Abstract
The main neurotransmitters in the brain—dopamine, γ-aminobutyric acid (GABA), glutamate, and opioids—are recognized to be the most important for the regulation of aggression and addiction. The aim of this work was to study differentially expressed genes (DEGs) in the main reward-related brain regions, including the ventral tegmental area (VTA), dorsal striatum (STR), ventral striatum (nucleus accumbens, NAcc), prefrontal cortex (PFC), and midbrain raphe nuclei (MRNs), in male mice with 20-day positive fighting experience in daily agonistic interactions. Expression of opioidergic, catecholaminergic, glutamatergic, and GABAergic genes was analyzed to confirm or refute the influence of repeated positive fighting experience on the development of “addiction-like” signs shown in our previous studies. High-throughput RNA sequencing was performed to identify differentially expressed genes in the brain regions of chronically aggressive mice. In the aggressive mice, upregulation of opioidergic genes was shown (Oprk1 in VTA, Pdyn in NAcc, Penk in PFC, and Oprd1 in MRNs and PFC), as was downregulation of genes Opcml and Oprk1 in STR and Pomc in VTA and NAcc. Upregulation of catecholaminergic genes in VTA (Ddc and Slc6a2) and in NAcc (Th and Drd2) and downregulation of some differentially expressed genes in MRNs (Th, Ddc, Dbh, Drd2, Slc18a2, and Sncg) and in VTA (Adra2c, Sncg, and Sncb) were also documented. The expression of GABAergic and glutamatergic genes that participate in drug addiction changed in all brain regions. According to literature data, the proteins encoded by genes Drd2, Oprk1, Oprd1, Pdyn, Penk, and Pomc are directly involved in drug addiction in humans. Thus, our results confirm our earlier claim about the formation of addiction-like signs following repeated positive fighting experience in mice, as shown previously in our biobehavioral studies.
1. Introduction
The main neurotransmitters—dopamine (DA), serotonin (5-HT), γ-aminobutyric acid (GABA), and glutamate—are recognized as the most important neurotransmitters for the mechanisms of aggression [1,2,3,4,5,6,7]. A role of endogenous opioids has been shown in offensive aggression [8]. Aggression is also closely associated with activation or inhibition of opioidergic receptors and is strongly influenced by drugs [1,9]. 5-HT is also considered an active player in the inhibition of control over aggression [10,11,12,13,14,15,16]. It is well known that all these neurotransmitter systems are involved in the development of reward and addiction.
In our early study, we found the activation of brain dopaminergic systems in male mice winning in daily agonistic interactions with unfamiliar conspecifics [17]. This activation was detected as elevated DOPAC (3,4-dihydroxyphenylacetic acid) levels or/and increased DOPAC/DA ratios in the amygdala, nucleus accumbens, and dorsal striatum in the winners as compared with controls.
Pharmacological data have also confirmed changes of dopaminergic activity during positive fighting experience. Haloperidol (an antagonist of dopaminergic receptors [18]) and D1 receptor antagonist SCH-23390 [19] effectively decreased aggression in male mice with 2-day experience of aggression and were ineffective in the same doses in mice with 20-day winning experience. These observations indicate the desensitization of DA receptors induced by chronic activation of dopaminergic systems after experience of repeated aggression. Experiments have also revealed development of tolerance to an opioid receptor antagonist (naltrexone) [20,21,22]: blockade of opioid receptors had no significant effect on the level of aggression in winners with 20-day positive fighting experience in contrast to 2-day winners. Activation of the opioidergic system induced by an agonist of mu-opioid receptors (morphine) [23] and by a kappa-opioid receptor agonist (U-50,488H) revealed desensitization of opioid receptors: in some experiments, the winners hardly responded to treatment with these drugs in comparison with a control, which responded to the drug injections by behavioral changes [24].
Using reverse transcription (RT) and then qPCR, we also found that 20-day repeated aggression is accompanied by upregulation of the Th gene encoding tyrosine hydroxylase, the Slc6a3 (Dat1) gene encoding DA transporter, and the Snca gene encoding α-synuclein [25,26] in the ventral tegmental area (VTA) of winners and downregulation of Comt in the midbrain [27]. Moreover, the expression of genes Th and Slc6a3 stays enhanced in VTA for at least 2 weeks after discontinuation of agonistic interactions and correlates with the level of aggressiveness of winners [26].
It has been suggested that positive fighting experience, like other basic behaviors, provides a permanent reward to the winners, hence a tendency to repeat acts of aggression. Aggressive motivation becomes generalized and dominant in any social situation. Chronically aggressive male mice, as a rule demonstrating unmotivated aggression, attack any conspecifics in any situations [28,29]. There is evidence that the winners demonstrate impaired communication and sociability [30] and prolonged persistence of changes in behaviors and emotional states after 2 weeks without agonistic interactions. Generally, our behavioral and pharmacological data have allowed us to hypothesize the development of an addiction-like state in the winners as a result of positive fighting experience in daily agonistic interactions [29]. Increased aggressiveness after a period of fighting deprivation [31] confirms the development of psychopathology in male mice following repeated aggression. Additionally, other authors have noticed signs of pathological aggression [5,32,33] and signs of “addiction-like” alterations in aggressive mice [34,35].
The aim of the present work was to analyze differential expression of well-known genes encoding proteins that are involved in the development of reward, drug abuse, addiction, and aggression [2,3,4,6,8,36,37]. Primarily, these are the opioidergic and catecholaminergic (CAergic) genes, and their mRNA expression was analyzed to confirm or refute the development of an addiction-like state under the influence of repeated positive fighting experience, as shown in our previous studies [29]. Furthermore, we took into consideration data showing that abnormalities in glutamatergic [38] and/or GABAergic neurotransmission may underlie resting-state functional deficits in drug addiction [39] and aggression [6].
Transcriptomic analysis was performed in the main reward-related brain regions that are involved in the mechanisms of positive reinforcement [40,41,42]: these are the VTA, dorsal striatum (STR), ventral striatum (NAcc), prefrontal cortex (PFC), and midbrain raphe nuclei (MRNs) in male mice with 20-day positive fighting experience in daily agonistic interactions. Only the mice that demonstrated permanent strongest, uncontrollable maniacal excessive aggression during a 20-day period were analyzed. The study is carried out within the framework of the direction we are developing “Functional neurogenomics of pathological conditions” with use of the sensory contact model [43], renamed later as the chronic social conflicts model, allows to obtain male mice with an aggressive type of behavior [44].
2. Results
2.1. Agonistic Behavior
Among male mice with daily experience of aggression for 20 tests (days), highly aggressive animals were identified (Table 1), which in daily agonistic interactions demonstrated a high level of aggressiveness, stopping during an attack only for a short rest, after which they again attacked the loser, thus demonstrating unmotivated aggression and attacking another male, regardless of its behavior (active, passive, or full defense). Other aggressive mice usually stopped attacking if the loser showed postures of complete submission or immobility. Moreover, changes in social behaviors and psychoemotional states after long positive fighting experience that never appeared in control animals were noted in the winners and included the demonstration of abnormal aggression, strong hostility, an anxious state, uncontrollable behavior, disturbances in social recognition, hyperactivity, and stereotypic and hyperkinetic reactions [28,29,44]. In any social situation, these aggressive mice demonstrated high motivation to attack, bite, and chase any partners.

Table 1.
Agonistic behavior of mice with different levels of aggression.
The total time of attacking behavior, the average time of one attack, and the total time of hostile behavior were significantly longer in highly aggressive mice (which were subjected to transcriptome analysis) than in less aggressive mice (Table 1). Behavioral observations showed that 50% of highly aggressive mice additionally displayed total extended time of unmotivated self-grooming behavior (61.3 ± 13.8 s), which may be regarded as stereotypical behavior. Of note, the number of jerks (winces) was significantly higher in the aggressive mice compared with highly aggressive mice (15.9 vs. 8.5 s), possibly as a result of ambivalence when a decision (to attack or not) is making in situation of agonistic interactions. Aggressive mice, as a rule, first sniffed the partner and only then attacked it.
2.2. Neurotranscriptomic Data
All DEGS in different brain regions are presented in Supplementary Table S1.
2.2.1. DEGs in the VTA of Highly Aggressive Mice
Analysis of the RNA-Seq database of the VTA [45,46] revealed DEGs among genes of interest that participate in the rewarding processes (Figure 1A; Supplementary Table S1).
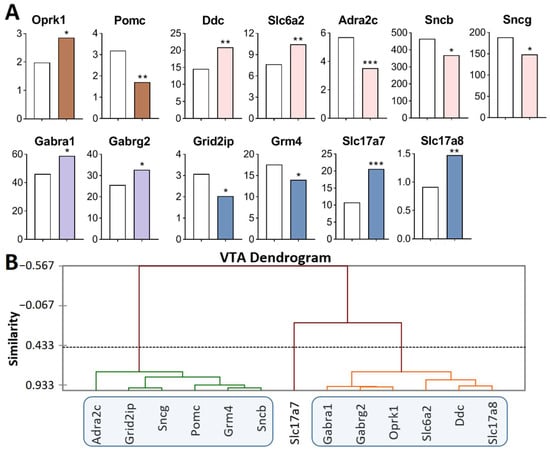
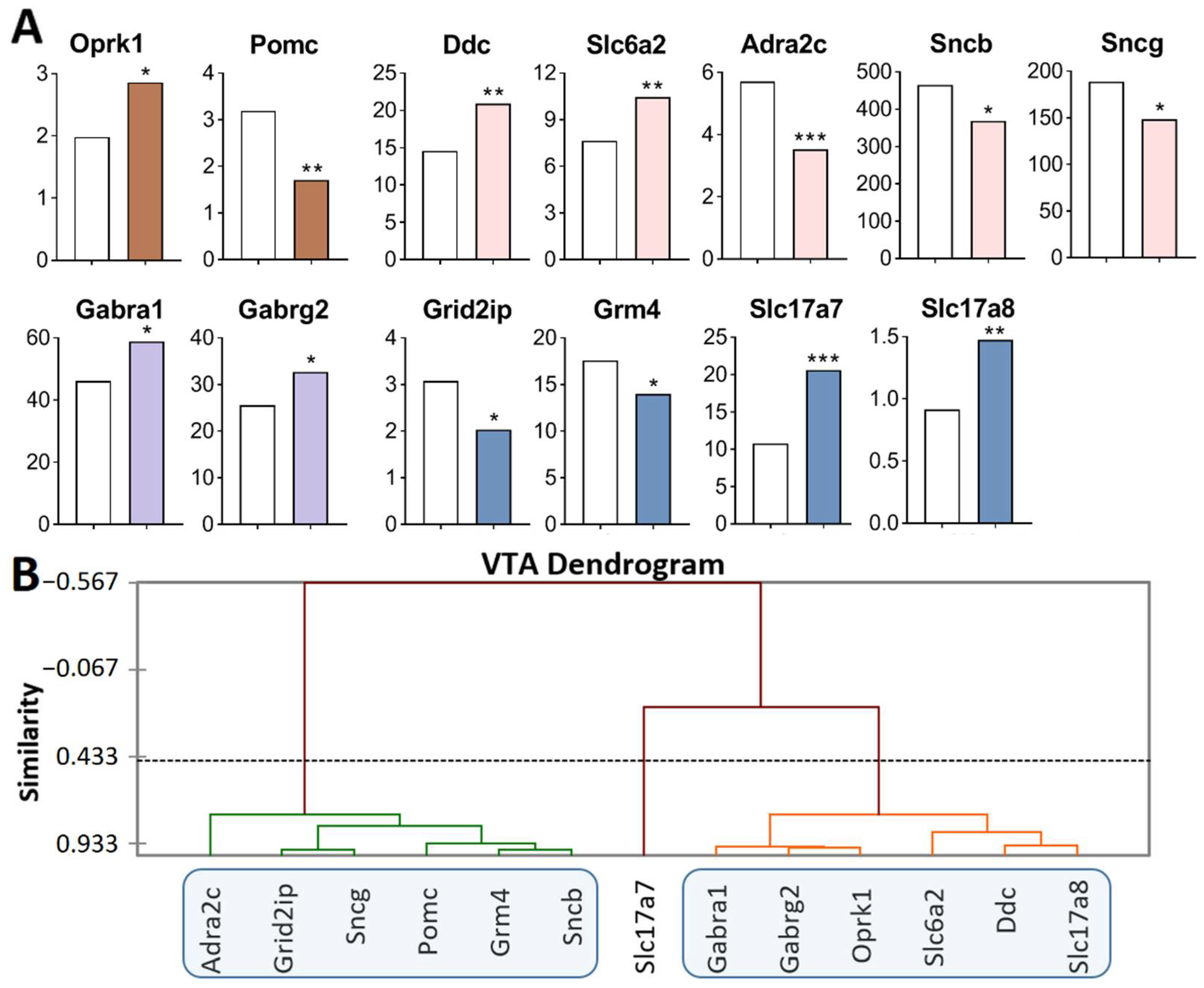
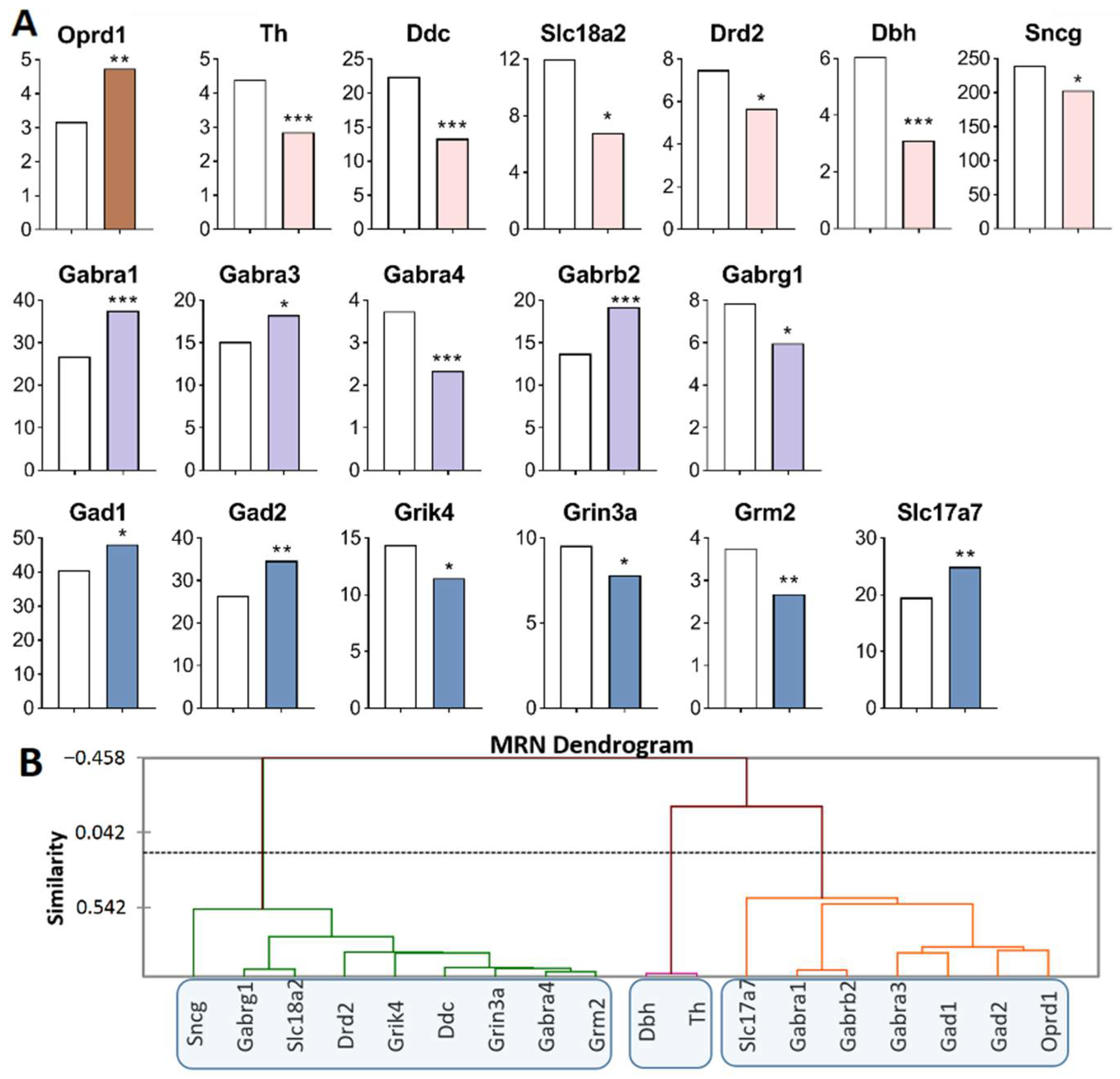
Figure 1.
