Sub-Genome Polyploidization Effects on Metabolomic Signatures in Triploid Hybrids of Populus
Abstract
1. Introduction
2. Materials and Methods
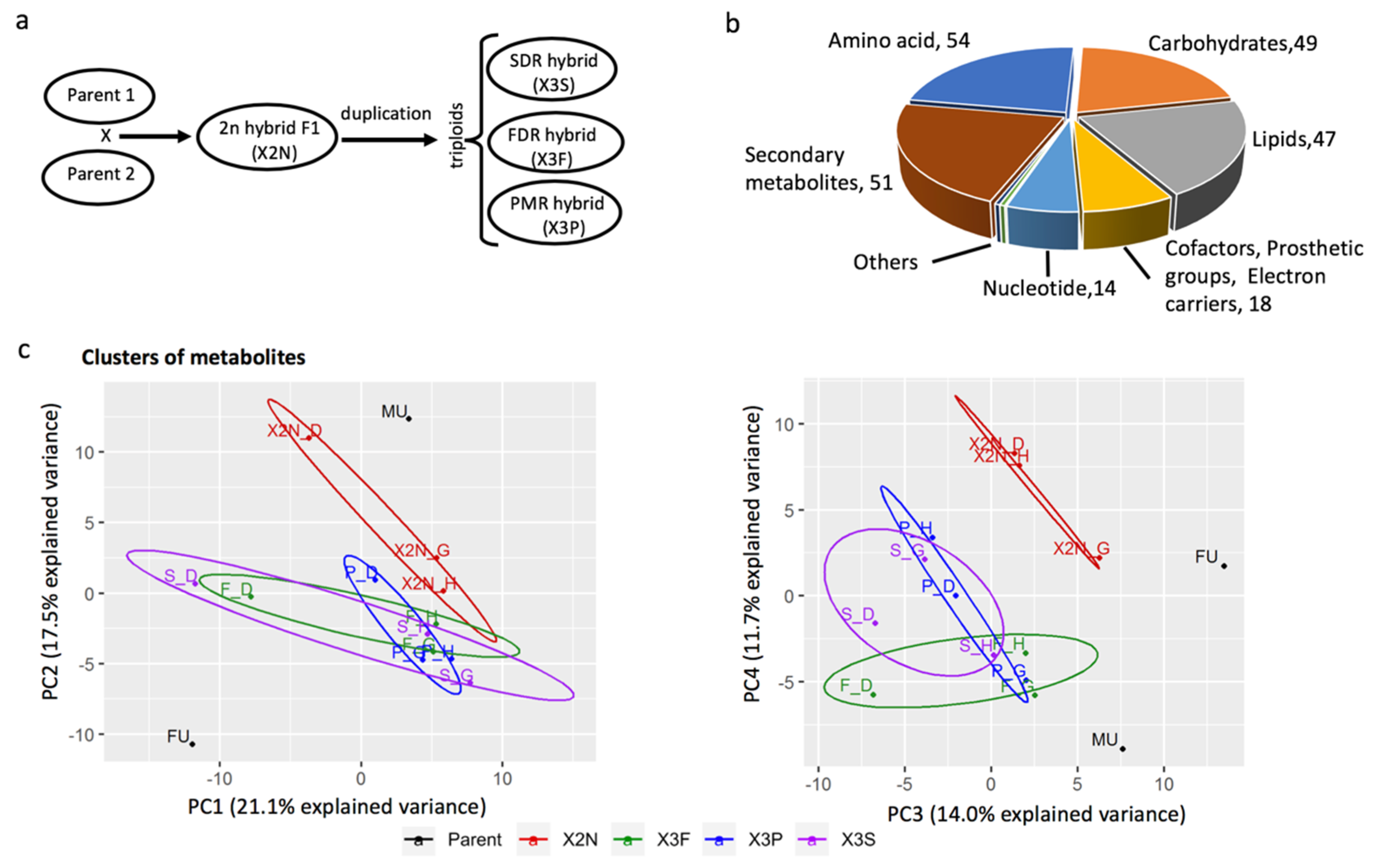
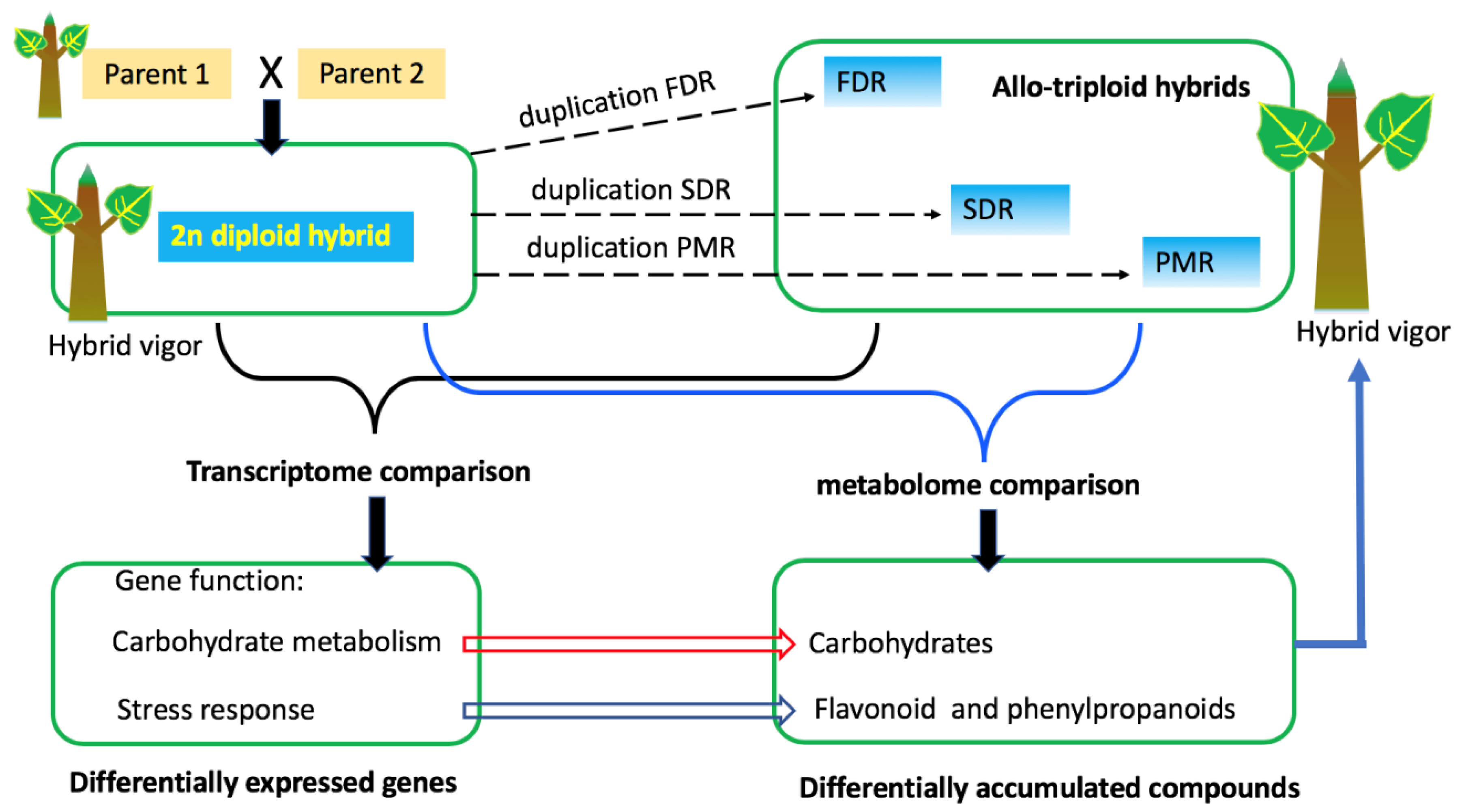
2.1. Plant Materials
2.2. Metabolites Extraction, Liquid Chromatography–Mass Spectrometry (LC-MS) Metabolomic Profiling, and Data Analysis
2.3. Transcriptome Data and Gene Ontology (GO) Analysis
3. Results
3.1. Identification and Clusters of Metabolites in Diploid and Triploid Populus
3.2. Alteration of Primary Metabolite Carbohydrate in Allo-Triploids
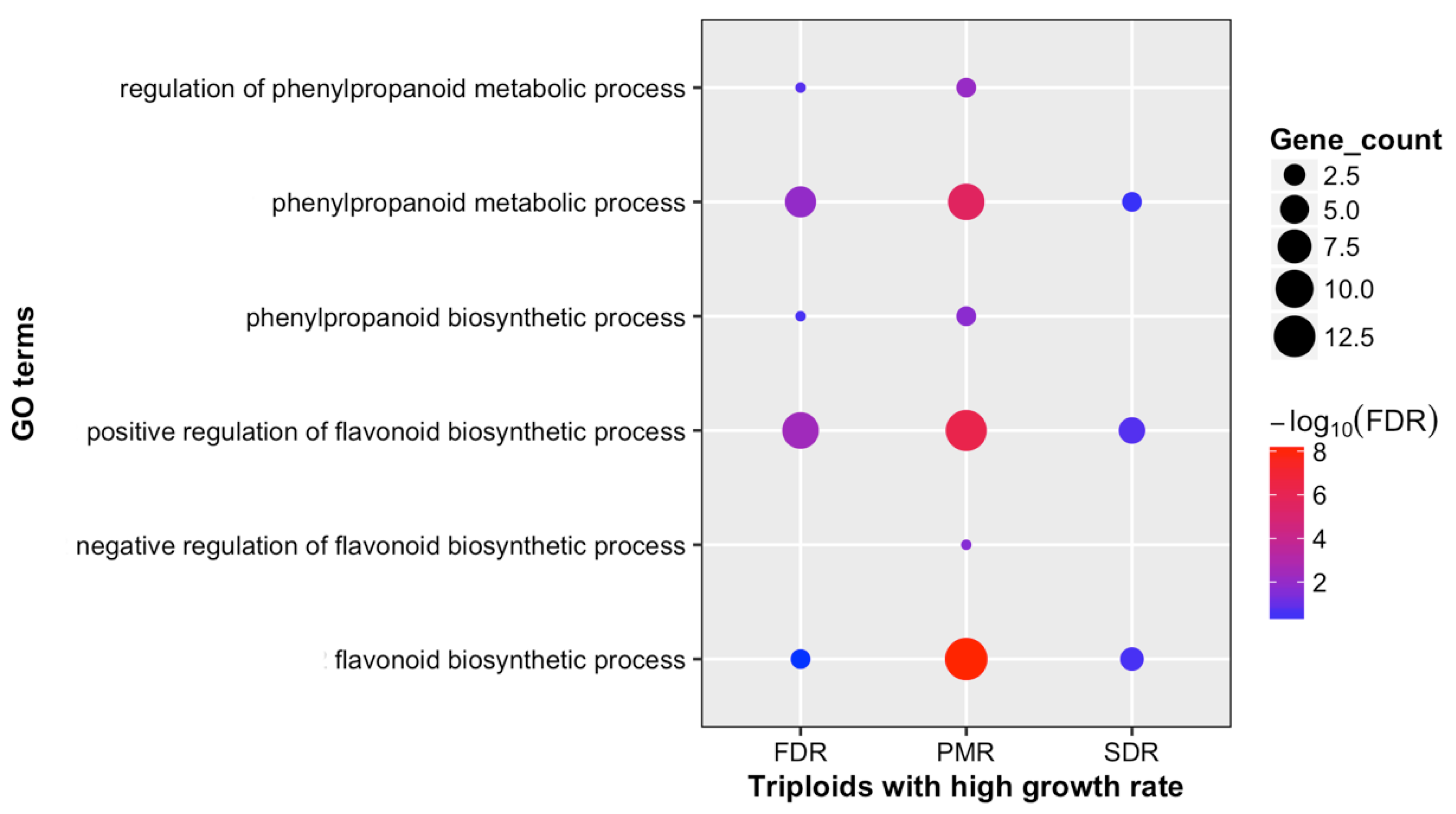
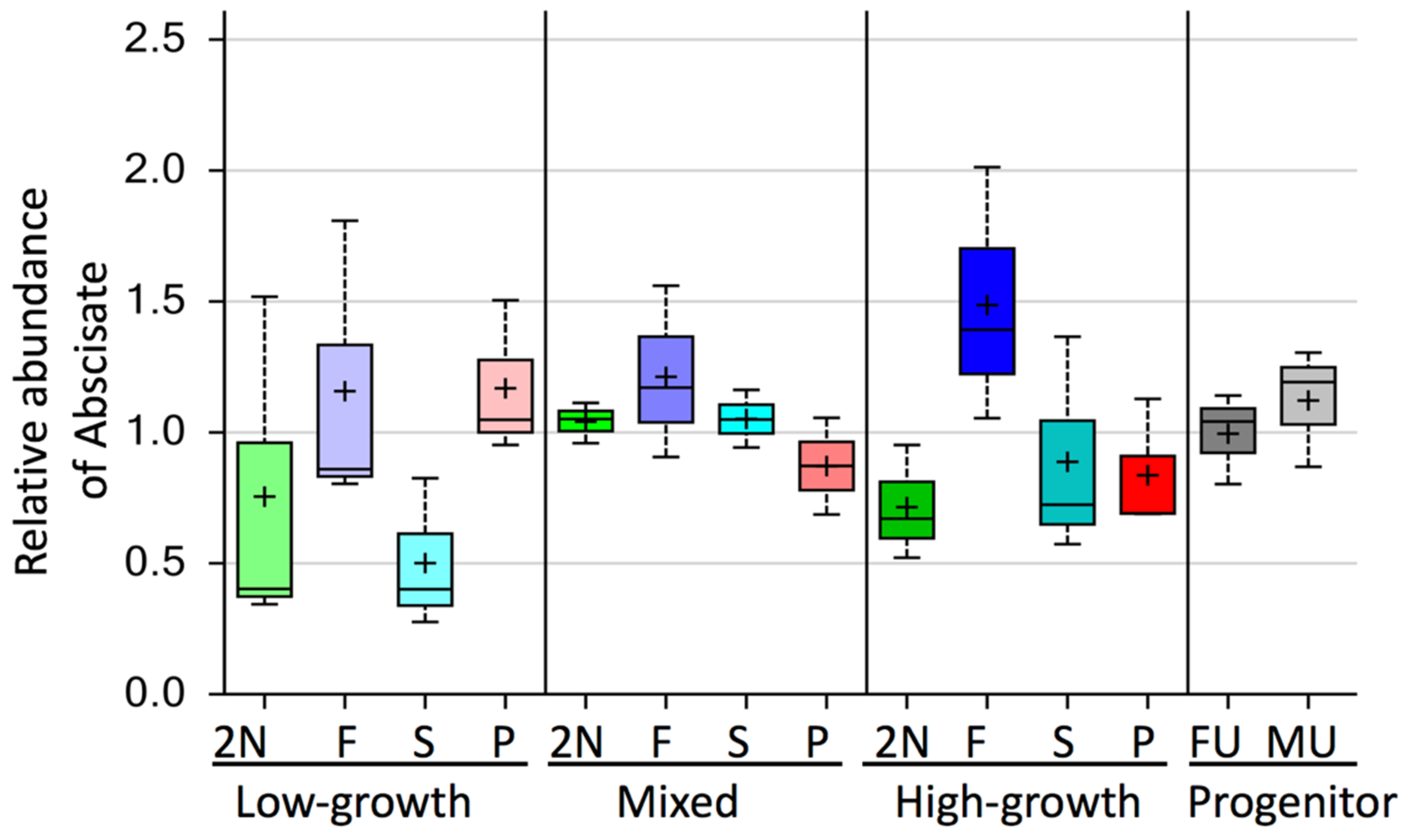
3.3. Alteration in Secondary Metabolites between Diploid and Triploid Populus
3.4. Difference in Lipids between Dipoid and Triploid Populus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Doyle, J.J.; Coate, J.E. Polyploidy, the Nucleotype, and Novelty: The Impact of Genome Doubling on the Biology of the Cell. Int. J. Plant Sci 2018, 180, 1–52. [Google Scholar] [CrossRef]
- Dong, C.-B.; Suo, Y.-J.; Kang, X.-Y. Assessment of the genetic composition of triploid hybrid Populus using SSR markers with low recombination frequencies. Can. J. For. Res. 2014, 44, 692–699. [Google Scholar] [CrossRef]
- Wang, J.; Kang, X.Y. Distribution of Microtubular Cytoskeletons and Organelle Nucleoids During Microsporogenesis in a 2n Pollen Producer of Hybrid Populus. Silvae Genet. 2009, 58, 220–226. [Google Scholar] [CrossRef][Green Version]
