Abstract
Eucalyptus camaldulensis Dehn. is one of the most morphologically and genetically variable Eucalyptus species. Growth, Leptocybe invasa Fisher & La Salle susceptibility, pilodyn penetration and other traits up to age 36 months were assessed in a seed source/family trial in China comprising 112 seedlots representing five natural stand and six exotic seed sources. Genetic diversity and population structure of this trial population were also analyzed using 48 simple sequence repeat (SSR) markers. The key objective was to examine whether the genomic data could provide value over information obtained from just quantitative trait data. Significant genetic variation was found among seed sources and among families within seed sources for most quantitative traits. The ratio of variance among seed sources to variance among families within seed sources, based on variances estimated from quantitative trait data, varied from 0.1% (height at 9 months) up to 75.2% (bark thickness). Equivalent ratios estimated from the AMOVA on SSR loci data were similar for height (ages 24 and 36 months) and also pilodyn penetration at 36 months, but not for 9-month height or 36-month bark thickness. From 48 SSR loci examined, the genetic differentiation coefficient (among seed sources) was 0.086, indicating low genetic differentiation among seed sources. While overall genetic diversity in the trial population examined was high, the levels within the different seed sources varied markedly. Prior to this study, genetic distances among families from the three exotic seed sources (from domesticated Indian populations) in the trial, along with their genetic distances from, and relatedness to, families from five natural stand seed sources (Australian) in the trial were unknown. The SSR loci data removed uncertainties and revealed that the exotic sources increased the breadth of genetic origins represented in the trial population—information that could not have been obtained from just the quantitative trait data.
1. Introduction
Eucalyptus camaldulensis Dehn. is one of the most morphologically variable species of the genus Eucalyptus. In natural stands its habit can vary from a single-stemmed, tall forest tree of up to 40 m height to a low spreading, multi-stemmed tree of less than 10 m in height [1]. It also has the notable distinction of having the widest geographical range of all Eucalyptus species found in Australia, occurring in all mainland States across an area of around 5 Mkm2 [2].
Across its natural range, E. camaldulensis encompasses substantial phenotypic and genetic variation with recent morphological and molecular genetic studies suggesting recognition of seven subspecies: subsp. acuta, subsp. arida, subsp. camaldulensis, subsp. minima, subsp. obtusa, subsp. refulgens and subsp. simulata [2,3]. Each subspecies is identifiable by both molecular genetics and key morphological features of the flowers, flower buds and leaves.
The species’ extensive variation has helped it achieve wide success in commercial and environmental plantings worldwide, with such variation providing a foundation for selection of subspecies and provenances for superiority in various economically important traits [1]. Indeed, E. camaldulensis is now one of the most widely cultivated eucalypts worldwide. In arid and semi-arid regions it is one of the most widely used trees for multi-purpose plantings whilst in somewhat higher rainfall tropical and subtropical regions, including in Southeast Asia, China, India, Mexico and Brazil, extensive commercial forest plantations have been established with this species [1]. The wood from such plantations is used as raw material for pulp, wood-based board industries, sawn timber production and a range of other end-uses [4].
Numerous studies using observations/measurements on quantitative (phenotypic) traits have been carried out worldwide on E. camaldulensis in statistically designed provenance and provenance/family field trials to examine genetic variability and diversity in adaptive and economically important growth, stem form and wood quality traits. Results of many such trials reported up to 1993 were reviewed in detail by Eldridge et al. [5] and a briefer, more recent review was provided by CABI [6]. As would be expected for a species from a very wide natural occurrence that extends over a broad range of habitats, E. camaldulensis has considerable natural genetic variation. Marked provenance and within-provenance variation has been recorded for at least growth rate, wood properties, tolerance to salinity and alkalinity, drought tolerance, frost tolerance, leaf oil content, polyphenols and pest resistance [1,5,6]. Better-performing tropical provenances like Petford and Katherine of subsp. obtusa are among the best-performing ones for growth and adaptation in seasonally dry tropical environments [6]. In other tropical environments where more resistance to leaf and shoot fungal blights is required, selected provenances of subsp. simulata are often superior for long-term growth [1], while provenances of subsp. camaldulensis from south-eastern temperate Australia, particularly from areas of north-western Victoria such as Lake Hindmarsh and Lake Albacutya, are some of the better ones for Mediterranean type climates [6].
Genetic variation and structure among and within natural populations of E. camaldulensis have also been examined using molecular genetics. Using 62 simple sequence repeat markers (SSRs) Arumugasundaram et al. [7] examined population structure of 40 E. camaldulensis accessions obtained from trees propagated from 11, or possibly more, natural stand provenances representing at least three subspecies that were growing in Indian field trials. They found that the magnitude of genetic variation among individuals (mother trees) was greater than that found among provenances/seed sources. Separately, Butcher et al. [2] employed 15 microsatellite loci to examine a more comprehensive sample of the species comprising of 990 trees from 97 natural stand populations spread across the species entire natural distribution. They found high levels of genetic diversity along with significant genetic differences among populations within river systems, a geographic structuring of molecular genetic variation and that historical factors seemed to have had more influence than selection on current patterns and distribution of genetic diversity.
More recently, Dillon et al. [8] investigated population-level genetic diversity of E. camaldulensis by examining single nucleotide polymorphism (SNP) variation in candidate gene loci sampled across 20 natural stand populations drawn from across the entire span of the species natural distribution. In contrast to Arumugasundaram et al.’s [7] and Butcher et al.’s [2] studies that used putatively neutral SSR markers, they found higher levels of genetic differentiation among populations (17% of total genetic variance) compared to that among individuals within populations (23%).
In China, E. camaldulensis is now of major economic importance to commercial forest plantation growers in southern, warmer regions of the country, not as a pure species but rather as a parental species of a number of successful commercial hybrid varieties [9]. In this latter role, it is valued for the typhoon tolerance, drought tolerance and vegetative propagation ability it can impart to hybrid progeny [1,10]. Typhoon tolerance is of particular economic importance for forest tree plantations in regions of closer proximity to China’s southern coast as typhoons impact this region almost annually.
Since the 1980s, a wide range of genetically diverse seedlots of E. camaldulensis have been imported to China for establishment of populations to support ongoing genetic improvement of this species [10,11,12]. One of the more recently established collections of such imported seedlots comprises a seed source-family trial established in 2012 in the south-west of Guangdong province, China [10]. This population includes 112 open-pollinated family and other seedlots sourced from natural stands in Australia and seed orchards in India and elsewhere. When this population was established it was unknown how the Indian-sourced families would compare to the Australian-sourced families for growth, survival and other key traits. Also unknown was how much genetic diversity this planted population of E. camaldulensis in China contained, and whether or not the Indian seed sources in this population brought genetically unique contributions and/or expanded the population’s genetic diversity.