DEGs in the VTA of highly aggressive mice. (A) FPKM of the DEGs in the VTA (Supplementary Table S1). White bars—controls; colored bars—winners: brown—opioidergic genes; pink—CAergic genes; lilac—GABAergic genes; violet—glutamatergic genes. * p < 0.05; ** p < 0.01; *** p < 0.001. (B) AHC based on 13 reference DEGs’ expression profiles (additional information: Supplementary Table S2). Similarity: Pearson’s correlation coefficient. Agglomeration method: unweighted pair-group average. The main clusters are highlighted with a blue rounded rectangle.
Opioidergic genes: Oprk1 was upregulated (p = 0.01) and Pomc was downregulated (p = 0.002); CAergic genes: Ddc (p = 0.003) and Slc6a2 (p = 0.007) were upregulated; Adra2c (p < 0.001), Sncb (p = 0.045), and Sncg (p = 0.03) were downregulated; GABAergic genes: Gabra1 and Gabrg2 were upregulated (p = 0.035 and 0.025, respectively); Glutamatergic genes: Slc17a7 (p < 0.001, q = 0.02) and Slc17a8 (p = 0.005) were upregulated, and Grm4 (p = 0.039) and Grid2ip (p = 0.013) were downregulated.
Three gene clusters were identified showing relationships among DEGs after repeated aggression (Figure 1B; Supplementary Table S2). The first cluster contains genes whose expression diminished: Pomc, proopiomelanocortin; CAergic genes Adra2c, Sncb, and Sncg; and glutamatergic genes Grm4 and Grid2ip (encoding metabotropic and ionotropic receptors). The expression and number of correlations with other genes (Supplementary Table S3) were the highest for the Sncg gene, which encodes a member of the synuclein family of proteins, which are believed to take part in the pathogenesis of neurodegenerative diseases together with Sncb (GeneCards). It is likely that it is the downregulation of this gene that leads to a decrease in the expression of the genes in this cluster.
The second cluster of genes contains the upregulated opioidergic Oprk1 gene encoding kappa-opioid receptors, CAergic genes Ddc and Slc6a2, glutamatergic gene Slc17a8, and genes Gabra1 and Gabrg2 coding for GABA receptors (Figure 1B, Supplementary Table S2). The expression and number of correlations with other genes (Supplementary Table S3) were the highest for genes Gabrg2 and Oprk1, which correlated with each other too (R = 0.957).
It can be assumed that the glutamatergic system is activated, as evidenced by high expression of genes of vesicular glutamate transporters Slc17a7 and Slc17a8, against the background of downregulation of the metabotropic glutamate Grm4 gene and Grid2ip gene encoding glutamate receptors. In this context, the expression of the gene encoding adrenergic receptor Adra2c declined. The lowest number of correlations was found for Slc17a7, Pomc, Slc6a2, and Adra2c (Supplementary Table S3).
Thus, the analysis of the DEGs of interest (Figure 1A) together with experimental data published by us earlier [17,25] indicates activation of CAergic systems that are accompanied by upregulation of genes encoding the main enzyme for the synthesis (Th gene), a gene coding for DA-DOPA-decarboxylase of aromatic amino acids (Ddc gene), and genes encoding noradrenaline and DA transporters (Slc6a2 and Slc6a3, respectively).
2.2.2. DEGs in the NAcc of Highly Aggressive Mice
It has been shown (Figure 2A, Supplementary Table S1) that: opioidergic genes Pdyn was upregulated (p = 0.022) and Pomc was downregulated (p = 0.027); CAergic genes: Th (p = 0.035) and Drd2 (p = 0.015) were upregulated; GABAergic genes: Gabrq was upregulated (p = 0.02), and Slc6a13 was downregulated (p < 0.001, q = 0.004); Glutamatergic genes: Grin3a was downregulated (p = 0.045), and Slc17a7 was upregulated (p = 0.007).
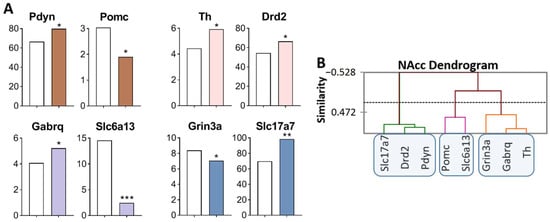
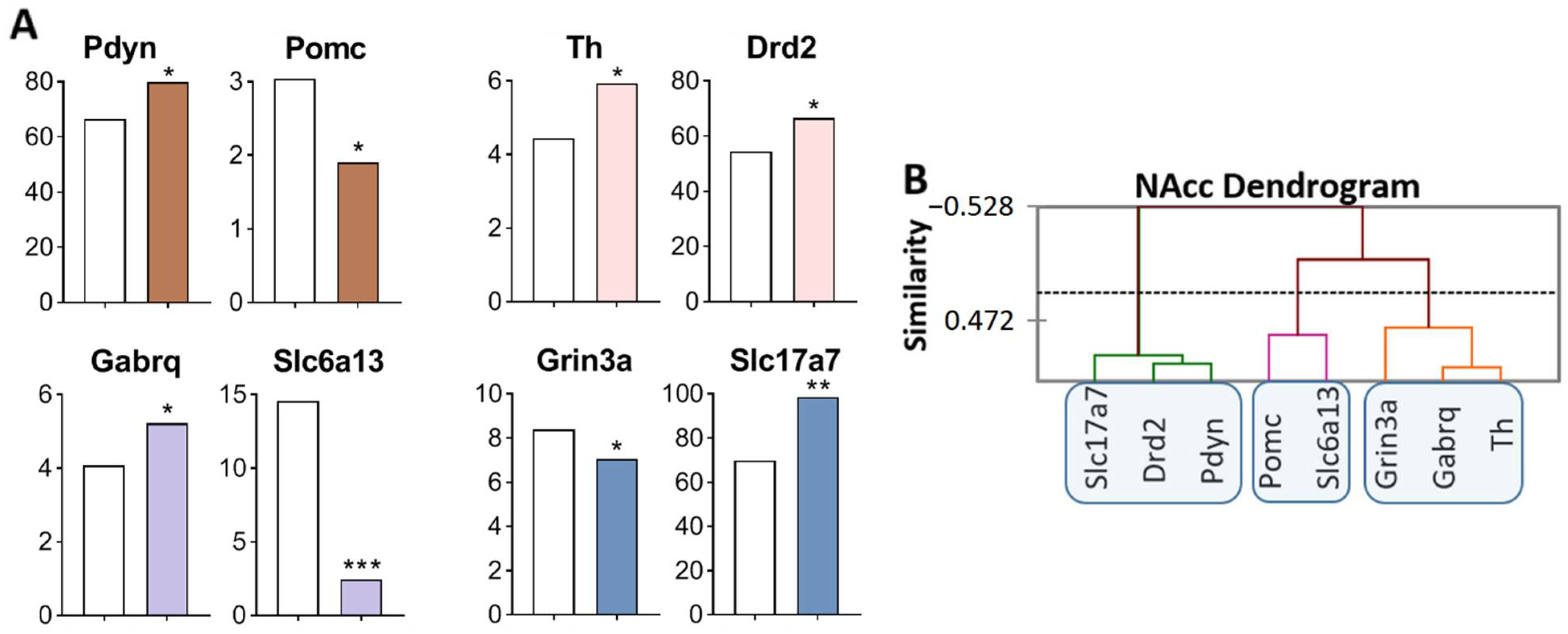
Figure 2.
DEGs in the NAcc of highly aggressive mice. (A) FPKM of the DEGs in the NAcc (Supplementary Table S1). White bars: controls; colored bars—winners: brown—opioidergic genes; pink - CAergic genes; lilac—GABAergic genes; violet—glutamatergic genes. * p < 0.05; ** p < 0.01; *** p < 0.001. (B) AHC based on expression profiles of eight reference DEGs (additional information: Supplementary Table S2). Similarity: Pearson’s correlation coefficient. Agglomeration method: Unweighted pair-group average. The main clusters are highlighted with a blue rounded rectangle.
Three gene clusters were identified showing a relationship among DEGs (Figure 2B). The first cluster contains genes whose expression increased (Slc17a7, Drd2, and Pdyn), and expression of the Drd2 gene positively correlated with that of genes Slc17a7 (R = 0.817) and Pdyn (R = 0.850) and negatively with the expression of Grin3a gene (R = −0.976; Supplementary Table S3). The second cluster comprises genes with decreased expression: Pomc and Slc6a13, encoding proopiomelanocortin and GABA transporter, respectively. There are no correlations in the expression of these genes with other genes. The third cluster combines genes with increased expression: Th and Gabrq (R = 0.876), encoding tyrosine hydroxylase and a GABA receptor, Grin3a, respectively. The observed strong correlation of gene expression levels indicates transcriptionally coordinated processes.
Similarly, with changes in the VTA, we can assume upregulation of dopaminergic genes Th and Drd2, of the opioidergic Pdyn gene, and of the glutamatergic Slc17a7 gene in the NAcc. The Slc17a7 gene codes for a vesicular glutamate transporter (VGLUT1), which mediates the uptake of glutamate into synaptic vesicles at presynaptic nerve terminals of excitatory neural cells and regulates the release of glutamate.
2.2.3. DEGs in the STR of Highly Aggressive Mice
Analysis of the RNA-Seq database of the STR [47] revealed DEGs of interest in this region that participate in rewarding processes (Figure 3A, Supplementary Table S1). Opioidergic genes were downregulated: Opcml (p = 0.034) and Oprk1 (p = 0.042); GABAergic genes encoding GABA receptors were downregulated: Gabra2 (p = 0.024), Gabra3 (p = 0.029), Gabrb2 (p = 0.042), Gabrg2 (p = 0.005), and Gabrg3 (p = 0.049); Glutamatergic transporter gene Slc17a7 was upregulated (p = 0.047).
