Abstract
The precision and reliability of reverse transcription quantitative polymerase chain reaction (RT-qPCR) depend mainly on suitable reference genes; however, reference genes have not yet been identified for Liriodendron chinense (Hemsl.) Sarg. In this study, the expression stability of 15 candidate reference genes, ACT7, ACT97, UBQ1, eIF2, eIF3, HIS, BIG, AGD11, EFG, GAPDH, CYP, RPL25, UBC, RPB1, and TUB, was tested across multiple organs of L. chinense using four algorithms, geNorm, NormFinder, BestKeeper, and RefFinder. To understand the difference between the selected reference genes and the unsuitable candidate reference genes, the expression level of a target gene, LcPAT7, was normalized across various plant samples. ACT97 and eIF3 represented the best combination across all samples tested, while AGD11 and UBQ1 were unsuitable for normalization in this case. In the vegetative organ subset, ACT97, ACT7, and GAPDH showed the highest expression stability. For floral organs, UBC and eIF3 were the most stable reference genes. Unsuitable reference genes underestimated the expression levels of a target gene, LcPAT7. This study identified two reference genes (ACT97 and eIF3) for the precise and reliable normalization of L. chinense RT-qPCR data across various organs. Our work provides an effective framework for quantifying gene expression in L. chinense.
1. Introduction
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) has been extensively applied in gene expression analysis due to its high accuracy, high throughput, and high sensitivity [1,2,3]. To perform relative quantification via RT-qPCR, normalization is essential to eliminate experimental errors between samples [1], and the selection of appropriate internal genes has a significant influence on the relative quantification results [2]. If the expression pattern of the reference gene is unstable, then the accuracy of the assay will be reduced, and small changes in the expression of the target genes will be impossible to detect [1]. Therefore, to obtain accurate and reliable results, internal control genes must be systematically evaluated to ensure their applicability under different experimental conditions and various species [4,5].
Reference genes usually have basic functions in cells or organisms and are normally expressed at relatively constant rates under different experimental conditions and across different organs [2,6]. Many genes, such as actin, beta-tubulin, elongation factor, polyubiquitin, eukaryotic translation initiation factor, glyceraldehyde-3-phosphate dehydrogenase, histone, ribosomal protein L, cyclophilin, and 18S RNA have been used as reference genes under different experimental conditions and in different species [2,6,7,8,9,10,11,12,13,14,15].
Different statistical algorithms have been developed to assess the expression stability of reference genes under certain experimental conditions for specific species; these algorithms include geNorm, BestKeeper, NormFinder, and RefFinder. These algorithms are widely employed to evaluate the expression stability of reference genes and identify reference genes that are consistent across various species such as Populus tomentosa Carr. [16], Bixa orellana L. [3] and Betula luminifera H.J.P. Winkl. [17]. At least two algorithms were suggested to be used for reference gene stability evaluation [18].
Liriodendron chinense (Hemsl.) Sarg, a broad-leaved deciduous tree species belonging to the magnolia family (Magnoliaceae) that is also called the Chinese tulip tree [19], is naturally distributed in mountainous areas of southern China and northern Vietnam at an elevation of 450–1800 m [20,21]. However, little molecular and gene expression data have been reported for L. chinense. Additionally, reference genes have not been properly characterized and identified for L. chinense, although a few studies have utilized a limited number of genes (including actin) in L. chinense without any systematic evaluation [22]. Therefore, the selection of suitable reference genes in L. chinense is very necessary for RT-qPCR analysis.
In this study, to identify suitable reference genes for RT-qPCR analysis in L. chinense, we used 15 genes, ACT7 (Actin 7), ACT97 (Actin 97), GAPDH (Glyceraldehyde-3-phosphate dehydrogenase), TUB (Tubulin beta), HIS (Histone H3), EFG (Elongation factor G), eIF2 (Eukaryotic translation initiation factor 2), eIF3 (Eukaryotic translation initiation factor 3), BIG (Auxin transport protein BIG), AGD11 (ADP-ribosylation factor GTPase-activating protein AGD11), UBQ1(Ubiquitin extension protein 1), CYP (Cyclophilin), RPL25 (50S ribosomal protein L25), RPB1 (RNA polymerase II subunit RPB1) and UBC (Ubiquitin conjugating enzyme ATG10), as candidate genes and tested their expression stability across multiple organs of L. chinense using the geNorm [23], BestKeeper [24], NormFinder [25], and RefFinder [26] programs. In addition, to compare the differences between the selected reference genes and unstable candidate reference genes, the LcPAT7 (Liriodendron chinese protein S-acyltransferase 7) gene was chosen as a target gene, and its expression level was determined by RT-qPCR analysis. In summary, this work aimed to identify appropriate reference genes for RT-qPCR analysis across various organs in L. chinense.
2. Materials and Methods
2.1. Plant Materials
Experimental materials were collected from an adult L. chinense tree in a provenance trial plantation in Xiashu, Jurong County, Jiangsu Province (1320 E, 78 N) [27]. The sample tree originated from the Danshan Nature Reserve, Xuyong County, Sichuan Province (2913 E, 1148 N) [19]. A total of 54 samples were classified into two organ subsets, vegetative organs and flower organs. The vegetative organ subset included leaves, roots, and twigs (see Appendix A: Figure A2a,b). Leaves were harvested at four stages: stage 1 (S1) corresponded to leaf buds, stage 2 (S2) corresponded to newly expanded leaves, stage 3 (S3) corresponded to larger leaves than those in S2, and stage 4 (S4) corresponded to mature leaves (see Appendix A: Figure A2). The flower organs consisted of floral buds, sepals, petals, pistils, and stamens. Flower developmental was also classified into four stages: floral buds (stage 1, S1), inflated buds (stage 2, S2), pre-anthesis (stage 3, S3), and opened flowers (stage 4, S4) (see Appendix A: Figure A2f). Sepals, petals, and pistils were harvested from the S2, S3, and S4 flowers (the S1 flowers were too small for the sepals, petals, stamens, and pistil to be completely separated) (see Appendix A: Figure A2c–e), and stamen materials were collected from the S2 and S3 flowers (see Appendix A: Figure A2c,d). Each sample included three biological replicates. A total of 54 samples were stored at −80 in a freezer until RNA extraction.
