, M.D.; Martin, F.N.; Chilvers, M.I. Development and application of qPCR and RPA genus- and species-specific detection of Phytophthora sojae and P. sansomeana root rot pathogens of soybean. Plant Dis. 2017, 101, 1171–1181. [Google Scholar] [CrossRef] [PubMed]
- Dai, T.T.; Yang, X.; Hu, T.; Xu, Y.; Zheng, X.B.; Jiao, B.; Shen, D.Y. Comparative evaluation of a novel recombinase polymerase amplification-lateral flow dipstick (RPA-LFD) assay, LAMP, conventional PCR, and leaf-disc baiting methods for detection of Phytophthora sojae. Front. Microbiol. 2019, 10, 1884. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Shen, D.; Dai, T.; Lu, X.; Xu, H.; Dou, D. Rapid and equipment-free detection of Phytophthora capsici using lateral flow strip-based recombinase polymerase amplification assay. Lett. Appl. Microbiol. 2019, 69, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Dai, T.T.; Hu, T.; Yang, X.; Shen, D.Y.; Jiao, B.B.; Tian, W.; Xu, Y. A recombinase polymerase amplification-lateral flow dipstick assay for rapid detection of the quarantine citrus pathogen in China, Phytophthora hibernalis. PeerJ 2019, 7, e8083. [Google Scholar] [CrossRef]
- James, A.; Macdonald, J. Recombinase polymerase amplification: Emergence as a critical molecular technology for rapid, low-resource diagnostics. Expert Rev. Mol. Diagn. 2015, 15, 1475–1489. [Google Scholar] [CrossRef]
- Daher, R.K.; Stewart, G.; Boissinot, M.; Bergeron, M.G. Recombinase polymerase amplification for diagnostic applications. Clin. Chem. 2016, 62, 947–958. [Google Scholar] [CrossRef]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.L.; Korzeniewski, F.; et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucl. Acids Res. 2014, 42, D699–D704. [Google Scholar] [CrossRef]
- Bendtsen, J.D.; Nielsen, H.; von Heijne, G.; Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef]
- Haas, B.J.; Kamoun, S.; Zody, M.C.; Jiang, R.H.Y.; Handsaker, R.E.; Cano, L.M.; Grabherr, M.; Kodira, C.D.; Raffaele, S.; Torto-Alalibo, T.; et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 2009, 461, 393–398. [Google Scholar] [CrossRef]
- Whisson, S.C.; Boevink, P.C.; Moleleki, L.; Avrova, A.O.; Morales, J.G.; Gilroy, E.M.; Armstrong, M.R.; Grouffaud, S.; van West, P.; Chapman, S.; et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 2007, 450, 115–118. [Google Scholar] [CrossRef]
- Jones, L.; Hamilton, A.J.; Voinnet, O.; Thomas, C.L.; Maule, A.J.; Baulcombe, D.C. RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 1999, 11, 2291–2301. [Google Scholar] [PubMed]
- Hellens, R.P.; Edwards, E.A.; Leyland, N.R.; Bean, S.; Mullineaux, P.M. pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 2000, 42, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Wise, A.A.; Liu, Z.; Binns, A.N. Three methods for the introduction of foreign DNA into Agrobacterium. In Agrobacterium Protocols; Wang, K., Ed.; Humana Press: Totowa, NJ, USA, 2006; pp. 43–54. [Google Scholar]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucl. Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.M.; Qutob, D.; Tedman-Jones, J.; Kuflu, K.; Wang, Y.C.; Tyler, B.M.; Gijzen, M. The Phytophthora sojae avirulence locus Avr3c encodes a multi-copy RXLR effector with sequence polymorphisms among pathogen Strains. PLoS ONE 2009, 4, e5556. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).