- Alix, K.; Gerard, P.R.; Schwarzacher, T.; Heslop-Harrison, J.S.P. Polyploidy and interspecific hybridization: Partners for adaptation, speciation and evolution in plants. Ann. Bot. 2017, 120, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2016, 243, 281–296. [Google Scholar] [CrossRef]
- Cheng, F.; Wu, J.; Cai, X.; Liang, J.; Freeling, M.; Wang, X. Gene retention, fractionation and subgenome differences in polyploid plants. Nat. Plants 2018, 4, 258–268. [Google Scholar] [CrossRef]
- Vergara, F.; Rymen, B.; Kuwahara, A.; Sawada, Y.; Sato, M.; Hirai, M.Y. Autopolyploidization, geographic origin, and metabolome evolution in Arabidopsis thaliana. Am. J. Bot. 2017, 104, 905–914. [Google Scholar] [CrossRef]
- Hochholdinger, F.; Hoecker, N. Towards the molecular basis of heterosis. Trends Plant Sci. 2007, 12, 427–432. [Google Scholar] [CrossRef]
- Chen, Z.J. Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet. 2013, 14, 471–482. [Google Scholar] [CrossRef]
- Jansson, S.; Douglas, C.J. Populus: A model system for plant biology. Annu. Rev. Plant Biol. 2007, 58, 435–458. [Google Scholar] [CrossRef]
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The Genome of Black Cottonwood, Populus trichocarpa. Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Huang, Z.; Suo, Y.; Wang, J.; Kang, X. Gene expression differences associated with growth vigor in Populus full-sib allotriploid progeny following manipulated first division restitution of the diploid maternal parent. Euphytica 2015, 203, 683–700. [Google Scholar] [CrossRef]