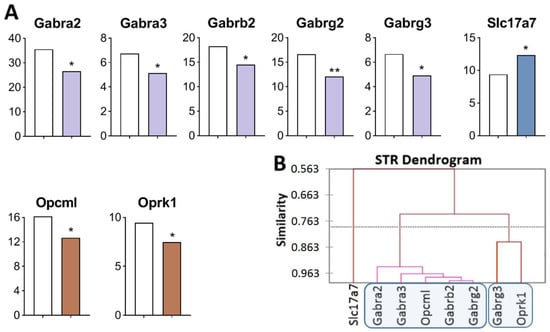
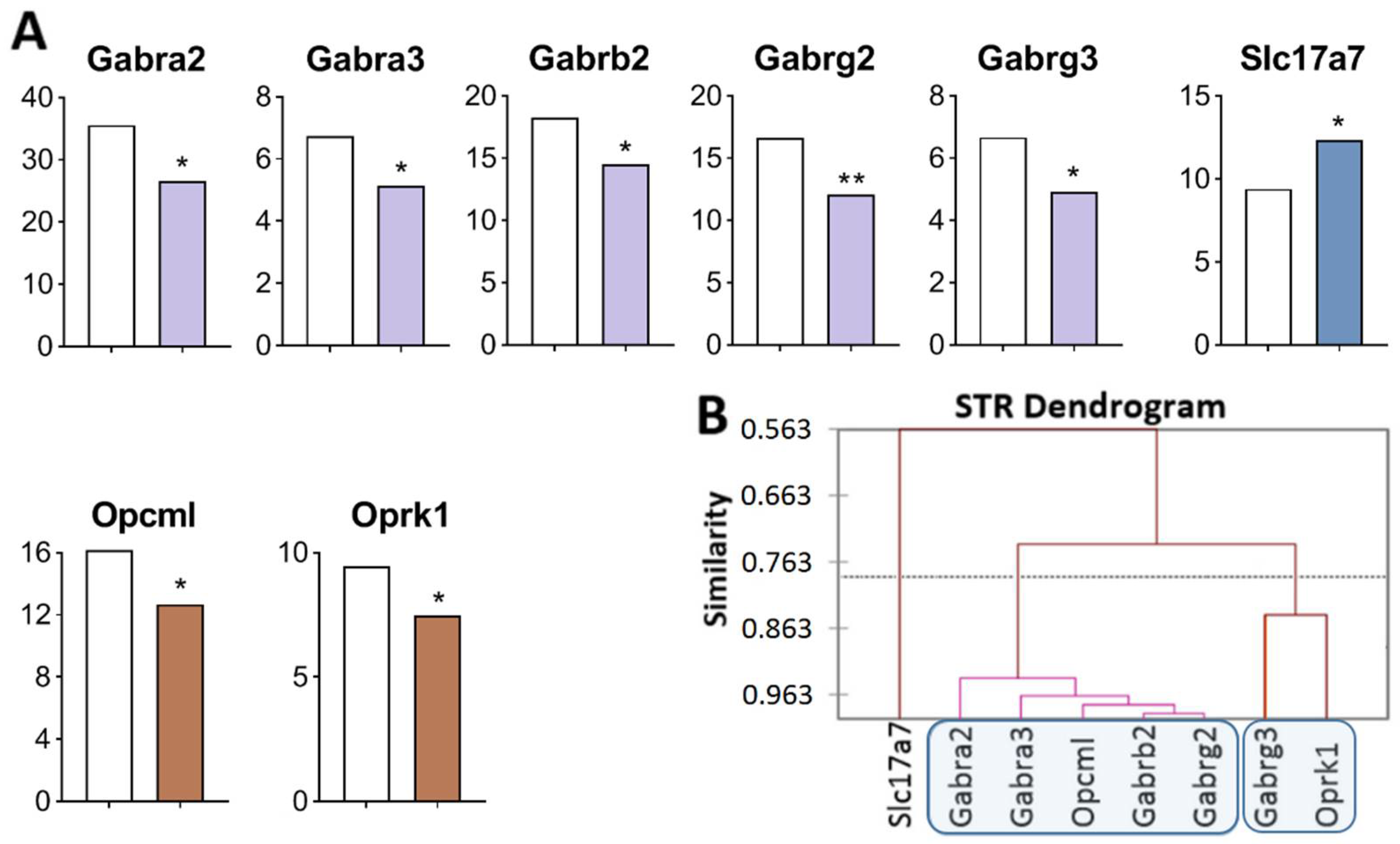
Figure 3.
DEGs in the STR of highly aggressive mice. (A) FPKM of opioidergic, GABAergic, and glutamatergic DEGs in the STR. White bars—controls; colored bars—winners: brown—opioidergic genes; lilac—GABAergic genes; violet—glutamatergic genes. * p < 0.05; ** p < 0.01 (Supplementary Table S1). (B) AHC based on expression profiles of eight reference DEGs (Supplementary Table S2). Similarity: Pearson’s correlation coefficient. Agglomeration method: Unweighted pair-group average. The main clusters are highlighted with a blue rounded rectangle.
Three gene clusters were identified showing a relationship or its absence among DEGs in aggressive mice (Figure 3B; Supplementary Table S2). The first cluster combines genes whose expression declined (Opcml encoding an opioid-binding protein, and genes encoding GABA receptors: Gabra2, Gabra3, Gabrb2, and Gabrg2), and the second cluster is composed of genes Gabrg3 and Oprk1. The third cluster consists of Slc17a7. Expression of all genes of interest was found to be downregulated except for the Slc17a7 gene, a single gene that was upregulated and did not correlate with other genes.
The largest number of correlations was found (Supplementary Table S3) between the Opcml gene (encoding an opioid-binding protein) and genes encoding GABA receptors: Gabra2 (R = 0.972), Gabra3 (R = 0.938); Gabrb2 (R = 0.974), Gabrg2 (R = 0.983,), and Gabrg3 (R = 0.826). The Gabra2 gene manifested the largest number of correlations.
2.2.4. DEGs in the PFC of Highly Aggressive Mice
In comparison with the control, opioidergic genes were upregulated (Figure 4A, Supplementary Table S1): Oprd1 (p = 0.028), encoding delta opioid receptors, and Penk (p < 0.001), encoding pentapeptide opioids Met-enkephalin and Leu-enkephalin. Glutamatergic genes: Grid2ip (p = 0.041) encoding glutamate receptor was upregulated, whereas Slc17a8 (p = 0.032) and Gad1 (p = 0.044), encoding glutamate transporter and glutamate decarboxylase, respectively, were downregulated.
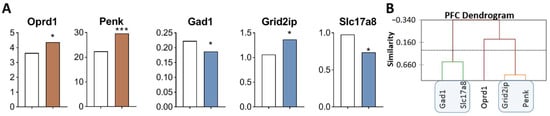
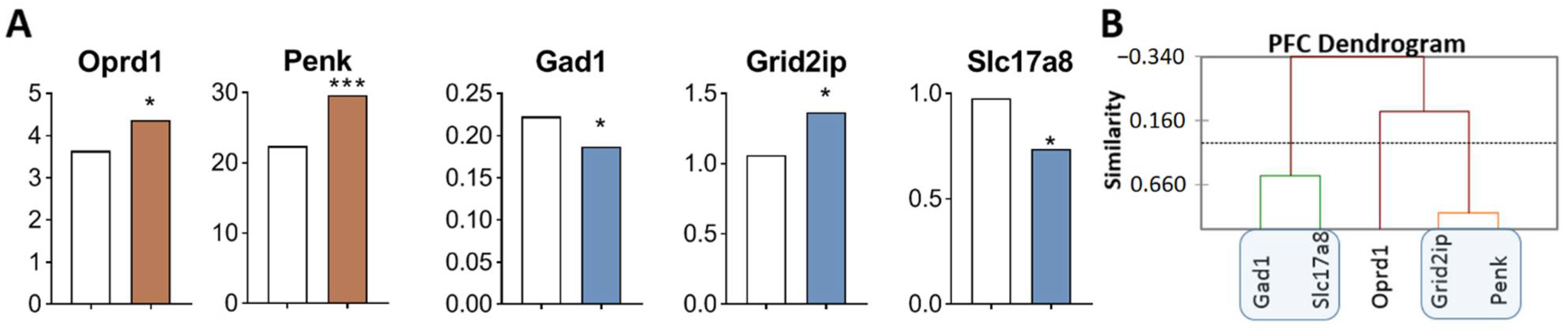
Figure 4.
DEGs in the PFC of highly aggressive mice. (A) FPKM of opioidergic and glutamatergic DEGs in the PFC of mice. White bars—controls; colored bars—winners: brown—opioidergic genes; violet—glutamatergic genes (Supplementary Table S1). * p < 0.05; *** p < 0.001. (B) AHC based on five reference DEGs’ expression profiles (Supplementary Table S2). Similarity: Pearson’s correlation coefficient. Agglomeration method: Unweighted pair-group average. The main clusters are highlighted with a blue rounded rectangle.
Three gene clusters were identified showing a relationship or its absence among DEGs in aggressive mice (Figure 4B; Supplementary Table S2). The first cluster combines genes whose expression declined (Gad1 and Slc17a8). The second cluster combines Grid2ip and Penk genes. The third cluster consists of Oprd1 gene. Expression of genes of second and third clusters was found to be upregulated.
Correlations of expression levels were found between the Grid2ip gene and Penk (R = 0.873) and Slc17a8 (R = −0.934) (Supplementary Table S3).
2.2.5. DEGs in the MRNs of Highly Aggressive Mice
Analysis of the RNA-Seq database of MRNs [48] revealed the largest number of DEGs of interest in this main region processes (Figure 5A, Supplementary Table S1). Opioidergic gene Oprd1 was upregulated (p = 0.009) and CAergic genes were downregulated: Th (p < 0.001; q = 0.035), Ddc (p < 0.001; q = 0.005), Dbh (p < 0.001; q = 0.005), Sncg (p < 0.046), Drd2 (p = 0.03), and Slc18a2 (p = 0.04). GABAergic genes encoding GABA receptors—Gabra1 (p < 0.001; q = 0.005), Gabrb2 (p < 0.001; q = 0.005), and Gabra3 (p = 0.024)—were upregulated, whereas Gabrg1 (p = 0.043) and Gabra4 (p < 0.001; q = 0.005) were downregulated. Glutamatergic genes: Grm2 (p = 0.003), Grin3a (p = 0.035), and Grik4 (p = 0.011) were downregulated, while Slc17a7 (p = 0.004), Gad1 (p < 0.032), and Gad2 (p = 0.001, q = 0.05) were upregulated.
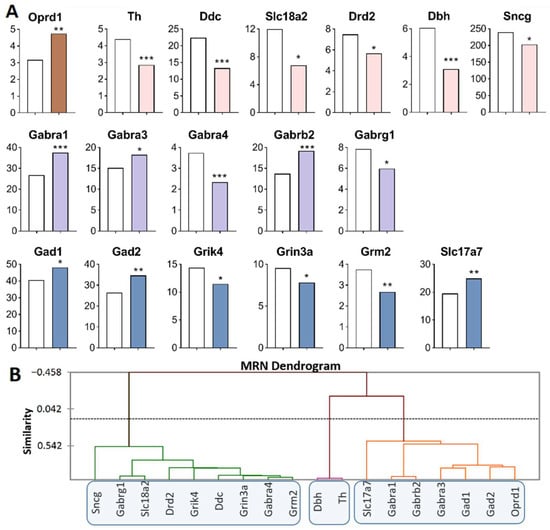
Figure 5.
DEGs in the MRNs of highly aggressive mice. (A) FPKM of opioidergic, CAergic, GABAergic and glutamatergic DEGs in the MRNs (Supplementary Table S1). White bars—controls; colored bars—winners: brown—opioidergic genes; pink—CAergic genes; lilac—GABAergic genes; violet—glutamatergic genes. * p < 0.05; ** p < 0.01; *** p < 0.001. (B) AHC based on expression profiles of 18 reference DEGs (Supplementary Table S2). Similarity: Pearson’s correlation coefficient. Agglomeration method: Unweighted pair-group average. The main clusters are highlighted with a blue rounded rectangle.
Three gene clusters were identified showing a relationship among DEGs in aggressive mice (Figure 5B; Supplementary Table S2). The first cluster combines genes whose expression declined (Sncg, Drd2, Ddc, Slc18a2, Gabrg1, Gabra4, Grik4, Grin3a and Grm2 genes), and the second cluster (Th and Dbh). The third cluster is composed of upregulated genes Slc17a7, Gabra1, Gabrb2, Gabra3, Gad1, Gad2, and Oprd1.
Our finding suggested that the Slc18a2 gene is associated with a greater risk of opioid dependence [49]. The expression of Oprd1 positively correlated with that of Gad2 (R = 0.829) and negatively with the expression of Grik4 (R = −0.814) in our experiment (Supplementary Table S3). The largest number of correlations with expression levels of various genes in the MRNs belongs to the Grik4 gene: correlations with Ddc (R = 0.862), Drd2 (R = 0.819), Gabra1 (R = −0.828), Gabra3 (R = −0.963), Gabra4 (R = 0.897), Gad2 (R = −0.827), Grin3a (R = 0.846), and Oprd1 (R = −0.814). A large number of expression correlations were detected for Gabra4: with Ddc (R = 0.959), Drd2 (R = 0.887), Gabra3 (R = −0.880), Grik4 (R = 0.897), Grin3a (R = 0.966), Grm2 (R = 0.970), and Slc18a2 (R = 0.887) (Supplementary Table S3).