2.2. Total RNA Extraction and cDNA Synthesis
Total RNA was extracted by using the RNAprep Pure Plant Kit (Tiangen, Beijing, China), the specific procedures can be found in the manufacturer’s instructions. The concentration and quality of the RNA samples were determined by a NanoDrop 2000 c spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and 1.0 percent (w/v) agarose gel electrophoresis. cDNA was synthesized from 1000 ng of total RNA in a 20 L reaction volume using PrimeScriptTM RT Master Mix (TaKaRa, Dalian, China) according to the manufacturer’s instructions.
2.3. Candidate Reference Genes and Primer Design
Based on previous studies of internal control genes for other plants and de novo transcriptome sequencing of L. chinense [28], 15 genes were chosen as candidates in this study. The candidate genes were ACT7, ACT97, UBQ1, eIF2, eIF3, HIS, BIG, AGD11, EFG, GAPDH, CYP, UBC, RPL25, RPB1 and TUB (Table 1). Using the Oligo 7 algorithm (https://en.freedownloadmanager.org/Windows-PC/OLIGO.html) to design RT-qPCR primers. The melting temperature (TM) of the primers was 58–65 , and their lengths were 20–25 bp. The GC contents of the primers were 40–60 percent, and the product lengths were 100–300 bp. The specificity of each amplifications was verified by 2.0 percent (w/v) agarose gel electrophoresis and dissociation curve analysis. The LinRegPCR 2014.x algorithm [29] was used to estimate the mean amplification efficiencies of the primer pairs, and the results are shown in Table 1.

Table 1.
Primer sequences for reverse transcription quantitative polymerase chain reaction (RT-qPCR).
2.4. RT-qPCR Assay
RT-qPCR was performed on a StepOnePlus System (Applied Biosystems) with a total volume of 10 L per reaction. Each 10 L reaction mixture contained 10 ng of cDNA, 10 pmol of each of the forward and reverse primers, 5 L of 2 × SYBR Premix Ex Taq and 0.2 L of 50 × ROX Reference Dye (TaKaRa, Dalian, China). The amplification program was as follows: 95 for 30 s and 40 cycles of 95 for 5 s and 60 for 34 s. After amplification, a thermal denaturing cycle was performed to obtain the dissociation curve of the amplification product to verify the primer’s amplification specificity. The dissociation stage was performed as follows: 95 for 15 s, 60 for 1 min, 95 for 15 s. All RT-qPCRs were carried out in three biological and three technical replicates.
2.5. Data Analysis
The expression stability of the 15 candidate genes was valued by four algorithms, geNorm, BestKeeper, NormFinder, and RefFinder. The cycle threshold (Ct) values were converted to relative expression quantities using the formula E (Ct = each corresponding Ct value—the minimum Ct value, E = mean amplification efficiency of the primer pairs) for geNorm and NormFinder. The geNorm algorithm uses the average expression stability value (M) to evaluate gene stability and uses the pairwise variation (V) to determine the optimal number of reference genes. The average pairwise variation of a specific candidate gene from all other candidate reference genes is called the M value, and the pairwise variation (V) is the standard deviation of the logarithmically transformed expression ratios of the combination of two candidate reference genes [23]. The NormFinder algorithm calculates the stability value of each candidate reference gene by combining intragroup and intergroup variations in gene expression [25]. For BestKeeper, the Ct values of candidate reference genes were used to calculate the standard deviation (SD) and the coefficient of variance (CV) [24]. On the basis of rankings from the Ct method, geNorm, NormFinder and BestKeeper, the RefFinder program ranks the reference genes comprehensively by calculating the geometric mean of the gene weights [30].
2.6. Comparison of Reference Genes and Unstable Reference Genes
To compare the expression stability of the selected genes to that of the unstable reference genes, the expression profile of the target gene LcPAT7 was analyzed by using the most stable and least stable reference genes after normalization in S1 leaves, S4 leaves, floral buds, S3 petals, S3 stamens, and S3 pistils. The primer pairs (forward: 5′–CTCGGTAGGCGGAGGTTTCATCA–3′ and reverse: 5′–CTATCTTGCTGCTGCTGCCACTG–3′) of LcPAT7 were used for RT-qPCR. The relative expression level was calculated by the 2 method and is shown as the fold change.
3. Results
3.1. Primer Specificity and PCR Amplification Efficiency Verification
After PCR amplification of the 15 candidate reference genes, 2.0 percent (w/v) agarose gel electrophoresis of each product showed that there was only one amplicon, and its size was consistent with the designed size. The melting curve analysis had the same results and exhibited a single peak in the template (see Appendix A: Figure A1). Furthermore, the mean amplification efficiency (E) of the primer pair ranged from 1.81 for BIG to 2.04 for CYP, and the correlation coefficient (R) of linear amplification was approximately 1 (0.99) (Table 1).