This present study was initiated in order to examine the above questions, with the specific objectives: (i) to evaluate the performance of all seed sources and families within seed sources in the population for growth and other key traits up to age 36 months; (ii) to examine genetic variance among and within seed sources for these growth and other phenotypically measured traits; (iii) analyze the genetic diversity and structure of the population using SSR markers; and (iv) compare the results obtained for genetic diversity from quantitative traits to results obtained using SSR markers.
2. Materials and Methods
2.1. Genetic Material
The field trial involved in this study was established as a key part of an E. camaldulensis breeding population to support an ongoing tree improvement program in China. It includes 112 seedlots comprising: 25 open-pollinated family seedlots from five natural stand sources in Australia; three multiple-parent bulked seedlots from improved sources (seedling seed orchards) in Australia, Zimbabwe and Thailand; and 84 open-pollinated family seedlots from two seedling seed orchards (SSOs) and one clonal seed orchard CSO) in India (see Table 1). The geographic locations of the natural stand seed sources are shown in Figure 1. These seedlots covered at least three subspecific taxa of the species (as recognized by McDonald et al. [3]).

Table 1.
Details of the Eucalyptus camaldulensis Dehn. families and seed sources established in the seed source-family trial in southwest Guangdong, China.
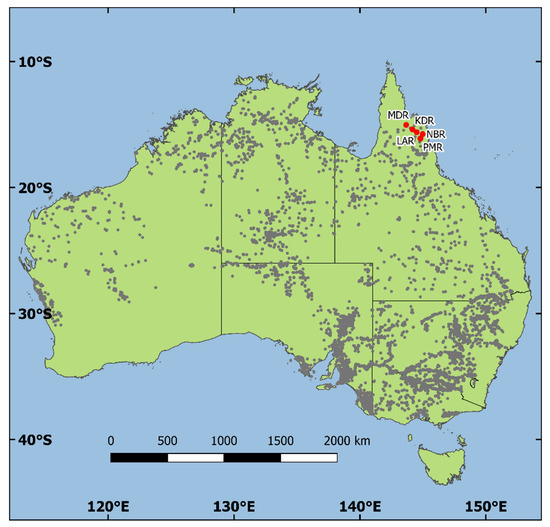
Figure 1.
Natural distribution of Eucalyptus camaldulensis Dehn. in Australia as represented by recorded locations (gray dots) of collections of seedlots and/or herbarium specimens, along with the geographic locations of the five E. camaldulensis natural stand seed sources in Australia that were included in the seed source–family trial in southwest Guangdong, China—seed source ID codes correspond to seed sources listed in Table 1.
2.2. Trial Design and Establishment
The trial was planted in August 2012, with four replicates in a randomized complete block design. Each family/seedlot was represented as a plot of a single row of four trees (four tree row plot) in each replicate, with 2.0 m between trees within rows and 3.0 m between rows—an initial stocking of 1667 tree ha−1. When planted, all seedlings of all seedlots were approximately the same height (shoot height around 30 cm). Complete details on seedling propagation, the trial site and trial establishment have been described previously by Luo et al. [10]. The numbers of families from Indian sources given in this report differ from those in Luo et al. [10] as data for a number of trial families of uncertain origins were omitted from that previous report; recent discovery of some misplaced records on source origins has enabled data from all families to be included in this study.
As stated by Luo et al. [10], all trees in this trial received 500 g tree−1 of NPK base fertilizer (N:P2O5:K2O = 6:12:6) at the time of planting (equating to 833.5 kg ha−1 of compound fertilizer). Additional fertilizer of 500 g tree−1 was then applied at 30 months after planting (N:P2O5:K2O = 15:5:9), in conjunction with directed herbicide application to control weeds. Though this amount of fertilization is markedly higher that used for plantation eucalypts in many other countries, it approximately conforms to standard eucalypt plantation silvicultural practice in China (see [13]).
2.3. Quantitative Trait Assessments
In May 2013 at approximately age 9 months, all living trees were assessed for total height, using height poles, and severity of susceptibility to attack by the gall wasp Leptocybe invasa Fisher & La Salle. The latter trait was assessed subjectively using the following scale (based on an infestation index used by Thu et al. [14]):
Score 0: healthy tree—no galls visible;
Score 1: low susceptibility—<25% of leaves and twigs of crown infested;
Score 2: low to moderate susceptibility—25%–50% leaves and twigs of crown infested;
Score 3: moderate susceptibility—51%–75% leaves and twigs of crown infested;
Score 4: highly susceptible—>75% leaves and twigs of crown infested.
At age 24 months all trees were assessed for height and diameter at breast height (DBH) measured at a height of 1.3 m, and then at age 36 months for height, DBH, pilodyn penetration and bark thickness.
Bark thickness was measured by removing one small square of bark (approximately 4 × 5 cm) from each tree at breast height (1.3 m), on the east side of the tree. The thicknesses of these samples were then measured directly using a metal ruler marked with 1 mm gradations. Pilodyn penetration was measured to provide an indirect, non-destructively obtained estimate of bole wood density. The pilodyn instrument (6-joule pilodyn fitted with a pin of 2 mm thickness) drives a steel pin into the tree with a precise force; the depth that the pin penetrates is inversely proportional to the density of the wood. Such pilodyn measures are known to be efficient for ranking trees for relative wood density [15,16]—lower pilodyn penetrations indicate higher wood density which is desirable for many end-uses. The pilodyn penetration reading on each tree was taken at the place where the bark sample had been removed for measuring bark thickness.
2.4. Analyses of Quantitative Trait Data
Survival, gall wasp (L. invasa) susceptibility, growth, bark and pilodyn penetration data were analyzed using the GLM and VARCOMP procedures of SAS statistical software (Version 8.01; SAS Cary, NC, USA) following procedures described by Williams et al. [17]. These analyses also provided estimates of trait means for each seed source and for each family within seed source, along with standard errors for differences between these means, and various genetic parameters.
Within seed source, individual tree heritabilities for gall wasp susceptibility, height, DBH, bark thickness and pilodyn penetrations were calculated according to the following formula:
where: r = coefficient of relationship;
hi2 = 1/r × σf2/σP2,
σf2 = variance between families within seed source;
σP2 = phenotypic variance = (σf 2 + σm2 + σt2);
σm2 = variance between plots;
σt2 = variance between trees within plots.