In the MRNs, genes encoding proteins that are involved in neurotransmitter systems—serotonergic [50] and dopaminergic—were downregulated, possibly as consequence of under expression of Ddc and Slc18a2, coding for the synaptic vesicle monoamine transporter common for the regulation of both systems (Figure 5A).
Table 2 presents the full list of DEGs in brain regions of the mice with positive fighting experience.

Table 2.
DEGs in brain regions of the mice with positive fighting experience.
2.3. PCA Based on CAergic and Opioidergic DEGs’ Expression Profiles in Brain Regions of Highly Aggressive Mice
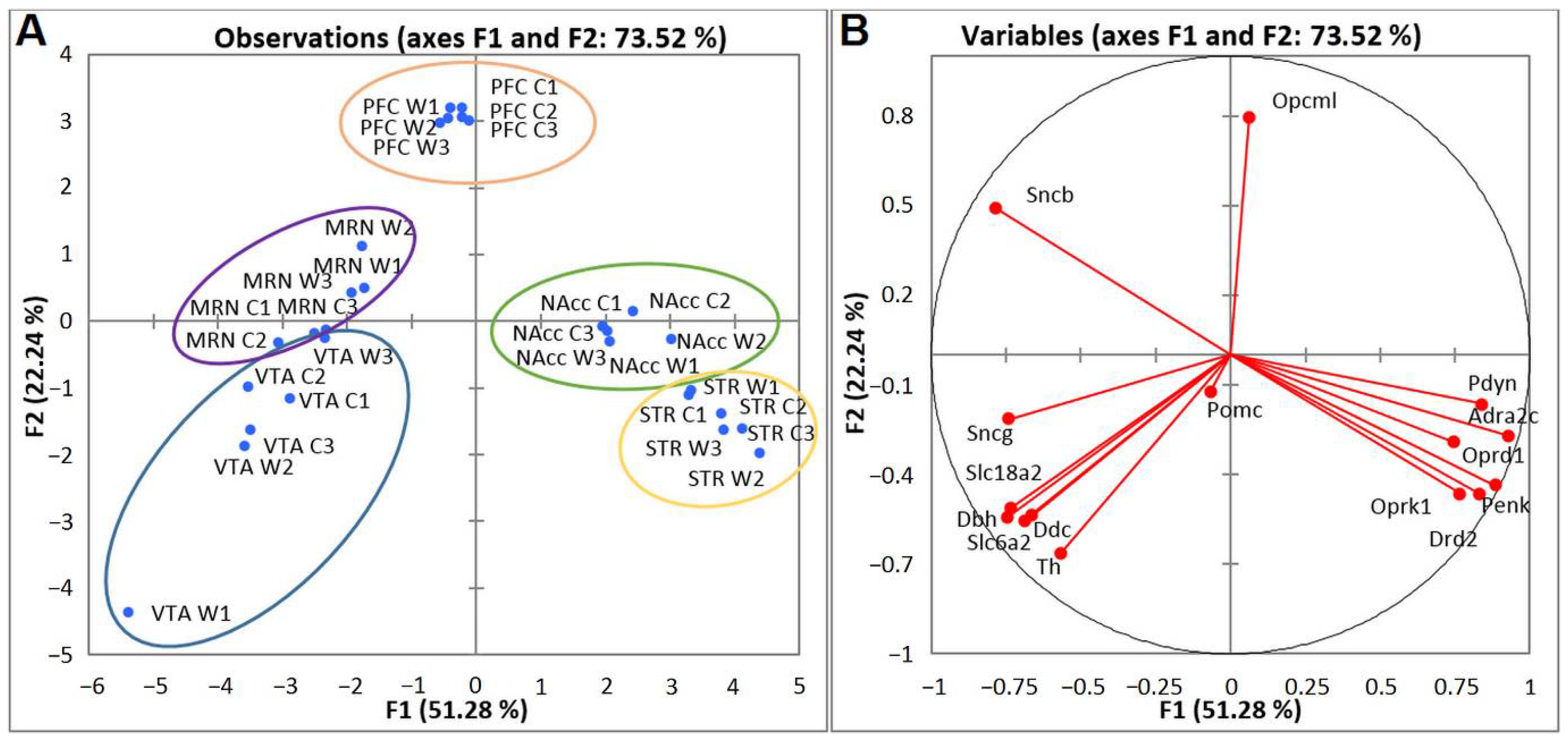
We next analyzed DEGs encoding proteins participating in the functioning of the main neurotransmitter systems of addiction—CAergic and opioidergic—in mice with positive fighting experience. Compact clustering of samples in the NAcc, STR, PFC, VTA, and MRNs on the basis of gene expression profiles can be observed (Figure 6A). Within the circled area, one can see the compact clustering of aggressive vs. control mice; thus, distinct expression patterns of the analyzed genes in each brain region and distinct clustering for the five brain regions were revealed. All genes in the blue cluster are CAergic and have much higher FPKM values in the VTA and MRNs. Most of the genes in the yellow cluster are opioidergic (except Adra2c and Drd2) and are much more strongly expressed in the STR and NAcc.
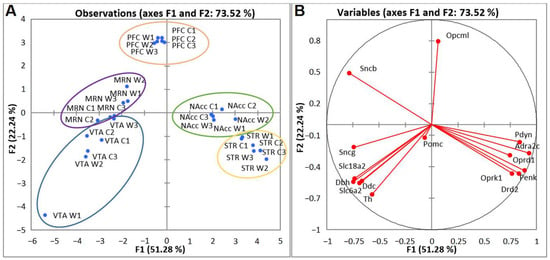
Figure 6.
PCA plots based on covariation of genes on the basis of expression profiles of 15 CAergic and opioidergic DEGs across 30 samples, which comprise RNA-Seq FPKM data for five brain regions. (A) Active observations. W1, W2, and W3: winners; C1, C2, and C3: controls; VTA: ventral tegmental area, NAcc: nucleus accumbens, MRN: midbrain raphe nuclei, STR: dorsal striatum, and PFC: prefrontal cortex. Ovals denote brain regions. (B) Active variables. The graph illustrates distinct clustering of DEGs.
PCA biplot analysis based on covariation of the gene expression profiles of six samples for each brain region (Figure 6B) uncovered distinct intergroup clustering of opioidergic and CAergic DEGs. Graphs represent the correlating clusters of DEGs directed oppositely, which means elevated and low levels of gene transcription.
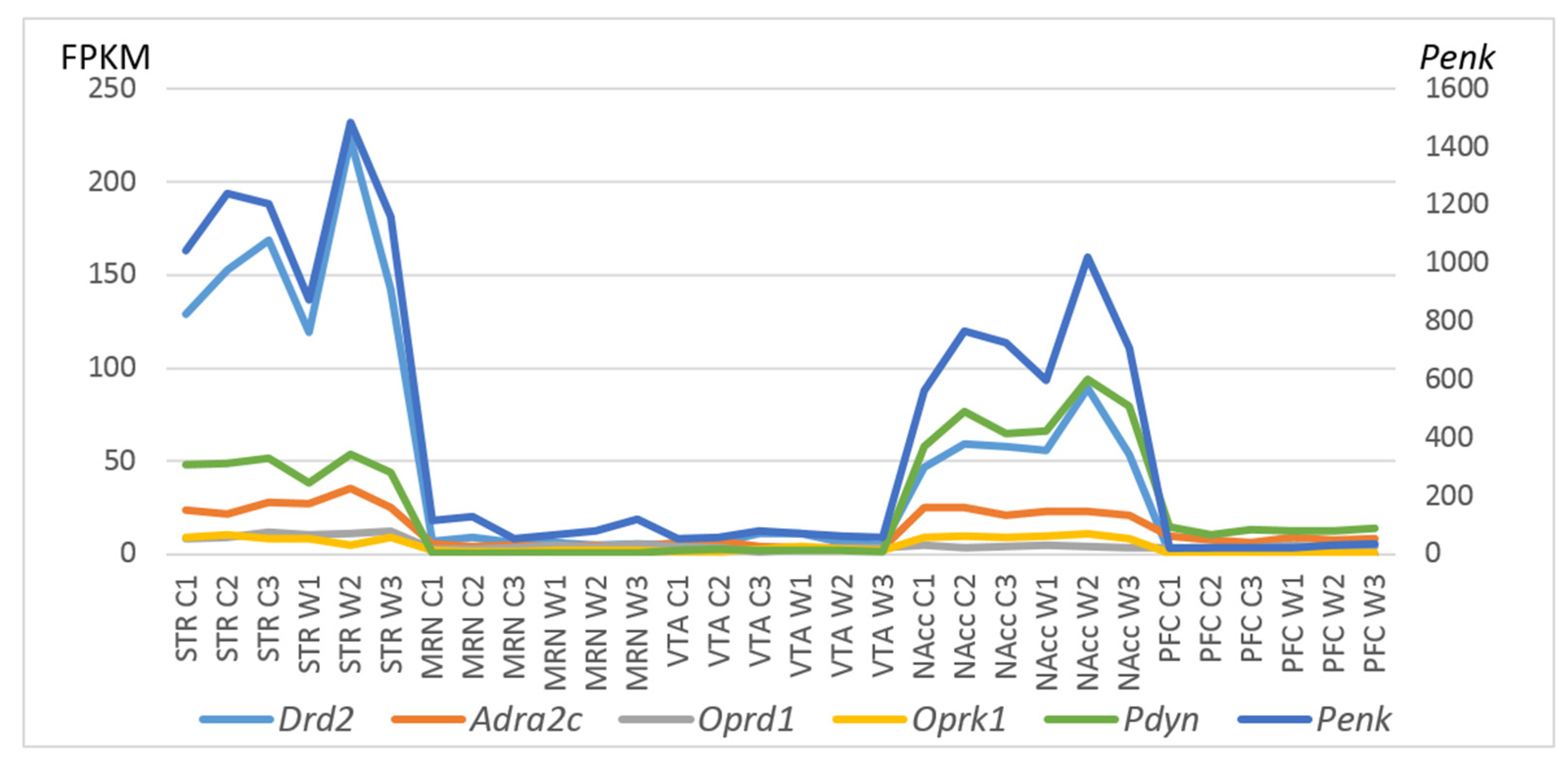
To gain further insight into CAergic–opioidergic interactions, we found the most overexpressed neuron-specific genes in the brain regions under study (Figure 7). Our genes of interest were the CA-specific Adra2c and Drd2 gene and opioid-associated genes Oprd1, Oprk1, Pdyn, and Penk for comparison, as presented in Figure 7. Both Drd2 and Penk in the STR and Penk, Pdyn, and Drd2 in the NAcc are the most highly expressed DEGs coding neuron-specific genes. These data suggest that all the main correlating events concerning the development of an addiction-like state take place in the STR and NAcc.
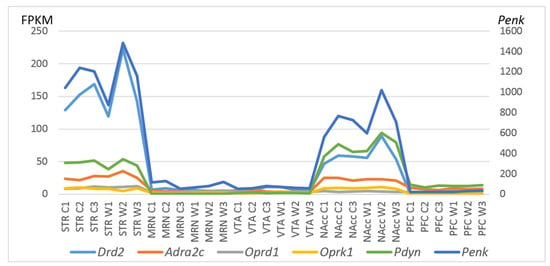
Figure 7.
FPKM values of five CAergic- and opioid-specific DEGs across 30 samples on the basis of the included RNA-Seq FPKM data for five brain regions. C1, C2, and C3: controls; W1, W2, and W3: winners; VTA: ventral tegmental area, NAcc: nucleus accumbens, MRN: midbrain raphe nuclei, STR: dorsal striatum, and PFC: prefrontal cortex.
3. Discussion
It is well known that any positive and long rewarding processes that are accompanied by positive emotions can lead to an addiction-like state, for example, drug abuse, sexual abuse, gaming abuse, etc. Earlier experimental data have led to the conclusion that aggression is rewarding and strongly influenced by experience of aggression: any positive reinforcement in agonistic interactions can cause a propensity to behave aggressively [28,51,52,53,54,55,56,57]. Our data revealed that repeated positive fighting experience in daily agonistic interactions leads to addiction-like signs in mice [29,31] according to criteria proposed for drug dependence and addictive states [58].
In this work and in our previous studies [25,26,45,46,47,48,59], we focus on DEGs encoding proteins that are related to reward-associated neurotransmitters in the brain: opioid-, catecholamine-, GABA-, and glutamatergic systems, which partake in the regulation of aggression in the VTA, STR, and NAcc (which are considered the main regions implicated in aggression stimulation) and in the MRNs and PFC, which as a rule, perform inhibitory functions toward aggressive behavior [6].
It has been shown earlier that repeated positive fighting experience is accompanied by overexpression of genes Th, Slc6a3, and Snca in the VTA [25,26]. Our RNA-seq data additionally revealed upregulation of the Ddc gene, encoding an enzyme of monoamine synthesis, and the Slc6a2 gene, encoding a noradrenaline transporter, and upregulation of glutamatergic genes Slc17a7 and Slc17a8, encoding vesicular glutamate transporters. Correlations were found between the expression of Oprk1 encoding opioid receptor kappa 1 (OPRK1) and Ddc and Slc17a8 expression levels (Supplementary Table S3). OPRK1’s function is connected with receptors of endogenous ligands, such as α-neoendorphins and dynorphins, as well as receptors of various synthetic opioids. According to the GeneCard database, diseases associated with the OPRK1 protein include morphine and alcohol dependence (https://www.genecards.org/cgi-bin/carddisp.pl?gene=OPRK1 (accessed on 3 July 2021).