3.2. Candidate Reference Genes Ct Values
We analyzed 54 samples by RT-qPCR assays. The Ct values of the 15 candidate reference genes varied (Figure 1), ranging between 16.47 (for HIS) and 29.25 (for AGD11), while the average Ct values varied between 17.84 (for HIS) and 26.31 (for UBC). Moreover, the ACT97 expression levels were the least variable (3.52 Ct, the maximum and minimum Ct values were 23.28 and 19.76, respectively), while the RPL25 expression levels were the most variable (5.92 Ct, the maximum and minimum Ct values were 27.80 and 21.88, respectively). Because Ct values are negatively correlated with expression levels, the results of this study indicated that HIS had the highest expression levels and that UBC had the lowest expression levels.
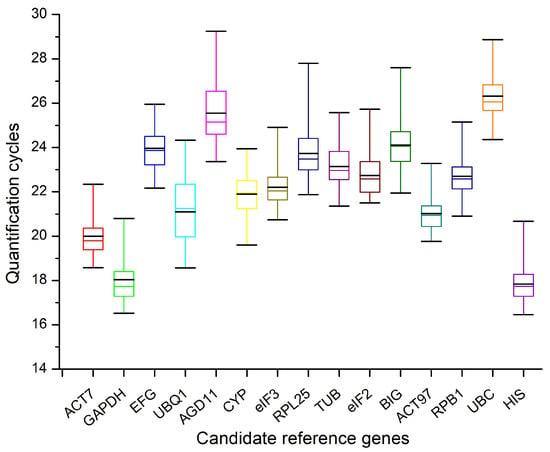
Figure 1.
Ct values of 15 candidate reference genes in 54 samples. Boxes indicate the 25th and 75th percentiles. Coloured lines across boxes represent the median Ct values and black lines across boxes indicate the mean Ct values. The maximum and minimum values are represented by whiskers.
3.3. Analysis of Gene Expression Stability
The geNorm analysis. The geNorm algorithm valuates the expression stability of the candidate reference gene by the M value. A smaller M value indicates higher expression stability, if the M value of a candidate reference gene is higher than 1.5, then the candidate reference gene is considered unstable. As measured by geNorm, ACT7 (M value 0.33) and GAPDH (M value 0.33) were the most suitable genes for normalization across all samples of L. chinense (Figure 2a). The third to fifth most suitable were eIF3 (M value 0.39), ACT97 (M value 0.40) and EFG (M value 0.44). In contrast, UBQ1 and AGD11 were the least stable reference genes. In the vegetative organ subset, the top five candidate reference genes were UBC (M value 0.35), HIS (M value 0.35), ACT97 (M value 0.55), RPB1 (M value 0.61), and BIG (M value 0.74) (Figure 2b). In the flower organ subset, HIS (M value 0.33) and eIF2 (M value 0.33) showed the most stable expression pattern, followed by UBC (M value 0.37), ACT97 (M value 0.39), and eIF3 (M value 0.41) (Figure 2c).
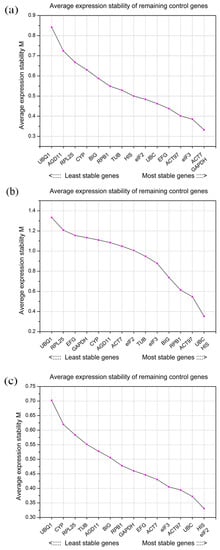
Figure 2.
Average expression stability (M) values and ranking of 15 candidate reference genes determined by geNorm. The least stable genes are on the left, and the most stable genes are on the right. (a) All tested samples; (b) vegetative organ subset; and (c) flower organ subset.
The pairwise variation (V) was used to determine the optimal number of internal control genes. If V values below 0.15, it was not required to include additional reference genes [23]. Across all samples and in the flower organ subset, the V value were less than 0.15 (0.126 and 0.119, respectively), which indicated that two reference genes were sufficient for normalization. In the vegetative organ subset, the V value was below 0.15, which suggested that three reference genes were the best choice for normalization (Figure 3).
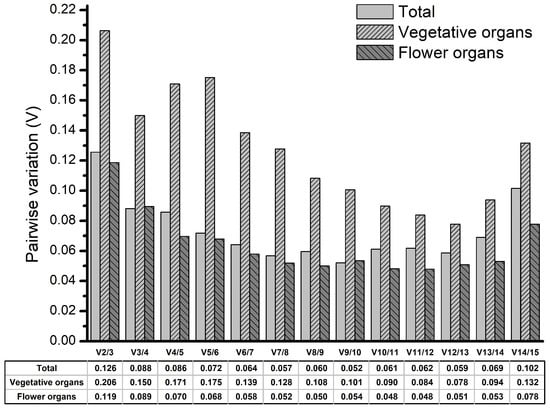
Figure 3.
Pairwise variation (V) of 15 candidate reference genes as measured by the geNorm algorithm.
NormFinder analysis. NormFinder ranks candidate reference genes based on their stability values. Stable reference gene expression has a low stability value. Among all the samples, eIF3, ACT97, UBC, ACT7 and GAPDH were the top five candidate reference genes. When the vegetative organ samples were evaluated, TUB, eIF3 and UBC were the most suitable genes for normalization, followed by ACT97 and ACT7. For the flower organ subset, eIF3, eIF2, ACT97, UBC and EFG were the top-ranked genes (Table 2). In contrast, UBQ1 and AGD11 were unsuitable for normalization across all samples, and UBQ1 was also the least stable gene evaluated within the vegetative organ samples and flower organ samples (Table 2).

Table 2.
Rankings of 15 candidate reference genes by the NormFinder algorithm.