The data used for heritability estimation was restricted to that for the natural stand families from Australia (25 families); the Indian families were excluded on account of uncertain relatedness among these families. The coefficient of relationship (r) used in computation of the genetic parameters for E. camaldulensis was taken as 0.30, which is slightly higher than that for true-half-sib families as this species is known to have high but imperfect rates of out-crossing in natural populations [18].
2.5. Sampling DNA
In May 2016, leaf samples were collected from the 109 single mother tree families representing 8 of the 11 seed sources of E. camaldulensis in the trial. From each family sampled, fresh leaf tissue was collected from one tree (i.e., samples obtained from one tree per family) in accordance with sampling methodologies used by Payn et al. [19]), Payn et al. [20], Tripiana et al. [21] and Lu et al. [22]. The three seed sources represented in the trial by only multiple-parent bulked seedlots were not sampled. The trees sampled were from the second replicate, with leaves taken from the first tree in the family plot; if the first tree was missing (dead), then the second tree was sampled.
The leaf samples were individually labeled and stored in liquid nitrogen. At the completion of sample collection in the field, these samples were taken to the laboratory and stored at −80 ℃ until required for DNA extraction.
2.6. DNA Extraction and SSR Marker Assay
Total genomic DNA was extracted from 300 mg of fresh leaf (from each tree sampled) using a modified CATB method [23]. After extraction, DNA quality and quantity were determined by applying 1.2% agarose gel electrophoresis and spectrophotometry, using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). The DNA solution was diluted to 30 ng/µL for subsequent PCR amplification.
To examine the DNA collected from the sample trees, 94 high polymorphism SSR marker loci were used. To carry this out, fluorescence dUTP (Fermentas, Burlington, Ont., Canada) was added to the 10 µL routine polymerase chain reaction (PCR), followed by the landing PCR amplification procedure. The Mg2+ concentration and annealing temperature of the PCR reaction were dependent on the SSR marker. The 1.5 µL PCR product was mixed with super pure formamide HiDi and molecular weight internal standard Liz-500 (Applied Biosystems, Foster City, CA, USA) according to the volume ratio of 9.34:0.18. Then, 8.5 µL was taken to the sample plate, denatured at 95 ℃ for 5 min and cooled at 4 ℃ for 5 min. An ABI 3130xl sequencer (Applied Biosystems, Foster City, CA, USA) was used for genotyping, following methodology described by Li et al. [24] with data collected according to the operation manual of the sequencer.
2.7. SSR Loci Data Analyses
Genepop version 4.2 software [25] was used to test for Hardy–Weinberg equilibrium (HWE) at each locus (p > 0.05, 1000 random checks). Then, from the from 94 high-polymorphism SSR markers, 48 neutral SSR markers (see Table S1) that conformed to HWE expectations (p > 0.05) were selected for use in genetic diversity and population structure (see Table S1).
Using loci data from these 48 selected SSR markers, the number of alleles (NA), expected heterozygosity (HE), observed heterozygosity (HO), Shannon information index (I) and fixation index (F) were calculated using the software GenAlEx version 6.4.1 [26]. The interpopulation differentiation coefficient (FST), interpersonal inbreeding coefficient (FIT) and intrapopulation inbreeding coefficient (FIS) were calculated separately for each locus, using software POPGENE version 1.32 [27]. The software PowerMarker version 3.25 [28] was used to calculate polymorphism information content (PIC).
Nei’s genetic distances (D) between families and between seed sources were calculated by using PowerMarker version 3.25 software [28]. A dendrogram of genetic distances between seed sources and then a consensus tree of genetic affinities between families were compiled using MEGA version 4 software [29] with the ‘unweighted pair group method with arithmetic mean’ (UPGMA). GenA1Ex version 6 software [26] was used to conduct an analysis of molecular variance (AMOVA) to examine the structure of the genetic variation within the field trial population.
3. Results
3.1. Variation among Seed Sources
Early results, to age 9 months, from the field trial involved in this study have reported previously by Luo et al. [10].
To age 36 months, significant differences (p < 0.01) were found among the E. camaldulensis seed sources in the field trial population for survivals (Table 2). However, survivals were generally high with only 2 of the 11 seed sources having average survivals below 80.0% (DCA and KDR). As with survival, significant differences (p < 0.01) were also found among seed sources for all other traits assessed, except for height at age 9 months. By the later assessments (i.e., 24 and 36 months), the best three seed sources for height and DBH were the seed orchard source from Zimbabwe, WPZ, along with two natural stand seed sources, NBR and LAR; all three of these seed sources also had above-average survivals (≥88%). Of the three Indian seed sources in the trial, only KUL had showed above-average height and DBH; this source ranked fourth overall for a combination of height and DBH at age 36 months.

Table 2.
Seed source means, coefficients of variation among family means within seed sources, and significance of various effects in analyses of variance, for the Eucalyptus camaldulensis Dehn. seed source-family trial in southwest Guangdong, China.
For bark thickness and pilodyn penetration, the latter an indicator of wood density, although differences among seed source means were significant, the magnitudes of these differences were relatively small. For the former trait, seed source means varied only from 0.51 to 0.59 cm and all but two of seed source means for pilodyn penetration were within the range 12.0 to 13.5 mm.
For susceptibility to the pest insect L. invasa, differences among sources were also significant with two India sources, KUL and DPK being the most susceptible (undesirable) with average scores of 1.44 and 2.09, respectively. The least susceptible seed sources were MDR and KDR, with average scores of 0.32 and 0.43, respectively, but both of these sources had some of poorest growth of all 11 seed sources (MDR ranked 10th and KDR ranked 9th, at age 36 months).
3.2. Variation among Families
Differences among families within seed sources were significant (p < 0.01) for all traits assessed in the trial, and there was marked variation among seed sources for the variability among family averages within sources, as shown by the coefficients of variation (CVs) among average means of families within seed sources (see Table 2). For instance, CVs for family averages for DBH at age 36 months varied from just 9% for LAR up to 28% for MPM. The lowest within-seed-source variability among family averages were for pilodyn penetration, with CVs among family means within seed sources ranging from just 3% for NBR up to 10% for KUL, while the trait with the highest CVs was susceptibility to L. invasa, and these ranged from 44% for PMR up to 105% for MPM.