Expression levels of vesicular glutamate transporters SLC17A6 and SLC17A7 are robustly raised by smoking: an effect that is reduced by alcohol coexposure [60]. Those authors proposed that glutamatergic transmission is crucial for the control of the VTA, and that enduring plasticity within the VTA may be a major molecular mechanism for the maintenance, for example, of smoking and alcohol addiction. DA- and cAMP-regulated neuronal phosphoprotein PPP1R1B and vesicular glutamate transporter SLC17A7 may be molecular targets for the treatment of substance abuse [61]. Thus, our findings about upregulation of Slc17a7 in the VTA, NAcc, STR, and MRNs are consistent with the involvement of this DEG in the development of an addiction-like state, in our case, induced by positive fighting experience accompanied by wins in daily agonistic interactions.
In the NAcc of the winners, there were revealed upregulation of dopaminergic genes (Th and Drd2) concurrently with upregulation of glutamatergic gene Slc17a7 and of opioid-related gene Pdyn (prodynorphin, DYN). Enhanced dopaminergic and glutamatergic signaling in the NAcc are thought to be hallmarks of addiction. Structural and functional alterations in dendritic MSNs within this region and their dopaminergic projections from the VTA are believed to facilitate addiction [62].
According to GeneCards data, the protein encoded by Pdyn gene is a preproprotein that is processed into secreted opioid peptides β-neoendorphin, dynorphin, leu-enkephalin, rimorphin, and leumorphin. These peptides are ligands for the kappa opioid receptor (KOR), and an association has been shown between prodynorphin gene polymorphisms and opioid dependence susceptibility [63]. It has been suggested that dysregulation of the DYN/KOR system and of DA signaling through both alterations in co-expression patterns of opioid genes and decreased Drd1 mRNA expression can contribute to an imbalance in the activity of D1- and D2-containing pathways thereby possibly causing a negative affective state in human alcoholics [64]. In contrast to these data, in this study, we found upregulation of Drd2 but not Drd1 mRNA expression in the winners. PDYN binds to κ-opioid receptors encoded by the Oprk gene and is known to regulate dopaminergic tone, making this system important for drug addiction. The DYN/KOR system, as supposed by the authors of ref. [65], is a powerful effector of stress-induced alterations in reward processing and dysphoric states. Thus, upregulation of genes Th, Drd2, Pdyn, and Slc17a7 in the NAcc and of Oprk1 in the VTA of mice with positive fighting experience is consistent with data indicating that this system is implicated in addiction.
NAcc pathways control cell-specific gene expression in the PFC [66], which is highly interconnected with other brain regions, including extensive connections with subcortical and other cortical structures, notably the thalamus, basal ganglia, hypothalamus, amygdala, and hippocampus, among others [67]. Several neurotransmitter systems are represented in the PFC, in particular dopaminergic, glutamatergic, and cholinergic ones [68,69]. Additionally, glutamatergic neurons from the PFC project to MSNs, providing an enriched target for the regulation of synaptic plasticity.
According to some authors [70,71], the VTA sends glutamate to the PFC. We can suppose the activation of opioid genes Oprd1 and Penk (proenkephalin), which are paralogs of the Pdyn gene. The Oprd1 gene encodes opioid receptor delta 1 with functions as a receptor for endogenous enkephalins and for a subset of other opioids. OPRD1 plays a role in the development of analgesic tolerance to morphine. The Penk gene codes for proteins, including pentapeptide opioids Met-enkephalin and to a lesser extent Leu-enkephalin, that are stored in synaptic vesicles and then released into the synapse, where they bind to mu- and delta-opioid receptors. PENK, according to the GeneCards database (https://www.genecards.org/cgi-bin/carddisp.pl?gene=PENK (accessed on 3 July 2021), participates in morphine, heroin, cannabis, and alcohol dependence. Diseases associated with PENK also include cannabis dependence. We found a significant correlation in expression levels between Penk and Grid2ip encoding glutamate receptor genes. There were no GABAergic and CAergic DEGs in the PFC.
Our earlier RT-PCR study has revealed under expression of the Th gene and overexpression of the Drd4 gene in the STR [72]. Moreover, the development of hyperactivity in the winners after long experience of aggression presented in our study may be induced by decreased activity of GABAergic systems, judging by downregulation of Gabra2, Gabra3, Gabrb2, Gabrg2, and Gabrg3 encoding GABA receptors and upregulation of the Slc17a7 gene, implicated in addiction.
The inhibitory influence of the MRNs on aggression supposedly is disturbed during repeated aggression because there is an overall under expression of serotonergic genes -Tph2 and Ddc encoding proteins, participating in the synthesis 5-HT, its transporters (Slc6a4 and Slc18a2), and receptors (Htr2a, Htr3a, and Htr5b) [50] and of dopaminergic genes—Th, Comt, Ddc, Dbh, Drd2, Slc18a2, and Sncg—encoding proteins involved in the synthesis and workings of receptors (our current data). This downregulation may be a consequence of inhibition of the neurotransmitter release because of upregulation of the Oprd1 gene, which is associated with glutamatergic transporters, as a result of overexpression of the Slc17a7 gene.
Our data are consistent with other studies, which indicate that GABAergic and glutamatergic systems are deeply involved in the development of addiction, for example, to amphetamine [73], alcohol, and cocaine [38]. Alcoholics and cocaine addicts show upregulation of three genes relative to controls: Gria4, Grik3, and Grm4. Expression of both Grm3 and Grin2d is high in alcoholics and low in cocaine addicts. These observations suggest that glutamate input into dorsal raphe nuclei is enhanced during escalated aggression, which causes a phasic increase in the 5-HT release from dorsal raphe nuclei 5-HT neurons into the NAcc and inhibits the reinforcing and motivational effects of cocaine, heroin, and ethanol [4].
Thus, our neurotranscriptomic data confirm the development of an addiction-like state during positive fighting experience in the winner. Some KEGG terms turned out to be significantly related to the DEGs associated with an addiction-like state in the VTA, NAcc, STR, MRNs, and PFC of the winners. These terms are nicotine, retrograde endocannabinoid signaling, morphine, cocaine, amphetamine, and alcoholism addiction (Table 3). This study shows that the DEGs that are implicated in the development of opioid dependence and addiction in the winners are Oprk1 in the VTA (upregulation) and STR (downregulation); Oprd1 (upregulation) in MRNs and Oprd1 and Penk (upregulation) in the PFC; Pdyn gene (upregulation) in the NAcc as well as Th, Ddc, Slc6a2, Slc6a3, and Snca in the VTA (upregulation) and Drd2 and Pdyn (upregulation) in the NAcc. As markers of changed function of glutamatergic and GABAergic systems in the STR, NAcc, and VTA, we registered overexpression of Slc17a7 and Slc17a8 in the VTA and the expression of genes involved in synthesis (except for the STR) of DA. In the MRNs, the expression of dopaminergic and serotonergic genes proved to be reduced or unchanged, but in both regions, the Oprd1 gene is upregulated, and the PFC is activated by the Penk gene.

Table 3.
KEGG terms related to the DEGs in the VTA, NAcc, STR, MRNs, and PFC.
Indirectly, all these genes and proteins encoded by them may induce neurotranscriptomic changes in the synaptic vesicle cycle, taste transduction, neuroactive ligand-receptor interaction, tyrosine metabolism, and glutamatergic, GABAergic, and dopaminergic synapses, or vice versa in some cases (Table 3).
Directly, our data prove changes in the expression of opioidergic genes which, according to GeneCards and MalaCards databases (https://www.malacards.org/card/opiate_dependence (accessed on 3 July 2021)), code for proteins implicated in opioid dependence and addiction: that is, OPRM1 (opioid dependence); OPRM1, OPRK1, OPRD1, and DRD2 (opioid addiction); PENK, PDYN, OPRM1, OPRK1, and OPRD1 (morphine dependence); and PDYN, OPRM1, OPRD1, and DRD2 (heroin dependence). The most studied candidate genes have included mu-opioid receptor (OPRM1), delta-opioid receptor (OPRD1), and dopamine D2 receptor (DRD2). Variants in these genes have been associated with relatively small but reproducible effects on addiction risk [74]. All these data support our notion of the development of an addiction-like state as a result of positive fighting experience in daily agonistic interactions in mice.
The term “addiction” describes a progressive loss of behavioral control leading to dependence on a drug and inability to stop without adverse consequences (American Psychiatric Association, 1994). All abused substances, tend to foster increasing use, threatening to cause dependence. This suggests that all reward stimuli have common properties that both depend on and contribute to altered functioning of the central nervous system. Replacing the word “drugs” with “aggression”, we can say that this quote also applies to the state induced by repeated positive fighting experience.
4. Methods and Materials
4.1. Animals
The experiment was carried out using 10–12-week-old C57BL/6J male mice. The mice were kept in the Conventional Vivarium (Federal Research Center Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia) under standard conditions at 22 ± 2 °C on a 12/12 h light–dark cycle (lights on at 8:00 AM) with dry laboratory feed and water available ad libitum. The mice lived in groups of 8–10 in plastic cages (36 × 23 × 12 cm). All procedures were carried out in compliance with the international regulations for animal experiments (Directive 2010/63/EU of the European Parliament and of the Council on the Protection of Animals Used for Scientific Purposes). The protocol for the study was approved by Scientific Council No. 9 of the Institute of Cytology and Genetics, SB RAS, of 24 March 2010, N 613 (Novosibirsk, http://spf.bionet.nsc.ru/ (accessed on 24 March 2010)).
4.2. The Sensory Contact Model for the Development of Pathological Aggression in Mice
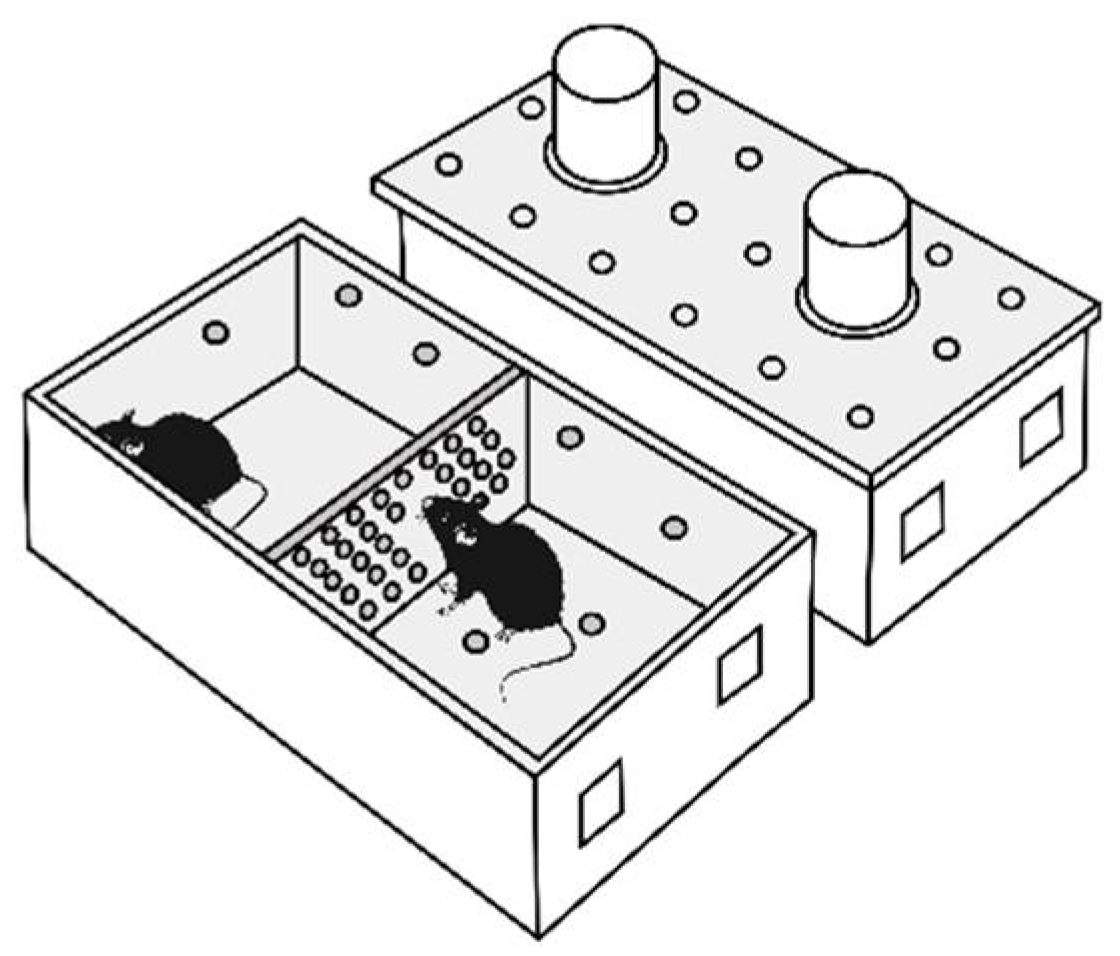
Repeated positive fighting experience, i.e., wins, in male mice were induced by daily agonistic interactions [43,44] in chronic social conflicts. Pairs of animals were each placed in a cage (28 × 14 × 10 cm) bisected by a transparent perforated partition allowing the animals to hear, see, and smell each other but preventing physical contact (Figure 8). The animals were left undisturbed for 2 days to adapt to the new housing conditions and sensory contact before they were exposed to agonistic encounters.