BestKeeper analysis. BestKeeper ranks candidate reference genes according to the CV and standard deviation (SD) of the average Ct values [24]. The most stable gene exhibits the lowest CV ± SD value. If the SD value was more than 1, then the candidate reference gene was regarded as unacceptable. Across all samples, ACT97 (2.93 ± 0.62) and HIS (3.45 ± 0.62) were identified as stable genes. In contrast, UBQ1 (5.90 ± 1.25) and AGD11 (4.40 ± 1.12) were unsuitable genes. In the vegetative organ samples, UBC (2.50 ± 0.67) and ACT97 (3.17 ± 0.68) were the top-ranked genes. In the flower organ subset, UBC (1.92 ± 0.50) and eIF3 (2.28 ± 0.50) were suitable genes for normalization (Table 3).

Table 3.
Rankings of 15 candidate reference genes by the BestKeeper algorithm.
The ranks determined by geNorm had a certain similarity to the ranks from NormFinder. Among all samples, GAPDH, ACT97, ACT7, and eIF3 showed better stability in both geNorm and NormFinder (Table 4). For the vegetative organs, ACT97 ranked in the top five in geNorm and NormFinder (Table 4). For the flower organ subset, eIF3 and ACT97 had better performance in geNorm and NormFinder (Table 4). The ranks from geNorm, NormFinder, and BestKeeper also showed some similarity. In all samples, eIF3 and ACT97 were always ranked in the top five by the above three algorithms (Table 4). In addition, ACT97 and eIF3 showed better stability in the vegetative organs and flower organs, respectively (Table 4).

Table 4.
Top five candidate reference genes ranked by the four algorithms.
3.4. Reffinder Analysis and Reference Gene Selection
Because of the differences in the evaluation results of the 15 candidate reference genes by geNorm, BestKeeper and NormFinder, we also used the RefFinder program to comprehensively evaluate the ranks of the candidate reference genes. To the best of our knowledge, the use of at least two reference genes represents a realistic calculation basis in a typical laboratory and the minimal number necessary for an accurate and reliable analysis of RT-qPCR [9,23,24]. Therefore, according to the optimal number of reference genes for normalization (Figure 3) and the results of the RefFinder analysis (Table 5), we selected eIF3 and ACT97 as the most suitable reference genes for RT-qPCR analysis across all samples. ACT97, ACT7 and GAPDH were collectively the best choice for normalization in the vegetative organ subset (Table 5). For the flower organ samples, eIF3 and UBC were the most stable reference genes (Table 5). In addition, AGD11 and UBQ1 were the least stable reference genes across all samples (Table 5).

Table 5.
The 15 candidate reference genes ranked by the RefFinder program.
We found that eIF3 and ACT97 ranked among the top four in the geNorm, NormFinder and BestKeeper algorithms for all samples (Table 4). For the vegetative organ subset, ACT97 showed better stability in the same three algorithms (Table 4). In the flower organs, eIF3 and UBC had better performance in the same three algorithms (Table 4). These results were similar to those of RefFinder.
3.5. Comparison of Reference Genes and Unstable Candidate Reference Genes
The expression level of LcPAT7 among the different samples was evaluated to compare the differences between the selected reference genes and unstable candidate reference genes. The results are shown in Figure 4. When the combination of ACT97 and eIF3 was used for normalization, the expression levels of LcPAT7 were upregulated in the floral buds, S3 petals, S3 stamens, and S3 pistils (Figure 4) but downregulated in S4 leaves. When the combination of AGD11 and UBQ1 was used as an internal control, LcPAT7 expression was underestimated (Figure 4). There were significant differences between the cases where the selected reference genes and the unstable candidate reference genes were used as internal controls for normalization in different samples of L. chinense. This result means that AGD11 and UBQ1 should be avoided when selecting internal controls for RT-qPCR in L. chinense.
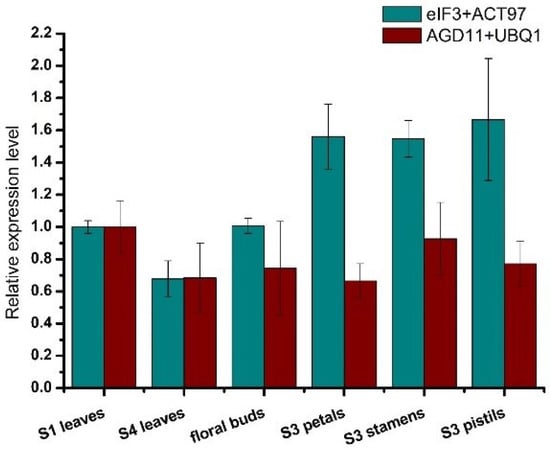
Figure 4.
Relative expression levels of LcPAT7. According to the results of analyses of 15 candidate reference genes using four programs, eIF3 and ACT97 were the selected reference genes, and AGD11 and UBQ1 were unstable candidate reference genes. Both sets were as internal control in six organs.
4. Discussion
As a high throughput and accurate method of gene expression level analysis, RT-qPCR has been extensively applied in studies of functional genes across various samples and under different experimental conditions [2,3]. However, the accuracy of RT-qPCR analyses relies on the selection of suitable genes as internal control, because if an unstable gene is chosen as the internal control, the analysis yields unreliable results [31,32]. However, no report exist of systematically selecting suitable reference genes for RT-qPCR analysis in L. chinense. Therefore, suitable internal control genes for RT-qPCR analysis must be carefully identified in L. chinense. To the best of our knowledge, this study is the first to systematically and comprehensively select internal control genes for RT-qPCR analysis in L. chinense.