Seed source mean performance for some key traits, namely L. invasa susceptibility, DBH (36 months) and pilodyn penetration, showed little relation to the potential of individual families from within sources to qualify for the groups of top ranked 10 and 20 families (Table 3). For L. invasa susceptibility 4 of the top ranked 10 families and 7 of the top ranked 20 families were from the poorest two seed sources (KUL and DPL). Similarly, for age 36 months DBH 4 of the top 10 families and 8 of the top 20 families came from two sources that had close to or well below average DBH (KUL and MPM). For pilodyn penetration, 6 of the top 10 and 10 of the top 20 families were from seed sources that had just average pilodyn penetration (KUL and MPM), though as emphasised above, variation among both seed source and family means for this trait was relatively low.

Table 3.
Seed source origins and means of the top ranked 10 and 20 families for Leptocybe invasa Fisher & La Salle susceptibility at age 9 months, and DBH at age 36 months and pilodyn penetration at age 36 months in the Eucalyptus camaldulensis Dehn. seed source–family trial in southwest Guangdong, China.
In concurrence with the both the significance of differences among means of families within seed sources and the magnitudes of variations in these means, individual within-seed-source (provenance) heritabilities ranged from low at 0.10 ± 0.15 for bark thickness at age 36 months up to moderate at 0.54 ± 0.15 for susceptibility to L. invasa, as shown in Table 4. From age 9 months to age 24 months and then to age 36 months the heritability for height increased progressively from 0.11 ± 0.15 to 0.24 ± 0.22 and then to 0.27 ± 0.19. However, that for DBH decreased from 0.37 ± 0.22 to 0.18 ± 0.15 from age 24 to 36 months.

Table 4.
Genetic variance and heritability estimates for various traits in an E. camaldulensis seed source-family trial in southwest Guangdong, China. The variance parameters were derived from data including all seedlots/families, while heritabilities were estimated just from data on families from natural stand seed sources.
To further examine sources of genetic variation, the ratios of the variance attributable to seed sources to the variance attributable to families within seed sources were computed for each trait; these are presented in Table 4. These ratios varied markedly, from a low of just 0.1% for height at age 9 months up to 75.2% for bark thickness at age 36 months. The only other trait for which this ratio exceeded 15% was susceptibility to L. invasa; the ratio of seed source variance to families within seed source variance for this trait was 50.8%.
3.3. Genetic Diversity by Molecular Markers
For the 48 SSR loci selected for conforming to HWE expectations, the average number of alleles per locus (NA) and the average effective number alleles per locus (NE) were 28.5 and 6.4, respectively (see Table 5), with the actual number of alleles (NA) per locus ranging from 8 to 42. Averaged across these loci, Shannon’s information index (I) and the polymorphism information content (PIC) were 1.84 and 0.84, respectively, while observed heterozygosity (HO) and expected heterozygosity (HE) both averaged 0.77, though the former had greater variation among loci (standard deviation = 0.22) than the latter (standard deviation = 0.14). The preceding values for these key genetic diversity parameters indicate high levels of genetic diversity in the field trial population studied here.

Table 5.
Loci genetic diversity indices for the population of Eucalyptus camaldulensis Dehn., presented as means across the 48 SSR loci that satisfied HWE criteria (standard deviations of these means given in brackets).
The inbreeding coefficient (FIS) averaged 0.05 across the 48 neutral SSR loci, implying a low level of inbreeding overall. Meanwhile, the coefficient of between-subpopulation differentiation (FST) averaged just 0.08 (here subpopulation is equivalent to seed source), indicating moderate genetic differentiation among subpopulations (according to criteria provided by Hartl and Clark [30] and Balloux and Lingnon-Moulin [31]). However, it must be noted that while the 48 SSRs were assumed to be neutral, they had been preselected (out of 94 SSR loci evaluated) for meeting HWE expectations; data from the other 46 SSR loci initially examined (but that did not conform with HWE expectations) were not involved in the computations of these parameters.
To examine the sources contributing to the total molecular genetic variation observed at the SSR loci, an AMOVA of hierarchical genetic variance within the E. camaldulensis trial population studied here was carried out (see Table 6). This revealed that 86% of the total variance was attributable to variation among loci within individuals, while only 1% of the total was attributable to variation among seed sources and 13% to variation among individuals within seed sources.

Table 6.
Analysis of molecular variance to partition genetic variation observed using SSR markers in the eight Eucalyptus camaldulensis Dehn. seed sources.
3.4. Genetic Diversity within Seed Sources
By individual seed source, the average number of alleles (NA) per loci ranged from 5.5 to 17.1, the number of effective alleles (NE) ranged from 4.5 to 10.3, and Shannon’s Information Index (I) ranged from 1.48 to 2.44 (Table 7). While these preceding values suggest reasonable levels of genetic diversity within each of the seed sources, it is noteworthy that the magnitudes of these parameters for each of the three Indian sources were markedly higher than for each of the five Australian sources, indicating that the former sources contain more genetic diversity. However, it must also be remembered that each of the three Indian sources was represented by 24 or more families while each of the Australian sources had just five families.

Table 7.
Genetic diversity parameters for the eight Eucalyptus camaldulensis Dehn. seed sources examined in this study.
Despite differences in the number of families per seed source between the Indian and Australian sources, observed heterozygosity (HO) ranged from 0.72 to 0.84, with all three of the former sources being very close to the average of the five Australian sources (0.77). Meanwhile the Indian sources stood apart from the Australian sources by fixation index (F), with the former all having positive values (0.03 to 0.08) while the latter had negative values (−0.05 to −0.11) with the exception of 10-PMR, for which F = 0.01. Even so, the relatively low absolute values found for fixation index (F) in all eight seed sources suggests inbreeding levels (within these sources) to be low.
3.5. Distribution and Structure of Genetic Variation
Genetic distances among the all pairwise groupings of the eight E. camaldulensis seed sources examined with SSR markers varied greatly, ranging from just 0.12 up to 0.61 (Table 8). The shortest genetic distances between pairs of seed sources were those among the three Indian sources, KUL, DPK and MPM; their pairwise distances were 0.21 or less. Among the five Australian seed sources, pairwise distances were at least double that of distances among the former and ranged from 0.42 to 0.61. The genetic distances between pairwise combinations of Indian and Australian seed sources were generally intermediate between distances among pairs of Indian seed source and among pairs of Australian seed sources, ranging from 0.29 up to 0.44.

Table 8.
Pairwise Nei’s genetic distances (GD) between the eight Eucalyptus camaldulensis Dehn. seed sources examined in this study.
These results for pairwise genetic distances among seed sources generally concurred with the patterns depicted in a dendrogram showing genetic distances and clustering among the 8 seed sources (Figure 2). This dendrogram reveals the three Indian seed sources as being clustered close together, while three of the five Australian sources, LAR, PMR and NBR, are also clustered close together as a group. Somewhat separate from the three foregoing Australian seed sources are the other two Australian ones, KDR and MDR, with the latter seed source appearing to be closest to three Indian ones.