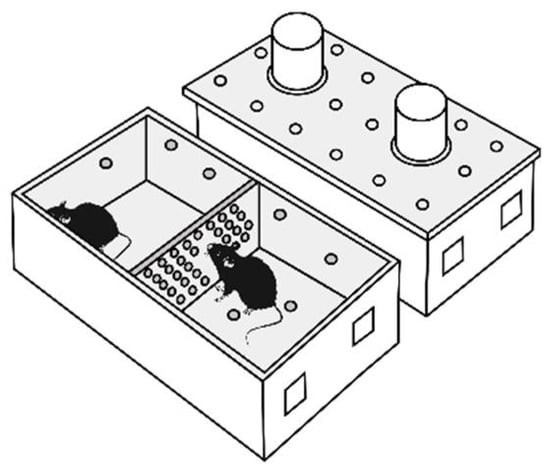
Figure 8.
The experimental cage.
Every afternoon (14:00–17:00 p.m. local time) the cage cover was replaced by a transparent one, and 5 min later (the period necessary for activation), the partition was removed for 10 min to encourage agonistic interactions. The superiority of one of the mice was established within two or three encounters with the same opponent. The superior (winning) mouse would be chasing, biting, and attacking another, who would be demonstrating only defensive behavior (e.g., upright or sideways postures and withdrawal). To prevent physical damage to the defeated mice, the aggressive interactions between males were discontinued by lowering the partition if the strong attacking behavior had lasted for 3 min (in some cases less). Each defeated mouse (loser) was exposed to the same winner for 3 days, whereas afterwards, each loser was placed after the fight in an unfamiliar cage with an unfamiliar winning partner behind the partition. Each aggressive mouse (winners, aggressive mice) remained in its own cage. This procedure was performed once a day for 20 days.
Two groups of animals were used in this study: (1) controls, i.e., mice without consecutive experiences of agonistic interactions; (2) winners, i.e., chronically aggressive mice. The winners with the most pronounced behavioral aggressive phenotypes were selected for the analysis. They demonstrated the largest number, the longest total attacking time, and hostile behavior during the 20-day experiment (Table 1). At 24 h after the last agonistic interactions, the control animals and the aggressive mice were simultaneously decapitated. Brain regions were dissected by the same experimenter according to the Allen Mouse Brain Atlas map [http://mouse.brain-map.org/static/atlas (accessed on 20 April, 2021)]. All tissue samples were placed in the RNAlater solution (Life Technologies, Waltham, MA, USA) and were stored at −70 °C until sequencing.
4.3. Analysis of the Winners’ Behavior during Agonistic Confrontation
Approximately 20–40% of C57BL/6J males displayed strong uncontrolled pathological aggression after the 20-day aggression experience. For analysis, only the winners demonstrating the most pronounced offensive aggression were selected.
Video recordings were used to describe the behavior of the winners in detail during the agonistic interactions. The Observer XT software (version 7.0; Noldus Information Technology, the Netherlands) was employed for manual registration of the behavioral indicators during the test. The following types of attacking behavior were analyzed: attacks, biting, and chasing a partner in agonistic interactions during 10 min, and the following parameters were measured: (1) latency to the first attack (seconds, s); (2) total time of attacks (s); (3) the number of attacks; (4) average time of one attack (total time/number) (s); (5) digging: here it means digging up and scattering the sawdust on the loser’s territory (kick digging or push digging the sawdust forward or backward by forepaws or hind paws): total digging time (s) and the number of diggings; (6) hostile behavior: the total time of attacking and digging behavior. All these activities were aimed at inflicting physical or psychological damage on the conspecific. Besides, stereotypic behaviors (jerks and unmotivated long self-grooming) were registered as parameters of pathological states.
4.4. High-Throughput RNA Sequencing (RNA-Seq)
The collected brain samples were sequenced at JSC Genoanalytica (www.genoanalytica.ru, Moscow, Russia) (accessed on 21 November 2017), and the mRNA was extracted using the Dynabeads mRNA Purification Kit (Ambion, Thermo Fisher Scientific, Waltham, MA, USA). cDNA libraries were constructed by means of the NEBNext mRNA Library PrepReagent Set for Illumina (New England Biolabs, Ipswich, MA USA) and were subjected to Illumina sequencing. More than 20 million reads were obtained from each sample. The resulting “fastq” format files were used to align all reads to the GRCm38.p3 reference genome in the TopHat aligner [75]. The Cufflinks software was utilized to estimate gene expression levels in FPKM (fragments per kilobase of transcript per million mapped reads) and to subsequently identify differentially expressed genes (DEGs) in the winner group and control group. Each brain area was analyzed in three versus three animals. Genes were considered differentially expressed at p ≤ 0.05, and these data were corrected for multiple comparisons at q < 0.05.
DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov (accessed on 20 July 2021) was used to describe DEGs’ functional relations and Gene Ontology terms. The Human Gene Database (http://www.genecards.org/, (accessed on 20 July 2021), Online Mendelian Inheritance in Man database (http://omim.org/, (accessed on 20 July 2021), and a human disease database (MalaCards, http://www.malacards.org, (accessed on 20 July 2021) were employed for the description and analysis of the data obtained.
4.5. Brain Regions That Are Responsible for the Regulation of Aggression and Development of Addiction during Reward
The VTA, which is a critical brain region involved in reward processes and processes of dependence on drugs, such as cocaine, heroin, and ethanol, contains 55–65% of dopaminergic cell bodies [70,76,77,78]. DA is a major neurotransmitter participating in the integration of afferent signals with inhibitory or excitatory inputs, and the VTA gives rise to the dopaminergic mesolimbic and mesocortical pathways that project to the NAcc and PFC, respectively [70,76,79], play an important part in the mediation of rewarding processes and are associated with many types of social behavior. VTA DA-releasing neurons are heterogeneous in their afferent and efferent connectivity and, in some cases, release GABA or glutamate in addition to DA [70,71]. DA is a key neurotransmitter in reward circuitry and reward-guided learning. Physiological activity of GABAergic and cholinergic interneurons is regulated by dopaminergic transmission in a complex manner [80]. It has been suggested that the VTA can act as a hub combining and integrating multimodal signals that contains dopaminergic neurons and transmits excitation to other regions with the help of DA [81], particularly, to the regions we are interested in.
The NAcc is a principal target of VTA dopaminergic neurons. Most of the neurons in the NAcc are GABAergic medium spiny neurons (MSNs), which express D1-type or D2-type DA receptors [82,83]; ~1–2% are cholinergic interneurons and another 1–2% are GABAergic interneurons. GABA is the predominant neurotransmitter in the NAcc, and GABA receptors are abundant there [84]. GABAergic MSNs play an important role in the processing of reward stimuli [85], and they are regulated by DA from the VTA. The VTA–NAcc circuit is a key detector of rewarding stimuli. This area is considered a critical brain region involved in reward and drug dependence processes [4,86]. Structural and functional alterations in MSNs within the NAcc and its dopaminergic projections from the VTA are believed to facilitate these behavioral sequelae related to neuroadaptations in NAcc MSNs from dopaminergic and glutamatergic pathways in opioid use disorder [62]. NAcc pathways control cell-specific gene expression in the medial PFC [66].
In the STR, MSNs constitute the bulk (95% in mice) of neurons and represent dopaminoceptive GABAergic neurons [87]. Intracellular signal transduction is crucial for the STR participation in motor and behavioral functions as well as all types of addiction. Via correlation analysis, we have succeeded in dissecting Drd1- and Drd2-dopaminoceptive neurons’ gene pathways in winners [47,88] with hyperactive behavior revealed by us before [72] in chronically aggressive mice.
The PFC is highly interconnected with other brain regions including extensive connections with subcortical and other cortical structures, notably the thalamus, basal ganglia, hypothalamus, amygdala, hippocampus, and others [67]. Several neurotransmitter systems are represented in the PFC, in particular dopaminergic, glutamatergic, and cholinergic systems [68,69]. Persistent strengthening of the PFC–NAcc pathway during cocaine-seeking behavior has been documented [86].
The MRNs have a vast impact upon the central nervous system. A large number of neurons in these nuclei are serotonergic [89]. It is reported that glutamate input into dorsal raphe nuclei is enhanced during escalating aggression, thereby causing a phasic increase of the 5-HT release from the 5-HT neurons. It is believed that the MRNs are involved in a variety of reward-related phenomena including drug addiction and mediate primary reinforcement via GABAA receptors [90]. Downregulation of serotonergic genes in the MRNs has been shown in our experiments on winners [50,59].
4.6. The Genes That Were Analyzed in Different Brain Regions
CAergic systems: Th, Ddc, Dbh, Maoa, Maob, Comt, Slc6a2, Slc6a3, Slc18a2, Snca, Sncb, Sncg, Ppp1r1b, Drd1, Drd2, Drd3, Drd4, Drd5, Adra1a, Adra1b, Adra1d, Adra2a, Adra2b, Adra2c, Adrb1, Adrb2, Adrb3, Adrbk1, and Adrbk2;
Opioidergic and cannabinoidergic systems: Pdyn, Penk, Pomc, Pnoc, Oprm1, Oprd1, Oprk1, Opcml, Ogfr, Ogfrl1, Cnr1, Cnr2, and Faah;
GABAergic system: Gabra1, Gabra2, Gabra3, Gabra4, Gabra5, Gabra6, Gabrb1, Gabrb2, Gabrb3 Gabrg1, Gabrg2, Gabrg3, Gabrd, Gabre, Gabrp, Gabrq, Gabbr1, Gabbr2, Gabrr1, Gabrr2, Gabrr3, Slc6a11, and Slc6a13;
Glutamatergic system: Gria1, Gria2, Gria3, Gria4; Grik1, Grik2, Grik3, Grik4, Grik5; Grin1, Grin2a, Grin2b, Grin2c, Grin2d, Grin3a, Grin3b, Grm1, Grm2, Grm3, Grm4, Grm5, Grm6, Grm7, Grm8; Grid1 u Grid2; Grid2ip, Gad1, Gad2, Slc17a6, Slc17a7, and Slc17a8;
Expression of serotonergic genes—Tph2, Ddc, Maoa, Maob, Htr1a, Htr1b, Htr2a, Htr2c, Htr3a, Htr4, Htr5b, Htr6, Htr7, Htr1d, Htr1f, Htr2b, Htr3b u Htr5a, Slc6a4, and Slc18a2—have been previously analyzed by us [70,71].
In Supplementary Table S1, DEGs in FPKM units in all brain regions are presented.
4.7. Statistical Analysis
For the transcriptome data, principal component analysis (PCA) was conducted in the XLStat software (www.xlstat.com, accessed on 31 March 2016). PCA was based on the Pearson product moment correlation matrix calculated from FPKM profiles of the analyzed DEGs. We also used Pearson’s correlation as a similarity metric for agglomerative hierarchical clustering (AHC). The agglomeration method was based on an unweighted pair-group average.
5. Conclusions
Previously, it has been suggested [28,29] that accumulation of positive fighting effects day in and day out is accompanied by significant dynamic changes in social behaviors as well as in brain neurotransmitter activity in experienced winners. Repeated manifestation of aggression accompanied by wins and graduate acquisition of winning experience induce multiple long-term changes in neurotransmitter systems of the brain. These alterations arise due to a rearrangement of brain regulatory mechanisms involving (consecutively or simultaneously) neurotransmitters’ synthesis, catabolism, receptors, and now we can say gene activity. We propose that as a result of long positive fighting experience, the balance between activities of the neurotransmitter systems is disturbed as excitation processes beginning to dominate over inhibitory processes in the winners. This imbalance is due to a reduced activity of the serotonergic system and an enhanced activity of dopaminergic systems in the brain. Under these circumstances, a low threshold for aggressive behavior gets established, which is one of the reasons for recurrent aggression (relapse) demonstrated in experiments in male mice [29,31]. Now, we have obtained evidence that numerous genes associated with major neurotransmitter systems in the reward-related brain regions of mice with positive fighting experience are involved in these processes.
6. Limitations
The same genes may be upregulated in one brain region and downregulated in others. As an example, the Oprk1 gene in the VTA (up) and in the STR (down). The Th gene was found to be upregulated in the VTA and NAcc and downregulated in the STR and MRNs. This probably means that the expression of genes may depend on a brain region’s function and cell surroundings. This state of affairs creates difficulties with finding the main target for drugs to cure the addictive state.
Conversely, the same gene may change its expression similarly in several regions: the Slc17a7 gene proved to be upregulated in the STR, VTA, NAcc, and MRNs, and Oprd1 is upregulated in the MRNs and PFC. Sncg is downregulated in the VTA and MRNs.
It can also be hypothesized that there are dynamic changes of gene expression depending on the duration of agonistic interactions, and these dynamics may be different and specific for every gene, brain region, and time point of analysis after exposure to an experimental factor. At each specific moment, we can look at a certain picture of alterations and relationships of genes, judging by a change in gene expression or its absence. Further studies are necessary to reveal dynamic changes and possible interconnection between pathological aggression and brain expression of opioidergic and other genes associated with neurotransmitter systems in order to understand the mechanisms underlying the role of endogenous opioids in the offense induced by addictive states.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms232113644/s1.