Fifteen candidate reference genes were selected for this study, and most of them had been studied previously in different plants such Oryza sativa L. [9], Pinus massoniana L. [11], citrus [33], Pisum sativum [34], Petunia hybrida [35], Linum usitatissimum L. [36], Nicotiana tabacum [37], Camellia sinensis [38], and Populus [39]. In terms of standardization and quality, amplification efficiencies of the candidate reference gene’s primer pairs varied from 1.81 to 2.04 (Table 1). The correlation coefficient (R) of linear amplification was approximately 1 (0.99) (Table 1). These results revealed that the primer pairs for the candidate reference genes were highly accurate, efficient, and sensitive.
Under any experimental conditions, an appropriate reference gene should exhibit a steady expression pattern across different samples [38]. However, these 15 candidate reference genes had variable expression in all samples (Figure 1). The variation (Ct− Ct) in the 15 candidate reference genes was between 3.52 Ct (for ACT97) and 5.92 Ct (for RPL25). This result strongly indicated that ACT97 and RPL25 were the most stably and the least stably expressed genes, respectively. The mean Ct values of candidate reference genes varied from 17.84 (for HIS) to 26.31 (for AGD11). In a study of the Vigna unguiculata L. [40], Setaria virdis [41], Euscaphis konishii Hayata [42] and Solanum tuberosum L. [43], candidate reference genes also had variable expression among tested materials, and the average Ct values of the candidate reference genes were between 15 and 27. These results suggested that it is necessary to evaluate the stability of reference genes systemically before selecting them as internal controls for RT-qPCR analysis [40].
Four methods (geNorm, NormFinder, BestKeeper and RefFinder) were used to evaluate the expression stability of the 15 candidate reference genes in our study. Among all samples, eIF3 and ACT97 ranked third and fourth in geNorm, first and second in NormFinder, and fourth and first in BestKeeper (Table 4). The different ranks of the candidate reference genes between geNorm and NormFinder and BestKeeper was expected because these three methods use different algorithms [23,24,25]. In addition, recent studies have noted that there is no universal internal control gene applicable to all experimental conditions and that using a single reference gene as internal control could introduce errors in the results [9,23,25,44]. According to the analysis results of RefFinder and the optimal number of reference genes given by geNorm analysis, we selected eIF3 and ACT97 as the most suitable reference genes for RT-qPCR analysis in L. chinense (Figure 3, Table 5).
In general, eIF3 and ACT97 showed the highest expression stability among all samples of L. chinense (Table 4 and Table 5). eIF (Eukaryotic translation initiation factor) and ACT (actin) also showed good expression stability in different plants across different sample sets. For example, eIF was chosen as the internal control gene for RT-qPCR analysis under drought stress in Lolium multiflorum [45], and it also showed the most stable expression pattern in Linum usitatissimum L. stems [36]. ACT was the most suitable internal control for RT-qPCR analysis in Pinus massoniana L. [11], and it was still the most stably expressed reference gene in Daucus carota L. leaves [7], stem segments of Populus tomentosa [16], and Glycine max L. Merr. [44]. In addition, in a study of Gossypium hirsutum, ACT presented a more stable expression pattern during flower development than other candidate reference genes [31]. However, in a study of Euscaphis konishii Hayata [42] and Linum usitatissimum L. [36], ACT was unsuitable for normalization. In contrast, UBQ1 and AGD11 were unstably expressed genes in L. chinense (Table 5). As a member of the UBQ family, UBQ1 did not have good expression stability in our study, and it also showed an unstable expression pattern in lettuce (Lactuca sativa) [8] and in leaves of Ilex paraguariensis under drought treatment [46]. However, UBQ1 had better expression stability in the parasitic life cycle of Striga hermonthica [10]. These results can be clarified by the fact that reference genes exhibited species-specific expression.
Generally, reference genes exhibit organ-specific expression as well as species-specific expression [12]. We classified 54 samples into two subsets: flower organs and vegetative organs. In the flower organ subset, eIF3 and UBC were the best choice for normalization (Table 5). Consistent with this result, UBC had the highest expression stability among different organs of the same development stage in Glycine max [L.] Merr. [44], and UBC was identified as a suitable reference gene across all Brachypodium distachyon samples [5]. In the vegetative organs, ACT97, ACT7, and GAPDH were the best combination for RT-qPCR analysis (Figure 3, Table 5). Similarly, ACT7 had the highest expression stability under heat treatment in Stipa grandis [47]. However, compared to ACT in a previous study in tea plants, ACT7 was regarded as an unsuitable internal control [40]. GAPDH is commonly used as a reference gene and has been chosen as an internal control gene in the cotyledons of Cunninghamia lanceolata [12], different samples of Vigna unguiculata L. [40], cold-treated and GA-treated leaves of Daucus carota L. [7], and cold-treated and salt-treated leaves of Carex rigescens [48]. However, GAPDH was unsuitable for normalization in Pisum sativum and Petunia hybrida [34,35].
In addition, RPL25, CYP, and TUB did not demonstrate high expression stability in L. chinense (Table 5). However, RPL25 demonstrated the highest expression stability across stress-treated samples of Nicotiana tabacum [37]. CYP was identified as the most stable internal control gene in Petunia hybrida [35], insect-resistant leaves of Pinus massoniana L. [11], and GA-treated samples of Stellera chamaejasme [2]. In a study of Pisum sativum, TUB was suitable for normalization under abiotic stress and biotic stress [34].