- Cheng, S.; Yang, J.; Liao, T.; Zhu, X.; Suo, Y.; Zhang, P.; Wang, J.; Kang, X. Transcriptomic changes following synthesis of a Populus full-sib diploid and allotriploid population with different heterozygosities driven by three types of 2n female gamete. Plant Mol. Biol. 2015, 89, 493–510. [Google Scholar] [CrossRef] [PubMed]
- Corneillie, S.; De Storme, N.; Van Acker, R.; Fangel, J.U.; De Bruyne, M.; De Rycke, R.; Geelen, D.; Willats, W.G.T.; Vanholme, B.; Boerjan, W. Polyploidy Affects Plant Growth and Alters Cell Wall Composition. Plant Physiol. 2019, 179, 74–87. [Google Scholar] [CrossRef]
- Yoo, M.J.; Szadkowski, E.; Wendel, J.F. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity (Edinb) 2013, 110, 171–180. [Google Scholar] [CrossRef]
- Powell, J.J.; Fitzgerald, T.L.; Stiller, J.; Berkman, P.J.; Gardiner, D.M.; Manners, J.M.; Henry, R.J.; Kazan, K. The defence-associated transcriptome of hexaploid wheat displays homoeolog expression and induction bias. Plant Biotechnol. J. 2017, 15, 533–543. [Google Scholar] [CrossRef]
- Serapiglia, M.J.; Gouker, F.E.; Hart, J.F.; Unda, F.; Mansfield, S.D.; Stipanovic, A.J.; Smart, L.B. Ploidy Level Affects Important Biomass Traits of Novel Shrub Willow (Salix) Hybrids. BioEnergy Res. 2015, 8, 259–269. [Google Scholar] [CrossRef]
- Ma, Y.; Xue, H.; Zhang, L.; Zhang, F.; Ou, C.; Wang, F.; Zhang, Z. Involvement of Auxin and Brassinosteroid in Dwarfism of Autotetraploid Apple (Malus × domestica). Sci. Rep. 2016, 6, 26719. [Google Scholar] [CrossRef]
