Abstract
The coast of the Bulgarian Black Sea is a popular summer holiday destination. The Dam of Iskar is the largest artificial dam in Bulgaria, with a capacity of 675 million m3. It is the main source of tap water for the capital Sofia and for irrigating the surrounding valley. There is a close relationship between the quality of aquatic ecosystems and human health as many infections are waterborne. Rapid molecular methods for the analysis of highly pathogenic bacteria have been developed for monitoring quality. Mycobacterial species can be isolated from waste, surface, recreational, ground and tap waters and human pathogenicity of nontuberculose mycobacteria (NTM) is well recognized. The objective of our study was to perform molecular analysis for key-pathogens, with a focus on mycobacteria, in water samples collected from the Black Sea and the Dam of Iskar. In a two year period, 38 water samples were collected—24 from the Dam of Iskar and 14 from the Black Sea coastal zone. Fifty liter water samples were concentrated by ultrafiltration. Molecular analysis for 15 pathogens, including all species of genus Mycobacterium was performed. Our results showed presence of Vibrio spp. in the Black Sea. Rotavirus A was also identified in four samples from the Dam of Iskar. Toxigenic Escherichia coli was present in both locations, based on markers for stx1 and stx2 genes. No detectable amounts of Cryptosporidium were detected in either location using immunomagnetic separation and fluorescence microscopy. Furthermore, mass spectrometry analyses did not detect key cyanobacterial toxins. On the basis of the results obtained we can conclude that for the period 2012–2014 no Mycobacterium species were present in the water samples. During the study period no cases of waterborne infections were reported.
1. Introduction
During the last two decades waterborne infections have contributed to the emergence of illnesses in Bulgaria, such as Q-fever and tularemia, which were previously rare or unknown [1,2,3]. Studies have confirmed that the sources of infections originated from the drinking water supply system. Contamination of water sources often occurs by accidental falls of animals into water reservoirs or contamination by fecal matter linked to agricultural activities. Possible microbial pollution along the Black Sea marine coastal zone, especially in sea resorts, is a serious ecological and public health concern. The Bulgarian Black Sea shore is a popular recreational and vacation destination for tourists. The coast is 378 km long of which 130 km are tourist beaches. Two large industrial seaports, Varna and Burgas exert anthropogenic pressure on the marine environment. The Dam of Iskar, with a capacity of 675 million m3, is part of the largest artificial lake in Bulgaria, and a popular place for aquatic leisure activities. Furthermore, the Dam of Iskar is the main source of tap water for the capital Sofia, electricity production and for irrigation of the surrounding valley.
There is a close relationship between the quality of aquatic ecosystems and human health. This relationship stems primarily from the consumption of water potentially being polluted by chemicals and/or contaminated by pathogenic organisms. Monitoring water quality is of paramount importance for prevention and safeguarding public health.
Although routine examination of drinking water and sea water for a number of chemical and microbiological parameters has been carried out by environmental protection authorities in Bulgaria, no published reports have evaluated microbial water quality in relation to the application of rapid molecular methods for analysis of highly pathogenic bacteria such as Mycobacterium spp., Vibirio spp., Listeria monocytogenes, Clostridium perfringens, Clostridium botulinum, Shigella spp., Salmonella spp., and Yersinia enterocolitica.
Nontuberculosis mycobacteria (NTM) are free-living saprophytes that are found in water, soil, animals and dairy products. NTM were not accepted as human pathogens until the 1950s, but human pathogenicity is now well recognized [4]. More than 150 NTM species have been described and new species continue to be identified [5]. In the last three decades it was confirmed that these bacteria are opportunistic pathogens for humans, animals, poultry and fish [5]. The main feature of mycobacterial cells which is responsible for their survival and wide distribution is the presence of a lipid-rich outer membrane. This hydrophobic membrane is responsible for adaptation, surface adherence and biofilm formation. The outer membrane is also responsible for antibiotic and disinfectant resistance. All mycobacterial species are able to grow at low carbon levels, a feature allowing them to survive, persist, grow and form biofilms in a low level nutrient environment such as drinking water [5]. Water is currently not routinely tested for mycobacteria and detection methods based on culturing require several days to get the results. In addition, water is often only tested retrospectively when other signs of contamination are present to alert the public when potentially already exposed to the health hazard. There is therefore a need for a rapid, inexpensive and sensitive method for the detection of mycobacteria from environmental sources.