LcPAT7 in L. chinense is a member of the Asp–His–His–Cys (DHHC) protein family, and it has a significant influence on development and morphogenesis [49]. To compare the differences between the selected reference genes and unsuitable candidate reference gene, the expression profile of LcPAT7 was investigated in different organs. The results showed that using unsuitable candidate reference genes as internal controls resulted in underestimates of expression(Figure 4), which indicated that suitable reference genes must be selected for RT-qPCR analysis in L. chinense.
5. Conclusions
In this work, we identified specific reference genes for RT-qPCR analysis of different samples in L. chinense. The most suitable combination for RT-qPCR analysis across all samples was eIF3 and ACT97. Notably, AGD11 and UBQ1 should be eliminated when selecting suitable internal control genes for RT-qPCR analysis in all samples of L. chinense. In the vegetative organs, ACT7, ACT97, and GAPDH were the most stably expressed reference genes. eIF3 and UBC were the most suitable reference genes for normalization in flower organs. In conclusion, this study provides an effective framework for quantifying reference gene expression levels in L. chinense.
Author Contributions
Experimental design: H.L. and Z.T.; plant materials collection and performing the experiments: Z.T. and W.Z.; data analysis: Z.T. and Z.H.; manuscript writing: Z.T. and H.L.; all authors approved the final draft.
Funding
This study was financially funded by the National Natural Science Foundation of China (31770718, 31470660) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Acknowledgments
We wish to thank Meng Xu (Nanjing Forestry University, Nanjing, China) for the suggestions about data analysis and writing, Jikai Ma (Nanjing Forestry University, Nanjing, China) for providing the transcriptome data, and Sian Liu (Nanjing Forestry University, Nanjing, China) for suggestions about selecting candidate reference genes. The authors would also like to thank colleagues of the laboratory, Mujun Liu, Shenghua Zhu, and Jiayu Li for RNA extraction and cDNA synthesis, and for their contributions to discussions regarding RT-qPCR experiments.
Conflicts of Interest
The authors declare that they have no competing interests.
Abbreviations
The following abbreviations are used in this manuscript:
| ACT7 | actin 7 |
| ACT97 | actin 97 |
| UBQ1 | ubiquitin extension protein1 |
| eIF2 | eukaryotic translation initiation factor 2 |
| eIF3 | eukaryotic translation initiation factor 3 |
| HIS | histone H3 |
| BIG | auxin transport protein BIG |
| TUB | tubulin beta |
| AGD11 | ADP-ribosylation factor activating protein AGD11 |
| EFG | elongation factor G |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| CYP | cyclophilin |
| RPL25 | 50S ribosomal protein L25 |
| UBC | ubiquitin conjugating enzyme ATG10 |
| RPB1 | RNA polymerase II subunit RPB1 |
Appendix A
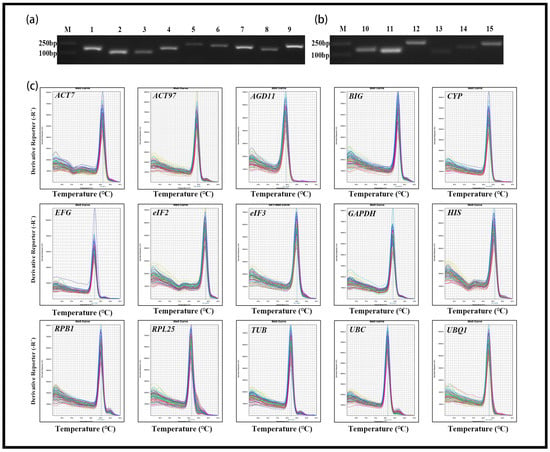
Figure A1.
Specificity of each candidate reference gene primer pair. (a,b) 2% agarose gel showing that all primer pairs have specificity and the size of the amplification products are consistent with the expected sizes. M, marker; 1, GAPDH; 2, ACT7; 3, UBQ1; 4, ACT97; 5, eIF2; 6, eIF3; 7, TUB; 8, EFG; 9, RBP1; 10, CYP; 11, HIS; 12, PRL25; 13, AGD11; 14, UBC; 15, BIG. (c) Melt curve analysis of the 15 candidate reference genes tested across different samples of L. chinense showed a single peak for each primer pair.

Figure A2.
Plant materials used in this experiment. (a) The leaf stages are leaf bud (stage 1, S1); young, newly expanded leaf (stage 2, S2); larger leaf than that in S2 (stage 3, S3); and mature leaf (stage 3, S4), ranging from left to right. (b) Twigs (left) and roots (right). (c) Materials from S2 flower. From top to bottom, materials are sepals, petals, stamens, and pistil. (d) Materials from S3 flower. From top to bottom, materials are sepals, petals, stamens, and pistil. (e) Materials from the S4 flower. From top to bottom, materials are sepals, petals, stamens, and pistil. (f) Flowers at four different developmental stages. From left to right, flowers are a floral bud (stage 1, S1), an inflated bud (stage 2, S2), pre-anthesis (stage 3, S3), and an opened flower (stage 4, S4).