- Evans, A.M.; DeHaven, C.D.; Barrett, T.; Mitchell, M.; Milgram, E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem. 2009, 81, 6656–6667. [Google Scholar] [CrossRef]
- Oliver, M.J.; Guo, L.; Alexander, D.C.; Ryals, J.A.; Wone, B.W.M.; Cushman, J.C. A Sister Group Contrast Using Untargeted Global Metabolomic Analysis Delineates the Biochemical Regulation Underlying Desiccation Tolerance in Sporobolus stapfianus. Plant Cell 2011, 23, 1231–1248. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Zhao, Y. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef]
- Walton, A.; Stes, E.; De Smet, I.; Goormachtig, S.; Gevaert, K. Plant hormone signalling through the eye of the mass spectrometer. Proteomics 2015, 15, 1113–1126. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Higashi, Y.; Nakabayashi, R. The Origin and Evolution of Plant Flavonoid Metabolism. Front. Plant Sci. 2019, 10, 943. [Google Scholar] [CrossRef]
- Mathesius, U. Flavonoid Functions in Plants and Their Interactions with Other Organisms. Plants (Basel, Switzerland) 2018, 7, 30. [Google Scholar] [CrossRef]
- Morreel, K.; Goeminne, G.; Storme, V.; Sterck, L.; Ralph, J.; Coppieters, W.; Breyne, P.; Steenackers, M.; Georges, M.; Messens, E.; et al. Genetical metabolomics of flavonoid biosynthesis in Populus: A case study. Plant J. 2006, 47, 224–237. [Google Scholar] [CrossRef]