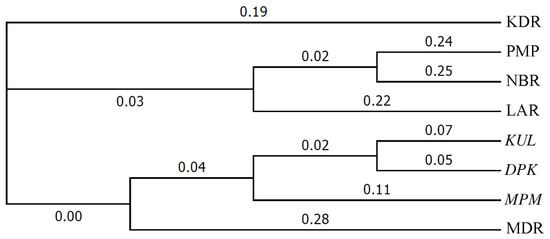
Figure 2.
Dendrogram showing genetic clustering, based on ‘unweighted pair group method with arithmetic mean’ (UPGMA) method, for the eight Eucalyptus camaldulensis Dehn. seed sources, sources from India are in italics—numbers placed along the branches represent Nei’s genetic distances.
A consensus tree based on estimated genetic distances among the 109 E. camaldulensis family seedlots reveals three clear groupings, as shown in Figure 3. The first of these groups, Group I, includes 21 families from just Indian sources; predominantly KUL and MPM. The second group is larger, having 54 families, and comprises two clear subgroups, one with 25 families and the other with 29 families. In the first subgroup, the families are predominantly from Indian seed sources (22 families), with just three Australian families originating from PMR and KDR seed sources. In the second subgroup, the families are also predominantly from Indian seed sources (26 families) with just three Australian families, which represented the three seed sources MDR, KDR and LAR. In contrast, the third group comprised 34 families that were dominated by Australian sources (19 families), with the other 15 families it contained being from a mix of the Indian seed sources (three families from KUL, 5 from DPK and 7 from MPM).
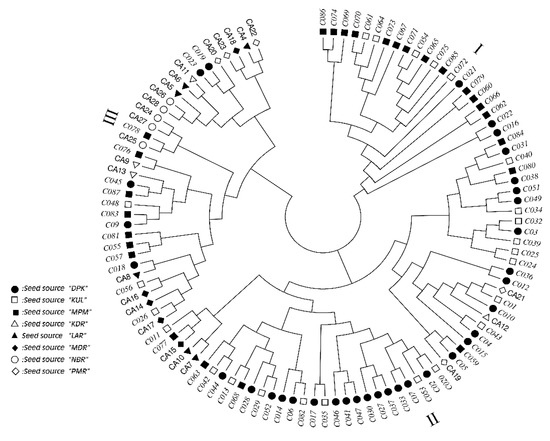
Figure 3.
A consensus tree based on genetic distances, estimated using SSR markers, among the 109 Eucalyptus camaldulensis Dehn. family seedlots examined in this study (families from India are shown in italics) that was constructed using UPGMA.
4. Discussion
Our results showed wide genetic variation within the E. camaldulensis trial population involved in this study, with individual tree within seed source (syn. provenance) heritabilities for growth traits ranging up to 0.37 ± 0.22 (DBH at 24 months) and for other traits up to 0.54 ± 0.40 (L. invasa susceptibility). High genetic variation in growth and other important traits in this species has been reported from an early age assessment of the field trial involved in this current study [10] as well as numerous other previous trials of this species [32,33].
Below-average height and DBH growth of some of the exotic (i.e., imported) improved seed sources in the trial, especially LKT (Thailand origin) and MPM (Indian origin) relative to the best natural stand seed sources observed in this E. camaldulensis trial, has analogies to previous results from trials of other eucalypt species in China and elsewhere. In an E. dunnii trial in Yunnan province established in 2008, one natural stand seed sources (from Australia) had age 5.5 years’ average tree volume above that of three out of five exotic improved seed sources [34]. Similarly, in a range of trials in Australia exotic improved E. grandis, E. saligna, E. tereticornis and E. urophylla seed sources, including ones from Brazil, South Africa and Zimbabwe, reportedly offered zero or negative margins in growth that ranged to 40% below the growth of the best natural stand seed sources [35,36].
A number of factors likely to contribute to suboptimal growth of exotic, improved seed source sources of E. camaldulensis in the trial reported here, including that genotypes selected for superior growth and adaptation in other (exotic) environments that differ from that of southwest Guangdong where the trial population of this study is located, might not be well adapted for superior performance in this environment. Such environmental differences will have been exacerbated by the high rates of fertilization applied in this study’s trial. While the rate of fertilizer rate used in the trial (totaling 1.0 kg g tree−1 of compound fertilizer, equivalent to 1667 kg ha−1) conforms with general eucalypt plantation silvicultural practice in China, the rate is a markedly higher rate than what is used for plantation eucalypts in many other countries (around 100 to 200 kg ha−1 of compound fertilizer; see [13]). Thus, the seedlots from seed orchards in India, Thailand, Zimbabwe and even Australia included in the trial had been selected based for performance under lower levels of fertilization (and different soil types) and could not necessarily be expected to maintain superior performance given silviculture that incorporated markedly higher fertilizer application.
In the field trial population examined in this study, the best families for various traits came from a wide range of seed sources including Indian ones. Having top-ranked families for growth originating from a wide range of seed sources that included some with the poorest average growth was also not without precedent. In a Sri Lankan E. grandis provenance-family trial involving 18 natural stand provenances, the second-best family at age 27 months by mean tree volume was from the provenance ranked third last for the same trait [37]. Similarly, in E. saligna trials in south-eastern China involving 84 families representing nine natural stand provenances from Australia, the top 10 ranked families by age 3.5 years’ tree volume represented up to 8 of the 9 provenances [38].
The AMOVA conducted with the SSR loci data found just 1% of total variance at SSR loci was attributable to variation among seed sources, compared to 13% attributable to variation among families within seed sources, providing a ratio of the variance attributable to seed sources to that attributable to family within seed sources of 7.7%. This concurs closely with the ratios of seed source variance to family within seed source variance estimated for some of the quantitative traits measured on the trees in the trial, namely height at ages 24 and 36 months and pilodyn penetration (ratios = 7.4%, 10.1% and 9.8%, respectively; see Table 4). However, some of the other quantitative traits had ratios that differed markedly from the 7.7% estimated from the AMOVA, namely L. invasa susceptibility, age 9 months’ height and bark thickness (ratios = 50.8%, 0.1%, and 75.2%, respectively).