References
- Dheda, K.; Huggett, J.F.; Chang, J.S.; Kim, L.U.; Bustin, S.A.; Johnson, M.A.; Rook, G.A.; Zumla, A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 2005, 344, 141–143. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Guan, H.; Song, M.; Fu, Y.; Han, X.; Lei, M.; Ren, J.; Guo, B.; He, W.; Wei, Y. Reference gene selection for qRT-PCR assays in Stellera chamaejasme subjected to abiotic stresses and hormone treatments based on transcriptome datasets. PeerJ 2018. [Google Scholar] [CrossRef] [PubMed]
- Moreira, V.S.; Soares, V.L.F.; Silva, R.J.S.; Sousa, A.O.; Otoni, W.C.; Costa, M.G.C. Selection and validation of reference genes for quantitative gene expression analyses in various tissues and seeds at different developmental stages in Bixa orellana L. Physiol. Mol. Biol. Plants 2018, 24, 369–378. [Google Scholar] [CrossRef]
- Lee, P.D.; Sladek, R.; Greenwood, C.M.T.; Hudson, T.J. Control genes and variability: Absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genome Res. 2002, 12, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.-Y.; Seo, P.J.; Yang, M.-S.; Xiang, F.; Park, C.-M. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 2008, 8, 112. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, H.; Liu, L.; Li, W.; Wei, Y.; Shi, S. Validation of Reference Genes for RT-qPCR Studies of Gene Expression in Preharvest and Postharvest Longan Fruits under Different Experimental Conditions. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Jiang, Q.; Wang, F.; Wang, G.L.; Xu, Z.S.; Xiong, A.S. Selection of suitable reference genes for qPCR normalization under abiotic stresses and hormone stimuli in carrot leaves. PLoS ONE 2015, 10, e0117569. [Google Scholar] [CrossRef] [PubMed]
- Sgamma, T.; Pape, J.; Massiah, A.; Jackson, S. Selection of reference genes for diurnal and developmental time-course real-time PCR expression analyses in lettuce. Plant Methods 2016, 12. [Google Scholar] [CrossRef]
- Jain, N.; Vergish, S.; Khurana, J.P. Validation of house-keeping genes for normalization of gene expression data during diurnal/circadian studies in rice by RT-qPCR. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Ferna’ndez-Aparicio, M.; Wickett, N.J.; Timko, M.P.; Huang, K.; Wafula, E.K.; dePamphilis, C.W.; Honaas, L.A.; Yoder, J.I.; Westwood, J.H. Application of qRT-PCR and RNA-seq analysis for the identification of housekeeping genes useful for normalization of gene expression values during Striga hermonthica development. Mol. Biol. Rep. 2013, 40, 3395–3407. [Google Scholar] [CrossRef]
- Chen, H.; Yang, Z.; Hu, Y.; Tan, J.; Jia, J.; Xu, H.; Chen, X. Reference genes selection for quantitative gene expression studies in Pinus massoniana L. Trees 2015, 30, 685–696. [Google Scholar] [CrossRef]
- Bao, W.; Qu, Y.; Shan, X.; Wan, Y. Screening and Validation of Housekeeping Genes of the Root and Cotyledon of Cunninghamia lanceolata under Abiotic Stresses by Using Quantitative Real-Time PCR. Int. J. Mol. Sci. 2016, 17, 1198. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Chen, C.; Cai, H.; Wu, L. Overexpression of PeHKT1;1 Improves Salt Tolerance in Populus. Genes 2018, 9, 475. [Google Scholar] [CrossRef]
- Liu, S.; Sun, Z.; Xu, M. Identification and characterization of long non-coding RNAs involved in the formation and development of poplar adventitious roots. Ind. Crops Prod. 2018, 118, 334–346. [Google Scholar] [CrossRef]
- Chen, C.; Zheng, Y.; Zhong, Y.; Wu, Y.; Li, Z.; Xu, L.; Xu, M. Transcriptome analysis and identification of genes related to terpenoid biosynthesis in Cinnamomum camphora. BMC Genom. 2016, 11, e0157370. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, Y.; Ding, L.; Zhang, J.; Wei, J.; Wang, H. Validation of Reference Genes for Gene Expression by Quantitative Real-Time RT-PCR in Stem Segments Spanning Primary to Secondary Growth in Populus tomentosa. PLoS ONE 2016, 11, e0157370. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, J.; Pan, Y.; Huang, H.; Lou, X.; Tong, Z. Identification and evaluation of reference genes for normalization in quantitative real-time PCR analysis in the premodel tree Betula luminifera. J. For. Res. 2016, 28, 273–282. [Google Scholar] [CrossRef]
- Jacob, F.; Guertler, R.; Naim, S.; Nixdorf, S.; Heinzelmann-Schwarz, V. Careful Selection of Reference Genes is Required for Reliable Performance of RT-qPCR in Human Normal and Cancer Cell Lines. PLoS ONE 2013, 8, e59180. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, M.; Luo, Q.; Wang, J.; Li, H. De novo transcriptome analysis of Liriodendron chinense petals and leaves by illumina sequencing. Gene 2014, 534, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, H.; Xie, T.; Xu, Y.; Zhao, L.; Tian, W. Effects of Climate Change on the Potentially Suitable Climatic Geographical Range of Liriodendron chinense. Forests 2017, 8, 399. [Google Scholar] [CrossRef]
- Hao, R.; He, S.; Tang, S.; Wu, S. Geographical distribution of Liriodendron chinense in China and its significance. J. Plant Resour. Environ. 1995, 4, 1–6, (Chinese with English Abstract). [Google Scholar]
- Xu, J.; Li, G. Cloning and primary functional analysis of LcPAT8 gene from Liriodendron chinense. Sci. Silvae Sin. 2017, 53, 10, (Chinese with English Abstract). [Google Scholar]