| Pathway ID 1 | Pathway | Metabolites |
|---|---|---|
| pop01100 | Metabolic pathways (general) | 134 |
| pop01110 | Biosynthesis of secondary metabolites | 73 |
| pop02010 | ABC transporters | 36 |
| pop01230 | Biosynthesis of amino acids | 33 |
| pop01210 | 2-Oxocarboxylic acid metabolism | 21 |
| pop01200 | Carbon metabolism | 20 |
| pop00970 | Aminoacyl-tRNA biosynthesis | 17 |
| pop00630 | Glyoxylate and dicarboxylate metabolism | 14 |
| pop00052 | Galactose metabolism | 13 |
| pop00260 | Glycine, serine and threonine metabolism | 11 |
| pop00250 | Alanine, aspartate and glutamate metabolism | 11 |
| pop00941 | Flavonoid biosynthesis | 11 |
| Pathway | Biochemical Name | Comparison within Low Growth Rate | Comparison within High Growth Rate | Comparison within Mixed Growth Rate | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| F_D/2N_D | S_D/2N_D | P_D/2N_D | F_G/2N_G | S_G/2N_G | P_G/2N_G | F_H/2N_H | S_H/2N_H | P_H/2N_H | ||
| Glycolysis | 1,3-dihydroxyacetone | 0.93 | 1.08 | 0.89 | 1.29 | 1.48 | 1.32 | 1.94 | 1.11 | 1.04 |
| 1,6-anhydroglucose | 0.7 | 0.93 | 0.64 | 0.86 | 0.8 | 0.89 | 0.69 | 0.74 | 0.95 | |
| fructose-6-phosphate | 1.44 | 1.69 | 1.67 | 1.23 | 1.45 | 1.37 | 1.29 | 1.29 | 1.29 | |
| glucose | 0.97 | 0.9 | 0.91 | 0.88 | 0.96 | 0.94 | 0.96 | 1 | 0.96 | |
| glucose-6-phosphate | 2.17 | 2.78 | 3.53 | 1.37 | 1.46 | 2.23 | 1.16 | 1.35 | 1.26 | |
| glycerate | 2.94 | 2.32 | 0.77 | 1.02 | 0.99 | 0.77 | 0.92 | 0.82 | 0.78 | |