Methods based on the amplification of organism-specific nucleic acids are faster than culturing methods and yield high precision and sensitivity to reliably and simultaneously detect waterborne pathogens. The aim of the present work was to conduct a two-year monitoring study to the water quality from the Dam of Iskar and the Black Sea. We focused on the application of rapid molecular tests based on the use of PCR and Real-Time PCR for detection of the key-pathogens Mycobacterium spp., Vibirio spp., Listeria monocytogenes, Clostridium perfringens, Clostridium botulinum, Shigella spp., Salmonella spp., and Yersinia enterocolitica. No previous studies have been carried out in Bulgaria using molecular analysis of pathogens including the detection of mycobacteria.
2. Experimental Section
2.1. Sampling
The monitoring period was from June 2012 to February 2014, yielding a total of 38 water samples—24 from the Dam of Iskar (one sample per month) and 14 from the Black Sea coastal zone (one sample per two months) (Figure 1, Appendix, Table A1). All samples consisted of 50 L taken with a bucket at from the surface at 0 m deep, brought to the laboratory the same day and immediately filtered to concentrate microorganisms using an ultrafiltration device (dialyser); a 1.8 m2 sterile, disposable, polysulfone hollow-fiber module (Etropal JSC, Etropole, Bulgaria). Water was pushed at a constant 15 psi pressure through the hollow fibers using a peristaltic pump. After filtration, the microorganisms trapped within the filter fibers were recovered by backflushing in the reverse direction (from the outside to the inside of the fibers) with one liter of eluting solution (phosphate buffered saline 0.01 M (Sigma, St. Louis, MO, USA), Tween 80 0.5% (Sigma), sodium hexametaphosphate 0.01% (Sigma) and antifoam B 0.1% (Sigma). The eluted concentrate was preserved at 4 °C for immediate analyses or at −80 °C for longer storage. An aliquot of 200 mL of the eluted concentrate, corresponding to 10 L of the initial volume, was filtered through 20, 5, 2, 0.8 and 0.45 µM filters to further concentrate the sample. Each filter was processed as follows: the 0.45 µM filter was processed with 1 mL of lysis buffer (20 mg/mL lysozyme, Sigma); 20 nM Tris HCl, pH 8.0 (Sigma); 2 mM EDTA (Sigma); 1.2% Triton (Sigma)) for 1 h at 37 °C by shaking at 500 rpm. (Eppendorf Thermomixer comfort, Eppendorf AG, Hamburg, Germany). DNA was extracted from 200 µL. Six µL of Proteinase K 20 mg/mL (QIAGEN, GmbH, Hilden, Germany) were added and incubated at 56 °C for 2 h. Then, 150 µL of Magnetic Bead suspension (Perkin Elmer Inc, Waltham, MA, USA) were added. DNA was purified by processing the sample with Prepito device (Prepito, Chemagen AG, Baesweiler, Germany) and dissolved in 50 µL TE buffer following manufacture’s recommendation. Of this, 5 µL were used in duplicate Real-Time PCR analysis. The initial step allowed 50× sample concentration. The second step allowed additional 200× concentration, thus the final sample concentration was about 104. From the 50× concentrate 40 mL were serially filtered as described above, including an end filtration through 0.1 µM filter for viral nucleic acids extraction.
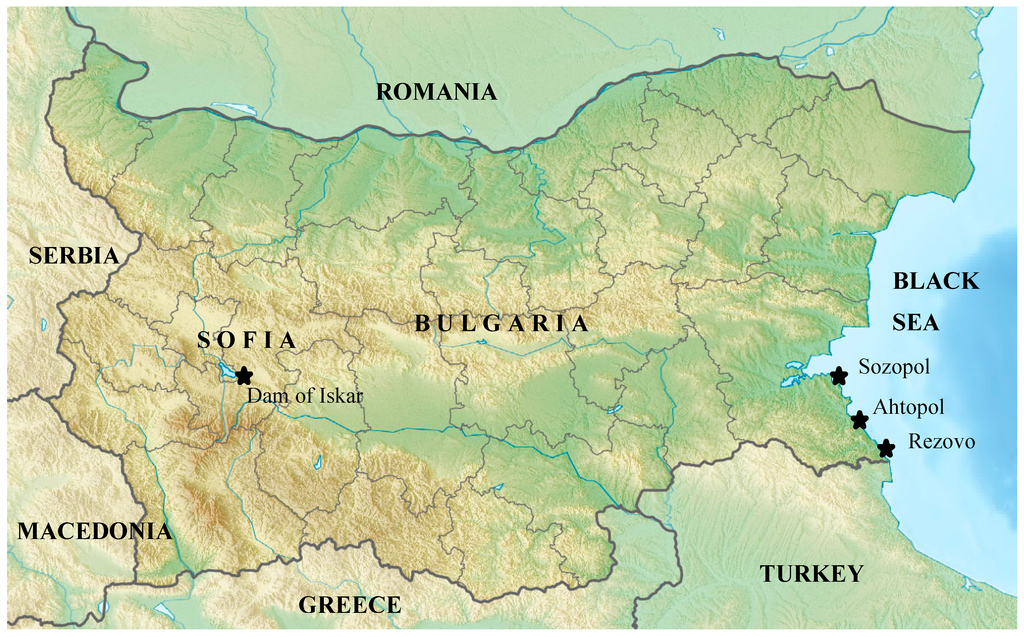
Figure 1.