Author Contributions
Conceptualization, N.N.K.; Data curation, D.A.S. and N.N.K.; Formal analysis, D.A.S. and N.N.K.; Funding acquisition, N.N.K.; Investigation, D.A.S., I.L.K. and A.G.G.; Methodology, N.N.K.; Project administration, N.N.K.; Resources, D.A.S., I.L.K. and A.G.G.; Supervision, N.N.K.; Visualization, D.A.S.; Writing—original draft, N.N.K. and D.A.S.; Writing—review & editing, D.A.S. and N.N.K. All authors gave final approval of the text of the manuscript. All authors have read and agreed to the published version of the manuscript. Informed consent was obtained from all co-authors involved in the study.
Funding
This research was funded by the Russian Science Foundation, grant No. 19-15-00026-∏ to N.N.K. The funding body had no role in the design of the study; in the collection, analysis, and interpretation of the data; and in the writing of the manuscript.
Institutional Review Board Statement
All procedures were conducted in compliance with European Communities Council Directive 210/63/EU of September 22, 2010. The study protocol was approved by Bioethical Commission (Scientific Council No. 9) at the Institute of Cytology and Genetics SB RAS of 24 March 2010, No. 613 (Novosibirsk, Russia).
Informed Consent Statement
Not applicable.
Data Availability Statement
The additional statistics on the obtained data used to support the findings of this study are available in Supplementary Table S1 (differentially expressed genes in FPKM units) and are cited at relevant places within the text. The other datasets generated during the current study are available from the corresponding author on reasonable request.
Acknowledgments
The authors are grateful to N.A. Shevchuk for correcting the English version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| 5-HT | serotonin |
| AHC | agglomerative hierarchical clustering |
| C | control |
| W | winners |
| CAergic | catecholaminergic |
| FPKM | fragments per kilobase of transcript per million mapped reads |
| GABA | γ-aminobutyric acid |
| MRNs | midbrain raphe nuclei |
| MSNs | medium spiny neurons |
| NAcc | nucleus accumbens |
| PCA | principal component analysis |
| PFC | prefrontal cortex |
| STR | dorsal striatum |
| VTA | ventral tegmental area |
References
- Miczek, K.A.; Faccidomo, S.P.; Fish, E.W.; DeBold, J.F. Neurochemistry and molecular neurobiology of aggressive behavior. In Handbook of Neurochemistry and Molecular Neurobiology: Behavioral Neurochemistry, Neuroendocrinology and Molecular Neurobiology; Lajtha, A., Blaustein, J.D., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 285–336. [Google Scholar]
- De Almeida, R.M.; Ferrari, P.F.; Parmigiani, S.; Miczek, K.A. Escalated aggressive behavior: Dopamine, serotonin and GABA. Eur. J. Pharmacol. 2005, 526, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Narvaes, R.; de Almeida, R.M.M. Aggressive behavior and three neurotransmitters: Dopamine, GABA, and serotonin—A review of the last 10 years. Psychol. Neurosci. 2014, 7, 601–607. [Google Scholar] [CrossRef]
- Takahashi, A.; Lee, R.X.; Iwasato, T.; Itohara, S.; Arima, H.; Bettler, B.; Miczek, K.A.; Koide, T. Glutamate input in the dorsal raphe nucleus as a determinant of escalated aggression in male mice. J. Neurosci. 2015, 35, 6452–6463. [Google Scholar] [CrossRef] [PubMed]
- Haller, J. Studies into abnormal aggression in humans and rodents: Methodological and translational aspects. Neurosci. Biobehav. Rev. 2017, 76, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Aleyasin, H.; Flanigan, M.E.; Russo, S.J. Neurocircuitry of aggression and aggression seeking behavior: Nose poking into brain circuitry controlling aggression. Curr. Opin. Neurobiol. 2018, 49, 184–191. [Google Scholar] [CrossRef] [PubMed]
- de Boer, S.F. Animal models of excessive aggression: Implications for human aggression and violence. Curr. Opin. Psychol. 2018, 19, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Tordjman, S.; Carlier, M.; Cohen, D.; Cesselin, F.; Bourgoin, S.; Colas-Linhart, N.; Petiet, A.; Perez-Diaz, F.; Hamon, M.; Roubertoux, P.L. Aggression and the three opioid families (endorphins, enkephalins, and dynorphins) in mice. Behav. Genet. 2003, 33, 529–536. [Google Scholar] [CrossRef]
- Benton, D.; Brain, P.F. The role of opioid mechanisms in social interaction and attachment. In Endorphins, Opiates and Behavioural Processes; Rodgers, R.J., Cooper, S.J., Eds.; John Wiley and Sons Ltd: London, UK, 1988; pp. 217–235. [Google Scholar]
- De Boer, S.F.; Buwalda, B.; Koolhaas, J.M. Untangling the neurobiology of coping styles in rodents: Towards neural mechanisms underlying individual differences in disease susceptibility. Neurosci. Biobehav. Rev. 2017, 74, 401–422. [Google Scholar] [CrossRef]
- Glick, A.R. The role of serotonin in impulsive aggression, suicide, and homicide in adolescents and adults: A literature review. Int J. Adolesc. Med. Health 2015, 27, 143–150. [Google Scholar] [CrossRef]
- Kulikov, A.V.; Osipova, D.V.; Naumenko, V.S.; Terenina, E.; Mormede, P.; Popova, N.K. A pharmacological evidence of positive association between mouse intermale aggression and brain serotonin metabolism. Behav. Brain Res. 2012, 233, 113–119. [Google Scholar] [CrossRef]
- Montoya, E.R.; Terburg, D.; Bos, P.A.; van Honk, J. Testosterone, cortisol, and serotonin as key regulators of social aggression: A review and theoretical perspective. Motiv. Emot. 2012, 36, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Popova, N.K.; Naumenko, E.V.; Kolpakov, B.G. Serotonin and behavior. Novosib. Nauka SB RAS 1978, 4, 12. [Google Scholar]
- Pavlov, K.A.; Chistiakov, D.A.; Chekhonin, V.P. Genetic determinants of aggression and impulsivity in humans. J. Appl. Genet. 2012, 53, 61–82. [Google Scholar] [CrossRef]
- Takahashi, A.; Quadros, I.M.; de Almeida, R.M.; Miczek, K.A. Brain serotonin receptors and transporters: Initiation vs. termination of escalated aggression. Psychopharmacology 2011, 213, 183–212. [Google Scholar] [CrossRef] [PubMed]
- Kudriavtseva, N.N.; Bakshtanovskaia, I.V. The neurochemical control of aggression and submission. Zh. Vyssh. Nerv. Deiat. Im. I P Pavlova 1991, 41, 459–466. [Google Scholar] [PubMed]
- Kudryavtseva, N.N.; Lipina, T.V.; Koryakina, L.A. Effects of haloperidol on communicative and aggressive behavior in male mice with different experiences of aggression. Pharmacol. Biochem. Behav. 1999, 63, 229–236. [Google Scholar] [CrossRef]
- Bondar, N.P.; Kudryavtseva, N.N. The effects of the D1 receptor antagonist SCH-23390 on individual and aggressive behavior in male mice with different experience of aggression. Neurosci. Behav. Physiol. 2005, 35, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Lipina, T.V.; Avgustinovich, D.F.; Koriakina, L.A.; Alekseenko, O.V.; Kudriavtseva, N.N. Differences in the effects of naltrexone on the communicative and aggressive behavi.iors of subjects with different experiences of social conquests. Eksp. Klin. Farmakol. 1998, 61, 13–18. [Google Scholar]
- Kudriavtseva, N.N.; Dolgov, V.V.; Avgustinovich, D.F.; Alekseenko, O.V.; Lipina, T.V.; Koriakina, L.A. Modifying effect of the repeated experience of agonistic confrontations on effect of naltrexone in male mice. Ross. Fiziol. Zh. Im. I. M. Sechenova 2001, 87, 227–238. [Google Scholar]
- Bondar, N.P.; Smagin, D.A.; Kudryavtseva, N.N. Effects of single and chronic naltrexone treatment on agonistic behavior of male mice with repeated experience of aggression. Psychopharmacol. Biol. Narcol. 2011, 11, 2688–2700. [Google Scholar]
- Kudryavtseva, N.N. Straub tail, the deprivation effect and addiction to aggression. In Motivation of Health Behavior; O’Neal, P.W., Ed.; NOVA Science Publishers: New York, NY, USA, 2007; pp. 97–110. [Google Scholar]
- Kudryavtseva, N.N.; Gerrits, M.A.; Avgustinovich, D.F.; Tenditnik, M.V.; van Ree, J.M. Modulation of anxiety-related behaviors by mu- and kappa-opioid receptor agonists depends on the social status of mice. Peptides 2004, 25, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Filipenko, M.L.; Alekseyenko, O.V.; Beilina, A.G.; Kamynina, T.P.; Kudryavtseva, N.N. Increase of tyrosine hydroxylase and dopamine transporter mRNA levels in ventral tegmental area of male mice under influence of repeated aggression experience. Brain Res. Mol. Brain Res. 2001, 96, 77–81. [Google Scholar] [CrossRef]
- Bondar, N.P.; Boyarskikh, U.A.; Kovalenko, I.L.; Filipenko, M.L.; Kudryavtseva, N.N. Molecular implications of repeated aggression: Th, Dat1, Snca and Bdnf gene expression in the VTA of victorious male mice. PLoS ONE 2009, 4, e4190. [Google Scholar] [CrossRef]
- Filipenko, M.L.; Beylina, A.G.; Alekseyenko, O.V.; Timofeeva, O.A.; Avgustinovich, D.F.; Kudryavtseva, N.N. Association between brain COMT gene expression and aggressive experience in daily agonistic confrontations in male mice. In Stress: Neural, Endocrine and Molecular Studies; McCarty, R., Aguilera, G., Sabban, E., Kvetnyansky, R., Eds.; Taylor & Francis: New York, NY, USA; London, UK, 2002; pp. 157–161. [Google Scholar]
- Kudryavtseva, N.N. Psychopathology of repeated aggression: A neurobiological aspect. In Perspectives on the Psychology of Aggression; Morgan, J.P., Ed.; NOVA Science Publishers, Inc.: New York, NY, USA, 2006; pp. 35–64. [Google Scholar]
- Kudryavtseva, N.N. Positive fighting experience, addiction-like state, and relapse: Retrospective analysis of experimental studies. Aggress. Viol. Behav. 2020, 52, 101403. [Google Scholar] [CrossRef]
- Kudryavtseva, N.N.; Kovalenko, I.L.; Smagin, D.A.; Galyamina, A.G.; Babenko, V.N. Abnormal social behaviors and dysfunction of autism-related genes associated with daily agonistic interactions in mice. In Molecular-Genetic and Statistical Techniques for Behavioral and Neural Research; Gerlai, R.T., Ed.; Academic Press: San Diego, CA, USA, 2018; pp. 309–344. [Google Scholar]
- Kudryavtseva, N.N.; Smagin, D.A.; Bondar, N.P. Modeling fighting deprivation effect in mouse repeated aggression paradigm. Prog. Neuropsychopharmacol. Biol. Psychiatry 2011, 35, 1472–1478. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.K.; Hassanein, N.M.A.; Ahmed, H.M.S. Psychopharmacological assessment of the sensory contact model as a possible model of mania. J. Glob. Biosci. 2016, 5, 3725–3741. [Google Scholar]
- Covington, H.E., 3rd; Newman, E.L.; Leonard, M.Z.; Miczek, K.A. Translational models of adaptive and excessive fighting: An emerging role for neural circuits in pathological aggression. F1000Research 2019, 8, 963. [Google Scholar] [CrossRef] [PubMed]
- Golden, S.A.; Shaham, Y. Aggression addiction and relapse: A new frontier in psychiatry. Neuropsychopharmacology 2018, 43, 224–225. [Google Scholar] [CrossRef] [PubMed]
- Golden, S.A.; Jin, M.; Heins, C.; Venniro, M.; Michaelides, M.; Shaham, Y. Nucleus accumbens Drd1-expressing neurons control aggression self-administration and aggression seeking in mice. J. Neurosci. 2019, 39, 2482–2496. [Google Scholar] [CrossRef]
- Rodriguez-Arias, M.; Navarrete, F.; Daza-Losada, M.; Navarro, D.; Aguilar, M.A.; Berbel, P.; Minarro, J.; Manzanares, J. CB1 cannabinoid receptor-mediated aggressive behavior. Neuropharmacology 2013, 75, 172–180. [Google Scholar] [CrossRef]