| Isobar 1 | 0.96 | 0.82 | 1.34 | 1.59 | 1.55 | 1.19 | 2.06 | 1.53 | 1.84 | |
| pyruvate | 0.77 | 0.92 | 0.77 | 0.6 | 0.73 | 0.63 | 0.82 | 0.86 | 0.87 | |
| Citric Acid Cycle TCA | alpha-ketoglutarate | 0.99 | 1.23 | 0.84 | 0.93 | 0.9 | 0.88 | 0.78 | 0.79 | 0.75 |
| cis-aconitate | 1.11 | 1.03 | 1.36 | 1.05 | 1.21 | 0.88 | 1.16 | 1.05 | 1.12 | |
| citrate | 1.13 | 1.23 | 1.41 | 0.94 | 1.06 | 1.01 | 1.11 | 1.12 | 1.04 | |
| fumarate | 0.77 | 0.78 | 0.85 | 0.79 | 0.86 | 0.99 | 1.3 | 1.1 | 1.13 | |
| isocitrate | 1.17 | 1.42 | 1.06 | 1.08 | 0.91 | 0.85 | 0.99 | 0.94 | 1.09 | |
| malate | 1.72 | 1.56 | 1.44 | 1.02 | 1.09 | 1.11 | 1.09 | 1.01 | 1.02 | |
| succinate | 0.68 | 0.74 | 1.25 | 0.98 | 0.81 | 1.33 | 0.72 | 0.84 | 0.81 | |
| Calvin Cycle and pentose phosphate | sedoheptulose-7-phosphate | 2.44 | 2.56 | 3.18 | 0.95 | 1.62 | 1.11 | 0.71 | 0.83 | 0.78 |
| Amino sugar and nucleotide sugar | arabinose | 1.3 | 1.29 | 1.17 | 1.02 | 1.13 | 1.12 | 0.93 | 1.04 | 1.04 |
| arabitol | 1.18 | 0.99 | 1.32 | 0.83 | 0.59 | 1.26 | 0.89 | 0.86 | 1.1 | |
| arabonate | 0.65 | 0.79 | 0.62 | 0.97 | 0.92 | 1.04 | 1.21 | 0.83 | 0.95 | |
| erythritol | 1.43 | 1.41 | 1.2 | 1.06 | 1.25 | 0.99 | 1.05 | 0.96 | 0.99 | |
| erythronate | 1.79 | 2.13 | 0.81 | 1.23 | 1.24 | 1.04 | 1.06 | 1.13 | 1.18 | |
| lyxose | 1.18 | 1.08 | 1.18 | 0.86 | 0.81 | 0.8 | 1.01 | 1.07 | 0.97 | |
| N-acetylglucosamine | 0.79 | 0.65 | 1.31 | 0.79 | 0.85 | 0.71 | 0.94 | 0.59 | 0.91 | |
| ribitol | 1.26 | 1.93 | 0.66 | 1.18 | 1.26 | 0.86 | 0.9 | 0.87 | 1.1 | |