- Vandesompele, J.; Preter, K.D.; Pattyn, F.; Poppe, B.; Roy, N.V.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal. Genome Biol. 2002, 3. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: Bestkeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Chen, L.; Feng, Y.; Yao, J.; Li, B.; Xu, M.; Li, H. High genetic diversity but limited gene flow among remnant and fragmented natural populations of Liriodendron chinense Sarg. Biochem. Syst. Ecol. 2014, 54, 230–236. [Google Scholar] [CrossRef]
- Ma, J.; Wei, L.; Li, J.; Li, H. The Analysis of Genes and Phytohormone Metabolic Pathways Associated with Leaf Shape Development in Liriodendron chinense via De Novo Transcriptome Sequencing. Genes 2018, 9, 577. [Google Scholar] [CrossRef] [PubMed]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.; Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef]
- Zhao, J.; Yang, F.; Feng, J.; Wang, Y.; Lachenbruch, B.; Wang, J.; Wan, X. Genome-Wide Constitutively Expressed Gene Analysis and New Reference Gene Selection Based on Transcriptome Data: A Case Study from Poplar/Canker Disease Interaction. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Artico, S.; Nardeli, S.M.; Brilhante, O.; Alves-Ferreira, M. Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol. 2010, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.; Mauriat, M.; Guenin, S.; Pelloux, J.; Lefebvre, J.F.; Louvet, R.; Rusterucci, C.; Moritz, T.; Guerineau, F.; Bellini, C.; et al. The lack of a systematic validation of reference genes: A serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 2008, 6, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Yuan, F.; Long, G.; Qin, L.; Deng, Z. Selection of reference genes for quantitative real-time RT-PCR analysis in citrus. Mol. Biol. Rep. 2012, 39, 1831–1838. [Google Scholar] [CrossRef] [PubMed]
- Die, J.V.; Roman, B.; Nadal, S.; Gonzalez-Verdejo, C.I. Evaluation of candidate reference genes for expression studies in Pisum sativum under different experimental conditions. Planta 2010, 232, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Mallona, I.; Lischewski, S.; Julia Weiss, B.H.; Egea-Cortines, M. Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol. 2010, 10, 414. [Google Scholar] [CrossRef] [PubMed]
- Rudy, H.; Simon, H.; Godfrey, N. Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.). BMC Plant Biol. 2010, 10, 71. [Google Scholar]
- Schmidt, G.W.; Delaney, S.K. Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol. Genet. Genom. 2010, 283, 233–241. [Google Scholar] [CrossRef]
- Wu, Z.J.; Tian, C.; Jiang, Q.; Li, X.H.; Zhuang, J. Selection of suitable reference genes for qRT-PCR normalization during leaf development and hormonal stimuli in tea plant (Camellia sinensis). Sci. Rep. 2016, 6, 19748. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, B.; Su, X.; Zhang, S.; Huang, M. Reference gene selection for quantitative real-time polymerase chain reaction in Populus. Anal. Biochem. 2011, 408, 337–339. [Google Scholar] [CrossRef]
- Amorim, L.L.B.; Ferreira-Neto, J.R.C.; Bezerra-Neto, J.P.; Pandolfi, V.; de Araujo, F.T.; da Silva Matos, M.K.; Santos, M.G.; Kido, E.A.; Benko-Iseppon, A.M. Cowpea and abiotic stresses: Identification of reference genes for transcriptional profiling by qPCR. Plant Methods 2018, 14, 88–105. [Google Scholar] [CrossRef]
- Nguyen, D.Q.; Eamens, A.L.; Grof, C.P.L. Reference gene identification for reliable normalisation of quantitative RT-PCR data in Setaria viridis. Plant Methods 2018, 14, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Zou, X.; Carballar-Lejarazu, R.; Wu, L.; Sun, W.; Yuan, X.; Wu, S.; Li, P.; Ding, H.; Ni, L.; et al. Selection and evaluation of reference genes for qRT-PCR analysis in Euscaphis konishii Hayata based on transcriptome data. Plant Methods 2018, 14, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Zhang, N.; Si, H.; Calderon-Urrea, A. Selection and validation of reference genes for RT-qPCR analysis in potato under abiotic stress. Plant Methods 2017, 13, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Jian, B.; Liu, B.; Bi, Y.; Hou, W.; Wu, C.; Han, T. Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol. Biol. 2008, 9, 59. [Google Scholar] [CrossRef]
- Liu, Q.; Qi, X.; Yan, H.; Huang, L.; Nie, G.; Zhang, X. Reference gene selection for quantitative real-time reverse-transcriptase pcr in annual ryegrass (Lolium multiflorum) subjected to various abiotic stresses. Molecules 2018, 23, 172. [Google Scholar] [CrossRef]
- Acevedo, R.M.; Avico, E.H.; Ruiz, O.A.; Sansberro, P.A. Assessment of reference genes for real-time quantitative PCR normalization in Ilex paraguariensis leaves during drought. Biol. Plant. 2018, 62, 89–96. [Google Scholar] [CrossRef]
- Wan, D.; Wan, Y.; Yang, Q.; Zou, B.; Ren, W.; Ding, Y.; Wang, Z.; Wang, R.; Wang, K.; Hou, X. Selection of Reference Genes for qRT-PCR Analysis of Gene Expression in Stipa grandis during Environmental Stresses. PLoS ONE 2017, 12, e0169465. [Google Scholar] [CrossRef]
- Zhang, K.; Li, M.; Cao, S.; Sun, Y.; Long, R.; Kang, J.; Yan, L.; Cui, H. Selection and validation of reference genes for target gene analysis with quantitative real-time PCR in the leaves and roots of Carex rigescens under abiotic stress. Ecotoxicol. Environ. Safety 2018, 168, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, H. Gene cloning and expression analysis of DHHC protein family genes from Liriodendron chinense. Guihaia 2015, 36, 1052–1060, (Chinese with English Abstract). [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).