The concurrence between the AMOVA and quantitative trait analyses for the relative contributions of among- and within-seed-source variation to the total genetic variation found in this current study for some traits contrasts to results found by Tripiana et al. [21] for E. urophylla. They reported genetic variance among populations (groupings of 2 to 19 proximal seed sources/provenances) of 21% to 50% for quantitative traits of height and bole circumference in field trials, while data on 10 microsatellite markers indicated that only about 4% of the total genetic variance (estimated for 10 microsatellite markers) was attributable to variation among populations and 96% was attributable to variation within populations.
Even so, the results from this study for both the AMOVA and some quantitative traits do concur with results from some previous E. camaldulensis studies. Butcher et al. [2], who genotyped 990 trees from 97 seed sources (synonymous with populations in their study) using 15 neutral microsatellite loci, found that 9% of the genetic variation could be attributed to differences among seed sources based on an AMOVA conducted using their data. In an earlier study, Butcher et al. [39] used 33 RFLP loci to examine over 250 trees representing 31 seed sources from across northern Australia and found 7.8% of genetic diversity attributable to differences among seed sources. In contrast, Dillon et al. [8] analyzed SNP variations in 12 candidate genes across 20 populations (synonymous with seed sources or provenances) approximately spanning the species’ natural distribution and found that 17% of the variance among SNP loci was attributable to among population variation, somewhat higher than estimates by Butcher et al. [39] and Butcher et al. [2] obtained using putatively neutral markers.
For the five Australian seed sources in this study, the average number of alleles per loci within seed sources (NA = 5.5 to 6.4) were higher and expected heterozygosities within seed sources (HE = 0.70 to 0.76) were generally lower than those reported previously for natural stand seed sources of similar subspecies of E. camaldulensis (i.e., subsp. obtusa, subsp. simulata and subsp. acuta) by Butcher et al. [2]. Differences between the estimates from the two studies are likely attributable in part to sample size differences—Butcher et al. [2] sampled 10 trees per seed source compared to this current study’s five families per seed source—and also that the two studies examined different sets of SSR loci. Even so, the average HE estimated by the current study across the trial population as a whole (0.77) is close to estimates reported for a range of other Eucalyptus species, including 0.80 for natural stand populations of E. tereticornis [40], 0.82 for natural stand populations of E. globulus population [41] and 0.74 for natural stand populations of E. urophylla [20].
Shannon’s information index (I) and number of alleles per locus (NA), which are indicative of genetic diversity, averaged across loci within each seed source in this study ranged from 1.48 to 2.44 and 5.5 to 17.1, respectively. While such values generally indicate high genetic diversity, it was notable that the three Indian seed sources had greater genetic diversity by these measures than all the five natural stand seed sources. Also, each of the natural stand seed sources included in the trial seemed lower in diversity by such parameters than what has been reported for most natural stand seed sources (populations) by both Butcher et al. [39] and Butcher et al. [2], while each of the Indian sources were higher. Higher diversity within each of the Indian sources was likely due to their diverse, mixed natural stand/seed source origins along with each of these sources being represented by greater numbers of families (from 24 to 34) compared to Australian sources (five families each) in the trial.
The average FST value (across all loci) estimated for the E. camaldulensis trial population (0.086) was indicative of moderate differentiation among subpopulations [30,31]. Even so, this level of differentiation among seed sources is lower than what has been reported for a number of other plantation Eucalyptus species, including: Corymbia citriodora (FST = 0.08) [42], E. globulus (FST = 0.09) [41] and E. pellita (FST = 0.09) [43], but higher than that reported for E. tereticornis (FST = 0.01) [40] and natural stand populations of E. urophylla (FST = 0.03) [20]. Previous studies have also shown the genetic differentiation between subpopulations (synonymous to natural stand seed sources) of E. camaldulensis to be relatively low with FST values of 0.026 to 0.054 [2]. That a greater portion of genetic variation was attributed to variation within rather than among subpopulations by the latter study is consistent with results obtained in this current study, both from the variance component analyses based on phenotypic trait data and the AMOVA conducted with SSR loci data.
That the five Australian sources clustered relatively close together in both the dendrogram and consensus tree generated by this study was not unexpected, all were from within a relatively small region (spanning latitude 15°02′ to 16°07′ S and longitude 143° 40′ to 144°59′ E) within the species’ vast natural range (spanning latitude 12°51′ to 37°14′ S and longitude 113°47′ to 150°51′ E). As discussed above, genetic differentiation between subpopulations (synonymous to natural stand seed sources) of this species has been found to be relatively low by other studies [2].
The consensus tree generated in this study using genetic distances among 109 E. camaldulensis families in this study also indicated that families from within any one of the Indian sources did not cluster as a single group. And, whilst some of the Indian families were found to be relatively close (by genetic distance) to some Australian families, most were quite distant, and 21 of the Indian families clustered in a group of exclusively Indian-sourced families. The range of distances among families from within each of the Indian sources could reflect their having either multiple origins (as in being composed of multiple seedlots/selected genotypes from multiple natural stand seed source origins in Australia) or that they were generated from parents representing a diversity of such seed source origins. These results, along with the seed source dendrogram, reveal that the three Indian sources have provided genotypes that are both genetically diverse and genetically different to those from the five Australian seed sources and thus provide unique genetic material that complements rather than duplicates the genotypes provided by the Australian seed sources. Also, importantly, the results provide reassurance that the Indian families as a whole, and even subgroupings of these, do not originate from narrow genetic bases.
The greater portion of genetic variation within the trial population involved in this current study was attributable to variation within seed sources, by both analyses of quantitative phenotypic trait data and analyses of SSR loci data. This was consistent with results previous studies on E. camaldulensis and concurs with what has generally been observed in many other plantation eucalypt species. Genetic variation within seed sources (provenances) often exceeds that among seed sources with individual genotypes and families showing superiority for key economic traits often found in seed sources of below-average performance for the same traits. All this suggests that genetic improvement of E. camaldulensis needs to focus on the selection and improvement of superior performing individual trees and families irrespective of seed source origins. Such a strategy of focusing on exploiting variation within seed sources/provenances has previously been recommended for E. grandis [44,45] and E. saligna [38].
5. Conclusions
In this study there was a general concurrence between structure of genetic diversity revealed by both analyses of molecular SSR loci data and analyses of that from many of the quantitative traits assessed on the E. camaldulensis population field trial population, and both revealed reasonable levels of genetic diversity. However, prior to commencement of this study, the genetic distances between families within and among seed sources, along with and their genetic distances from, and possible relatedness to, the families from the five natural stand seed sources was unknown. The quantitative trait data collected could not have provided answers to these critical questions, in the way that the SSR loci were able to; perhaps that is the greatest value of the work done to obtain the latter data. The SSR loci data removed uncertainties and confirmed that the Indian sources increased the breadth of genetic origins of families and seed sources included in the trial (i.e., proving that the Indian families were not inter-related to the Australian-sourced ones), providing confidence that that the trial population has a broad genetic base.