| ribose | 0.97 | 0.51 | 0.71 | 1.16 | 0.98 | 0.93 | 1.24 | 1.12 | 1.05 | |
| ribulose | 1.13 | 1.01 | 0.74 | 1.46 | 1.29 | 1.05 | 1.41 | 1.05 | 0.84 | |
| UDP-glucose | 0.89 | 1.11 | 1.72 | 1.09 | 1.38 | 1.35 | 1.39 | 1.07 | 1.54 | |
| xylonate | 1.08 | 0.92 | 0.99 | 0.94 | 0.93 | 0.89 | 1 | 1 | 1 | |
| xylose | 1.55 | 1.34 | 1.24 | 1.13 | 1.18 | 1.11 | 1.05 | 1.12 | 1.22 | |
| xylulose | 0.95 | 0.94 | 1.19 | 1.15 | 1.31 | 1.17 | 1.43 | 1.42 | 1.35 | |
| UDP-galactose | 0.58 | 0.59 | 1.31 | 0.72 | 1.08 | 0.99 | 0.88 | 0.84 | 1.31 | |
| Inositol metabolism | inositol 1-phosphate (I1P) | 0.99 | 0.79 | 1.23 | 0.89 | 0.94 | 0.8 | 0.86 | 0.85 | 0.83 |
| myo-inositol | 1.78 | 1.05 | 1.5 | 0.98 | 0.93 | 0.87 | 0.91 | 0.97 | 1.06 | |
| Sucrose, glucose, fructose metabolism | 3-deoxyoctulosonate | 1.12 | 0.92 | 1.41 | 1.1 | 1.06 | 1.06 | 1.04 | 1.07 | 1 |
| fructose | 0.81 | 0.84 | 0.82 | 0.91 | 0.85 | 0.84 | 0.82 | 0.84 | 0.84 | |
| galactinol | 9.52 | 5.62 | 3.27 | 3.92 | 2.9 | 2.18 | 1.91 | 1.54 | 1.9 | |
| galactitol (dulcitol) | 1.5 | 1.86 | 1.38 | 0.92 | 0.8 | 0.86 | 0.69 | 0.92 | 0.96 | |
| galacturonate | 1.17 | 0.84 | 0.97 | 1.36 | 1.1 | 1.06 | 0.82 | 0.83 | 1.02 | |
| glucarate 1,4-lactone | 0.89 | 0.8 | 0.81 | 0.7 | 1.39 | 0.58 | 0.82 | 0.88 | 1.25 | |
| mannitol | 1.38 | 0.96 | 0.96 | 0.93 | 0.85 | 0.93 | 0.94 | 0.96 | 1.01 | |
| mannose-6-phosphate | 1.28 | 1.31 | 1.97 | 0.85 | 1.02 | 1.46 | 1 | 0.95 | 1.15 | |