- Miczek, K.A.; Takahashi, A.; Gobrogge, K.L.; Hwa, L.S.; de Almeida, R.M. Escalated aggression in animal models: Shedding new light on mesocorticolimbic circuits. Curr. Opin. Behav. Sci. 2015, 3, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Enoch, M.A.; Rosser, A.A.; Zhou, Z.; Mash, D.C.; Yuan, Q.; Goldman, D. Expression of glutamatergic genes in healthy humans across 16 brain regions; altered expression in the hippocampus after chronic exposure to alcohol or cocaine. Genes Brain Behav. 2014, 13, 758–768. [Google Scholar] [CrossRef] [PubMed]
- Moeller, S.J.; London, E.D.; Northoff, G. Neuroimaging markers of glutamatergic and GABAergic systems in drug addiction: Relationships to resting-state functional connectivity. Neurosci. Biobehav. Rev. 2016, 61, 35–52. [Google Scholar] [CrossRef]
- Cooper, S.J. Interaction between endogenous opioids and dopamine: Implications for reward and aversion. In The Mesolimbic Dopamine System: From Motivation to Action; Willner, P., Scheel-Kruger, J., Eds.; John Wiley and Sons Ltd: London, UK, 1991; pp. 331–366. [Google Scholar]
- Van Ree, J.M.; Gerrits, M.A.; Vanderschuren, L.J. Opioids, reward and addiction: An encounter of biology, psychology, and medicine. Pharmacol. Rev. 1999, 51, 341–396. [Google Scholar]
- Volkow, N.D.; Morales, M. The brain on drugs: From reward to addiction. Cell 2015, 162, 712–725. [Google Scholar] [CrossRef] [PubMed]
- Kudryavtseva, N.N. The sensory contact model for the study of aggressive and submissive behaviors in male mice. Aggress. Behav. 1991, 17, 285–291. [Google Scholar] [CrossRef]
- Kudryavtseva, N.N.; Smagin, D.A.; Kovalenko, I.L.; Vishnivetskaya, G.B. Repeated positive fighting experience in male inbred mice. Nat. Protoc. 2014, 9, 2705–2717. [Google Scholar] [CrossRef]
- Redina, O.; Babenko, V.; Smagin, D.; Kovalenko, I.; Galyamina, A.; Efimov, V.; Kudryavtseva, N. Gene expression changes in the ventral tegmental area of male mice with alternative social behavior experience in chronic agonistic interactions. Int. J. Mol. Sci. 2020, 21, 6599. [Google Scholar] [CrossRef]
- Babenko, V.N.; Smagin, D.A.; Kovalenko, I.L.; Galyamina, A.G.; Kudryavtseva, N.N. Differentially expressed genes of the Slc6a family as markers of altered brain neurotransmitter system function in pathological states in mice. Neurosci. Behav. Physiol. 2020, 50, 199–209. [Google Scholar] [CrossRef]
- Babenko, V.N.; Galyamina, A.G.; Rogozin, I.B.; Smagin, D.A.; Kudryavtseva, N.N. Dopamine response gene pathways in dorsal striatum MSNs from a gene expression viewpoint: cAMP-mediated gene networks. BMC Neurosci. 2020, 21, 12. [Google Scholar] [CrossRef]
- Redina, O.E.; Babenko, V.N.; Smagin, D.A.; Kovalenko, I.L.; Galyamina, A.G.; Kudryavtseva, N.N. Correlation of expression changes between genes controlling 5-HT synthesis and genes Crh and Trh in the midbrain raphe nuclei of chronically aggressive and defeated male mice. Genes 2021, 12, 1811. [Google Scholar] [CrossRef] [PubMed]
- Randesi, M.; van den Brink, W.; Levran, O.; Blanken, P.; van Ree, J.M.; Ott, J.; Kreek, M.J. VMAT2 gene (SLC18A2) variants associated with a greater risk for developing opioid dependence. Pharmacogenomics 2019, 20, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Kudryavtseva, N.N.; Smagin, D.A.; Kovalenko, I.L.; Galyamina, A.G.; Vishnivetskaya, G.B.; Babenko, V.N.; Orlov, Y.L. Serotonergic genes in the development of anxiety/depression-like state and pathology of aggressive behavior in male mice: RNA-seq data. Mol. Biol. 2017, 51, 288–300. [Google Scholar] [CrossRef]
- Lagerspetz, K.M.J. Studies on the aggressive behavior of mice. Ann. Acad. Sci. Fenn. B 1964, 131, 1–131. [Google Scholar]
- Scott, J.P. Agonistic behavior of mice and rats: A review. Am. Zool. 1966, 6, 683–701. [Google Scholar] [CrossRef]
- Scott, J.P. Theoretical issues concerning the origin and causes of fighting. In The Physiology of Aggression and Defeat; Eleftheriou, B.E., Scott, J.P., Eds.; Plenum New-York: New York, NY, USA, 1971; pp. 11–42. [Google Scholar]
- Brain, P.F.; Kamal, K.B.H. Effects of prior social experience on individual aggressiveness in laboratory rodents. Rassegna Psicol. 1989, 6, 37–43. [Google Scholar]
- Ramirez, J.M. Aggression: Causes and functions. Hiroshima For. Psychol. 1996, 17, 21–37. [Google Scholar]
- Fish, E.W.; de Bold, J.F.; Miczek, K.A. Aggressive behavior as a reinforcer in mice: Activation by allopregnanolone. Psychopharmacology 2002, 163, 459–466. [Google Scholar] [CrossRef]
- Kudryavtseva, N.N. Lorenz was right! Or does aggressive energy accumulate? Russ. J. Genet. 2004, 40, 656–662. [Google Scholar] [CrossRef]
- Robinson, T.E.; Berridge, K.C. Addiction. Ann. Rev. Psychol. 2003, 54, 25–53. [Google Scholar] [CrossRef]
- Smagin, D.A.; Boyarskikh, U.A.; Bondar, N.P.; Filipenko, M.L.; Kudryavtseva, N.N. Reduction of serotonergic gene expression in the midbrain raphe nuclei under positive fighting experience. Adv. Biosci. Biotechnol. 2013, 4, 36–44. [Google Scholar] [CrossRef]
- Flatscher-Bader, T.; Zuvela, N.; Landis, N.; Wilce, P.A. Smoking and alcoholism target genes associated with plasticity and glutamate transmission in the human ventral tegmental area. Hum. Mol. Genet. 2008, 17, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Farris, S.P.; Harris, R.A.; Ponomarev, I. Epigenetic modulation of brain gene networks for cocaine and alcohol abuse. Front. Neurosci. 2015, 9, 176. [Google Scholar] [CrossRef] [PubMed]
- Thompson, B.L.; Oscar-Berman, M.; Kaplan, G.B. Opioid-induced structural and functional plasticity of medium-spiny neurons in the nucleus accumbens. Neurosci. Biobehav. Rev. 2021, 120, 417–430. [Google Scholar] [CrossRef]
- Wang, C.W.; Ma, M.; Lu, W.G.; Luo, R.Q. Association between prodynorphin gene polymorphisms and opioid dependence susceptibility: A meta-analysis. BMC Psychiatry 2019, 19, 281. [Google Scholar] [CrossRef]
- Bazov, I.; Sarkisyan, D.; Kononenko, O.; Watanabe, H.; Yakovleva, T.; Hansson, A.C.; Sommer, W.H.; Spanagel, R.; Bakalkin, G. Dynorphin and kappa-opioid receptor dysregulation in the dopaminergic reward system of human alcoholics. Mol. Neurobiol. 2018, 55, 7049–7061. [Google Scholar] [CrossRef]
- Yuanyuan, J.; Rui, S.; Hua, T.; Jingjing, C.; Cuola, D.; Yuhui, S.; Shuguang, W. Genetic association analyses and meta-analysis of dynorphin-kappa opioid system potential functional variants with heroin dependence. Neurosci. Lett. 2018, 685, 75–82. [Google Scholar] [CrossRef]
- Hikida, T.; Yao, S.; Macpherson, T.; Fukakusa, A.; Morita, M.; Kimura, H.; Hirai, K.; Ando, T.; Toyoshiba, H.; Sawa, A. Nucleus accumbens pathways control cell-specific gene expression in the medial prefrontal cortex. Sci. Rep. 2020, 10, 1838. [Google Scholar] [CrossRef]
- Alvarez, J.A.; Emory, E. Executive function and the frontal lobes: A meta-analytic review. Neuropsychol. Rev. 2006, 16, 17–42. [Google Scholar] [CrossRef]
- Steketee, J.D. Neurotransmitter systems of the medial prefrontal cortex: Potential role in sensitization to psychostimulants. Brain Res. Brain Res. Rev. 2003, 41, 203–228. [Google Scholar] [CrossRef]
- Del Arco, A.; Mora, F. Neurotransmitters and prefrontal cortex-limbic system interactions: Implications for plasticity and psychiatric disorders. J. Neural. Transm. 2009, 116, 941–952. [Google Scholar] [CrossRef] [PubMed]
- Morales, M.; Margolis, E.B. Ventral tegmental area: Cellular heterogeneity, connectivity and behaviour. Nat. Rev. Neurosci. 2017, 18, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Sheen, W.; Morales, M. Glutamatergic neurons are present in the rat ventral tegmental area. Eur. J. Neurosci. 2007, 25, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Smagin, D.A.; Galyamina, A.G.; Kovalenko, I.L.; Babenko, V.N.; Tamkovich, N.V.; Borisov, S.A.; Tolstikova, T.G.; Kudryavtseva, N.N. Altered expression of neurotransmitter genes in the dorsal striatum of male mice with psychomotor disturbances. Zh. Vyssh. Nerv. Deiat. Im. I. P. Pavlova 2018, 68, 227–249. [Google Scholar]
- Jiao, D.; Liu, Y.; Li, X.; Liu, J.; Zhao, M. The role of the GABA system in amphetamine-type stimulant use disorders. Front. Cell. Neurosci. 2015, 9, 162. [Google Scholar] [CrossRef]
- Crist, R.C.; Reiner, B.C.; Berrettini, W.H. A review of opioid addiction genetics. Curr. Opin. Psychol. 2019, 27, 31–35. [Google Scholar] [CrossRef]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef]
- Margolis, E.B.; Lock, H.; Hjelmstad, G.O.; Fields, H.L. The ventral tegmental area revisited: Is there an electrophysiological marker for dopaminergic neurons? J. Physiol. 2006, 577, 907–924. [Google Scholar] [CrossRef]
- Sesack, S.R.; Grace, A.A. Cortico-basal ganglia reward network: Microcircuitry. Neuropsychopharmacology 2010, 35, 27–47. [Google Scholar] [CrossRef]
- Trutti, A.C.; Mulder, M.J.; Hommel, B.; Forstmann, B.U. Functional neuroanatomical review of the ventral tegmental area. NeuroImage 2019, 191, 258–268. [Google Scholar] [CrossRef]
- Walsh, J.J.; Han, M.H. The heterogeneity of ventral tegmental area neurons: Projection functions in a mood-related context. Neuroscience 2014, 282, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Clarke, R.; Adermark, L. Dopaminergic regulation of striatal interneurons in reward and addiction: Focus on alcohol. Neural. Plast. 2015, 2015, 814567. [Google Scholar] [CrossRef] [PubMed]
- Nair-Roberts, R.G.; Chatelain-Badie, S.D.; Benson, E.; White-Cooper, H.; Bolam, J.P.; Ungless, M.A. Stereological estimates of dopaminergic, GABAergic and glutamatergic neurons in the ventral tegmental area, substantia nigra and retrorubral field in the rat. Neuroscience 2008, 152, 1024–1031. [Google Scholar] [CrossRef]
- Robison, A.J.; Nestler, E.J. Transcriptional and epigenetic mechanisms of addiction. Nat. Rev. Neurosci. 2011, 12, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Calipari, E.S.; Bagot, R.C.; Purushothaman, I.; Davidson, T.J.; Yorgason, J.T.; Pena, C.J.; Walker, D.M.; Pirpinias, S.T.; Guise, K.G.; Ramakrishnan, C.; et al. In vivo imaging identifies temporal signature of D1 and D2 medium spiny neurons in cocaine reward. Proc. Natl. Acad. Sci. USA 2016, 113, 2726–2731. [Google Scholar] [CrossRef]
- Meredith, G.E.; Pennartz, C.M.; Groenewegen, H.J. The cellular framework for chemical signalling in the nucleus accumbens. Prog. Brain Res. 1993, 99, 3–24. [Google Scholar] [CrossRef]
- Olsen, C.M. Natural rewards, neuroplasticity, and non-drug addictions. Neuropharmacology 2011, 61, 1109–1122. [Google Scholar] [CrossRef]
- Luis, C.; Cannella, N.; Spanagel, R.; Kohr, G. Persistent strengthening of the prefrontal cortex—nucleus accumbens pathway during incubation of cocaine-seeking behavior. Neurobiol. Learn. Mem. 2017, 138, 281–290. [Google Scholar] [CrossRef]
- Yager, L.M.; Garcia, A.F.; Wunsch, A.M.; Ferguson, S.M. The ins and outs of the striatum: Role in drug addiction. Neuroscience 2015, 301, 529–541. [Google Scholar] [CrossRef]
- Babenko, V.; Redina, O.; Smagin, D.; Kovalenko, I.; Galyamina, A.; Babenko, R.; Kudryavtseva, N. Dorsal striatum transcriptome profile profound shift in repeated aggression mouse model converged to networks of 12 transcription factors after fighting deprivation. Genes 2021, 13, 21. [Google Scholar] [CrossRef]
- Azmitia, E.C.; Segal, M. An autoradiographic analysis of the differential ascending projections of the dorsal and median raphe nuclei in the rat. J. Comp. Neurol. 1978, 179, 641–667. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.H.; Ikemoto, S. The midbrain raphe nuclei mediate primary reinforcement via GABA(A) receptors. Eur. J. Neurosci. 2007, 25, 735–743. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).