In combination with this population’s overall genetic richness (implied diversity of natural seed source origins), its genetic diversity will provide a good foundation for future recurrent cycles of selection and breeding to provide gains in key economic traits. And, given that most of the genetic variation within the trial population examined in this study existed within seed sources (among individuals) rather than among seed sources, the number of (unrelated) individual families represented in this trial population will be a key factor in providing for genetic gains.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4907/10/12/1090/s1, Table S1: Summary of the 48 loci that conformed with Hardy–Weinberg equilibrium (HWE; p > 0.05) expectations.
Author Contributions
X.S. and Z.W. conceived and designed the study; J.L. and P.Z. organized and conducted the trial sampling and managed the samples; X.S., N.Z. and G.L. carried out the laboratory analyses; X.S. and R.J.A. collated, managed and analyzed data; X.S., J.L. and R.J.A. wrote the paper. All authors read and approved the final manuscript.
Funding
This study was financially supported by Fundamental Research Funds for National Natural Science Foundation of China (Number: 31570615), the Central Non-profit Research Institution of Chinese Academy of Forestry (Project Number: CAFYBB2017ZX001-5), and Fundamental Research Funds for the Central Non-profit Research Institution of Chinese Academy of Forestry (Project Number: CAFYBB2019MA006).
Acknowledgments
We are grateful to Paul Macdonell from Queensland, Australia, for assistance with preparation of Figure 1.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Arnold, R.J.; Luo, J.Z. Eucalyptus camaldulensis. In Trees for Life in Oceania: Conservation and Utilisation of Genetic Diversity; ACIAR Monograph No. 201; Thomson, L., Doran, J., Clarke, B., Eds.; Australian Centre for International Agricultural Research: Canberra, Australia, 2018; pp. 94–99. [Google Scholar]
- Butcher, P.A.; McDonald, M.W.; Bell, J.C. Congruence between environmental parameters, morphology and genetic structure in Australia’s most widely distributed eucalypt Eucalyptus Camaldulensis. Tree Genet. Genomes 2009, 5, 189–210. [Google Scholar] [CrossRef]
- McDonald, M.W.; Brooker, M.I.H.; Butcher, P.A. A taxonomic revision of Eucalyptus camaldulensis (Myrtaceae). Aust. Syst. Bot. 2009, 22, 257–285. [Google Scholar] [CrossRef]
- Luo, J.Z.; Xie, Y.J.; Cao, J.G.; Lu, W.H.; Ren, S.Q. Genetic variation in 2-year eucalypt hybrids’ growth and typhoon resistance. J. Grass Ind. 2009, 18, 91–97. [Google Scholar]
- Eldridge, K.; Davidson, J.; Harwood, C.; Van Wyk, G. Eucalypt Domestication and Breeding; Oxford University Press: Oxford, UK, 1993; p. 312. [Google Scholar]
- CABI. Eucalyptus camaldulensis. In The Forestry Compendium: Global Module; CAB International: Oxon, UK, 2000. [Google Scholar]
- Arumugasundaram, S.; Ghosh, M.; Veerasamy, S.; Ramasamy, Y. Species discrimination, population structure and linkage disequilibrium in Eucalyptus camaldulensis and Eucalyptus tereticornis using SSR Markers. PLoS ONE 2011, 6, e28252. [Google Scholar] [CrossRef] [PubMed]
- Dillon, S.; McEvoy, R.; Baldwin, D.S.; Rees, G.N.; Parsons, Y.; Southerton, S. Characterisation of adaptive genetic diversity in environmentally contrasted populations of Eucalyptus camaldulensis Dehnh. (River Red Gum). PLoS ONE 2014, 9, e103515. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.Z.; Zhou, G.; Wu, D.; Chen, D.; Cao, J.; Lu, W.H.; Pegg, R.E.; Arnold, R.J. Genetic variation and age-age correlations Eucalyptus grandis in Southern China. Aust. For. 2010, 73, 67–80. [Google Scholar] [CrossRef]
- Luo, J.Z.; Arnold, R.J.; Lu, W.H.; Lin, Y. Genetic variation in Eucalyptus camaldulensis and E. tereticornis for early growth and susceptibility to the gall wasp Leptocybe invasa in China. Euphytica 2014, 196, 397–411. [Google Scholar] [CrossRef]
- Turnbull, J.W. Development of sustainable forestry plantations in China: A review. In ACIAR Impact Assessment Series Report No. 45; Australian Centre for International Agricultural Research: Canberra, Australia, 2007. [Google Scholar]
- Arnold, R.J.; Luo, J.Z.; Lu, W.H.; Wang, C.B.; Lin, Y. Co-operative Improvement of Key Eucalypt Species in China. In Scientific Cultivation and Green Development to Enhance the Sustainability of Eucalypt Plantations; Proceedings of IUFRO Eucalypt Conference, Zhanjiang, China, 21–24 October 2015; China Eucalypt Research Centre: Zhanjiang, China.