| raffinose | 1.91 | 1.31 | 1.19 | 1.97 | 1.8 | 1.92 | 1.7 | 1.4 | 1.43 | |
| rhamnose | 1.09 | 1.16 | 1.08 | 0.92 | 0.95 | 0.76 | 0.83 | 0.88 | 0.88 | |
| sorbitol | 1.11 | 1 | 1.09 | 0.86 | 0.84 | 0.99 | 0.92 | 0.91 | 0.99 | |
| stachyose | 2.85 | 1.78 | 0.77 | 2.64 | 2.91 | 2.22 | 2.39 | 1.68 | 2.02 | |
| sucrose | 2.38 | 2.04 | 2.16 | 1.12 | 1.03 | 1.16 | 1.22 | 1.19 | 1.13 | |
| trehalose | 1.06 | 0.91 | 1.25 | 1.01 | 1.28 | 1.02 | 1.07 | 1.03 | 1.03 | |
| methyl-beta-glucopyranoside | 0.45 | 0.48 | 0.66 | 0.76 | 0.82 | 1.03 | 0.83 | 1 | 0.75 | |
| C5 branched dibasic acid metabolism | citramalate | 1.06 | 0.99 | 0.94 | 0.84 | 0.77 | 0.95 | 0.88 | 0.94 | 0.97 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, S.; Zong, Y.; Wang, X. Sub-Genome Polyploidization Effects on Metabolomic Signatures in Triploid Hybrids of Populus. Forests 2019, 10, 1091. https://doi.org/10.3390/f10121091
Cheng S, Zong Y, Wang X. Sub-Genome Polyploidization Effects on Metabolomic Signatures in Triploid Hybrids of Populus. Forests. 2019; 10(12):1091. https://doi.org/10.3390/f10121091
Chicago/Turabian StyleCheng, Shiping, Yuxia Zong, and Xuewen Wang. 2019. "Sub-Genome Polyploidization Effects on Metabolomic Signatures in Triploid Hybrids of Populus" Forests 10, no. 12: 1091. https://doi.org/10.3390/f10121091
APA StyleCheng, S., Zong, Y., & Wang, X. (2019). Sub-Genome Polyploidization Effects on Metabolomic Signatures in Triploid Hybrids of Populus. Forests, 10(12), 1091. https://doi.org/10.3390/f10121091