- Harwood, C.E.; Nambiar, E.K.S. Sustainable Plantation Forestry in South-East Asia. In ACIAR Technical Reports No. 84; Australian Centre for International Agricultural Research: Canberra, Australia, 2014; p. 100. [Google Scholar]
- Thu, P.Q.; Dell, B.; Burgess, T.I. Susceptibility of 18 eucalypt species to the gall wasp Leptocybe invasa in the nursery and young plantations in Vietnam. Sci. Asia 2009, 35, 113–117. [Google Scholar]
- Hansen, C.P. Application of the Pilodyn in Forest Tree Improvement; DFSC Series of Technical Notes TN55; Danida Forest Seed Centre: Humlebaek, Denmark, 2000. [Google Scholar]
- Ilic, J.; Boland, D.; McDonald, M.; Downes, G.; Blakemore, P. Wood Density Phase 1—State of Knowledge; National Carbon Accounting System–Technical Report No. 18; Australian Greenhouse Office Press: Canberra, Australia, 2000. [Google Scholar]
- Williams, E.R.; Matheson, A.C.; Harwood, C.E. Experimental design and analysis for use in Tree Improvement, 2nd ed.; CSIRO: Melbourne, Australia, 2002. [Google Scholar]
- Butcher, P.A.; Williams, E.R. Variation in outcrossing rates and growth in Eucalyptus camaldulensis from the Petford region, Queensland; evidence of outbreeding depression. Silvae Genet. 2003, 51, 6–12. [Google Scholar]
- Payn, K.G.; Dvorak, W.S.; Myburg, A.A. Chloroplast DNA phylogeography reveals the island colonisation route of Eucalyptus urophylla (Myrtaceae). Aust. J. Bot. 2007, 55, 673–683. [Google Scholar] [CrossRef]
- Payn, K.G.; Dvorak, W.S.; Janse, B.J.H.; Myburg, A.A. Microsatellite diversity and genetic structure of the commercially important tropical tree species Eucalyptus urophylla endemic to seven islands in eastern Indonesia. Tree Genet. Genomes 2008, 4, 519–530. [Google Scholar] [CrossRef]
- Tripiana, V.; Bourgeois, M.; Verhaegen, D.; Vigneron, P.; Bouvet, J.M. Combining microsatellites, growth, and adaptive traits for managing in situ genetic resources of Eucalyptus urophylla. Can. J. For. Res. 2007, 37, 773–785. [Google Scholar] [CrossRef]
- Lu, W.H.; Arnold, R.J.; Zhang, L.; Luo, J.Z. Genetic diversity and structure through three cycles of a Eucalyptus urophylla S.T. Blake breeding program. Forests 2018, 9, 372. [Google Scholar]
- Gan, S.; Shi, J.; Li, M.; Wu, K.M.; Wu, J.Y.; Bai, J.Y. Moderate-density molecular maps of Eucalyptus urophylla S.T. Blake and E. tereticornis Smith genomes based on RAPD markers. Genetica 2003, 118, 59–67. [Google Scholar] [CrossRef]
- Li, F.; Gan, S. An optimised protocol for fluorescent-dUTP based SSR genotyping and its application to genetic mapping in Eucalyptus. Silvae Geneica 2011, 60, 18–25. [Google Scholar] [CrossRef]
- Rousset, F. Genepop’007: A complete reimplementation of the Genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Yeh, F.C.; Yang, R.; Boylet, T. POPGENE Version 1.32: Software Microsoft Window-Based Freeware for Population Genetic Analysis; University of Alberta: Edmonton, AB, Canada, 1997. [Google Scholar]
- Liu, K.; Muse, S.V. Powermarker: Integrated analysis environment for genetic marker data. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Hartl, D.L.; Clark, A.G. Principles of Population Genetics, 3rd ed.; Sinauer Associates, Inc. Publishers: Sunderland, MA, USA, 1997. [Google Scholar]
- Balloux, F.; Lugon-Moulin, N. The estimation of population differentiation with microsatellite markers. Mol. Ecol. 2002, 11, 155–165. [Google Scholar] [CrossRef]
- Bush, D.; Marcar, N.; Arnold, R.; Crawford, D. Assessing genetic variation within Eucalyptus camaldulensis for survival and growth on two spatially variable saline sites in southern Australia. For. Ecol. Manag. 2013, 306, 68–78. [Google Scholar] [CrossRef]
- Mahmod, K.; Marcar, N.E.; Naqvi, M.H.; Arnold, R.J.; Crawford, D.F.; Iqbal, S.; Aken, A.K. Genetic variation in Eucalyptus camaldulensis Denh. for growth and stem straightness in a provenance-family trial on saltland in Pakistan. For. Ecol. Manag. 2003, 176, 405–416. [Google Scholar] [CrossRef]
- Shi, T.Y.; Arnold, R.J.; Kang, W.L.; Duan, F.W.; Qian, Y.X.; Xie, H.; Xu, J.M. Genetic variation and gains for two generations of Eucalyptus dunnii in China. Aust. For. 2016, 79, 15–24. [Google Scholar] [CrossRef]
- Arnold, R.; Bush, D.; Stackpole, D. Genetic variation and tree improvement. In New Forests—Wood Production and Environmental Services; Nambiar, S., Ferguson, I., Eds.; CSIRO Publishing: Melbourne, Australia, 2005; pp. 25–49. [Google Scholar]
- Lee, D.J.; Huth, J.R.; Osborne, D.O.; Hogg, B.W. Selecting hardwood taxa for wood and fibre production in Queensland’s subtropics. Aust. For. 2010, 73, 106–114. [Google Scholar] [CrossRef]
- Bandara, K.M.A.; Arnold, R.J.; Aken, K.M. Genetic variation in a Eucalyptus grandis seedling seed orchard in the up-country of Sri Lanka. Sri. Lankan For. 2000, 25, 21–36. [Google Scholar]
- Lan, H.S.; Huang, X.M.; Luo, J.Z.; Arnold, R.J. Genetic variation in growth and stem straightness in Eucalyptus saligna trials in Fujian. Aust. For. 2012, 75, 163–174. [Google Scholar]
- Butcher, P.A.; Otero, A.; McDonald, M.W.; Moran, G.F. Nuclear RFLP variation in Eucalyptus camaldulensis Dehnh. From northern Australia. Heredity 2002, 88, 402–412. [Google Scholar] [CrossRef]
- Song, Z.J.; Yang, H.Y.; Weng, Q.J.; Zhou, C.P.; Li, F.G.; Li, M.; Lu, W.H.; Luo, J.Z.; Gan, S.M. Genetic diversity and selective loci in Eucalyptus tereticornis populations. Sci. Silvae Sin. 2016, 52, 40–47. [Google Scholar]
- Steane, D.A.; Potts, B.M.; Mclean, E.; Prober, S.M.; Srock, W.D.; Vaillancout, R.E.; Byrne, M. Genome-wide scans detect adaptation to aridity in a widespread forest tree species. Mol. Ecol. 2014, 23, 2500–2513. [Google Scholar] [CrossRef]
- Liu, S.R.; Lin, Y.; Liu, X.H.; Luo, J.Z. Genetic diversity and genetic structure of Corymbia citriodora based on SSR analysis. Mol. Plant. Breed. 2016, 14, 1923–1929. [Google Scholar]
- Li, C.R.; Xiong, T.; Chen, D.L.; Deng, Z.Y.; Chen, S.K.; Lan, B.B.; Liang, J.X.; Lan, J. Analysis on genetic diversity of Eucalyptus pellita based on SSR markers. J. South. Agric. 2016, 47, 7–12. [Google Scholar]
- Burgess, I.P. Provenance trials of Eucalyptus grandis and E. saligna in Australia. Silvae Genet. 1988, 37, 221–227. [Google Scholar]
- Matheson, A.C.; Mullin, L.J. Variation among neighbouring and distant provenances of Eucalyptus grandis and E. tereticornis in Zimbabwean field trials. Aust. For. Res. 1987, 17, 233–250. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).