Map of Bugaria, Black Sea and the sampling sites. Sampling sites are highlighted with a  .
.
 .
.
2.2. Bacterial and Viral Detection
Fifty µL of the concentrated 50× water sample was stained by Ziehl-Neelsen and examined by microscopy for primary screening for mycobacteria [6]. Bacterial genomic DNA was extracted from the 0.45 µM filter which was processed with 1 mL of TRIZOLTM reagent. 200 µL were used for DNA extraction applying a standard procedure [7]. All samples have been tested for 15 different pathogens including species of Mycobacterium spp., Vibirio spp., Listeria monocytogenes, Campilobacter spp., Pseudomonas spp., Shigella spp., Salmonella spp., pathogenic Escherichia coli, Legionella spp., Yersinia enterocolitica and Rotaviruses.
For the detection and identification of Mycobacterium spp., separate amplifications of three markers were applied. Amplification of the partial rpoB gene (360 bp) was performed using primers Rpo5′ (5′-TCAAGGAGAAGCGCTACGA-3′) and Rpo3′ (5′-GGATGTTGATCAGGGTCTGC-3′), as described by [8]. The second marker targeted was part of the 16S-23S spacer, amplified with primers Sp1 (5′-ACC TCC TTT CTA AGG AGC ACC-3′) and Sp2 (5′-GAT GCT CGC AAC CAC TAT CCA-3′) [9]. These primers amplified a 200–350 bp fragment specific to all mycobacteria The amplification was accomplished by an initial denaturation at 94 °C for 5 min, and 30 cycles at 94 °C for 30 seconds, 56 °C for 1 min and 72 °C for 40 seconds, followed by an extension at 72 °C for 10 min. The third marker was a species specific IS6110 for M. tuberculosis applying primers Tb294 (5′-GGACAACGCCGAATTGCGAAGGGC-3′) and Tb850 (5′-TAGGCGTCGGTGACAAAGGCCACG-3′) [10]. The selected markers have been widely used and tested for specificity and sensitivity [8,9,10]. Real-Time PCR tests for the detection of all other bacterial and viral markers were obtained from Primerdesign (Southampton, UK). Viral nucleic acid extraction (rotavirus) was performed by filtering 40 mL of ultrafiltered sample on 0.1 μM filter (Merck Millipore, Billerica, MA, USA). Viral DNA was extracted from 200 µL applying a standard procedure [11]. Internal amplification controls provided with the kits were added to each sample. Target bacteria were quantified by comparing the sample’s amplification curve to the standard curves (four consecutive 10-fold dilutions of 2 × 105 copies/µL) of control DNA.
3. Results and Discussion
Microscopic and PCR analyses demonstrated that no mycobacterial were present in the water samples (Table 1). In six water samples originating from the Black Sea, Vibrio spp. were detected. Rotavirus A was also identified in four samples from the Dam of Iskar. The detection of rotaviruses in the samples from the Dam of Iskar was prevalent during the summer months. In our study the majority of the tested microbial water pathogens in both sampling sites were positive during summer and early autumn. In seven samples from both locations we identified toxigenic Escherichia coli with markers for stx1 and stx2 at concentrations ranging from 50–3 × 102 copies/100 mL. The presence of Norovirus and enterohemorrhagic E. coli (EHEC) in recreational waters could be considered as an epidemiological risk for gastrointestinal illnesses [12]. To our knowledge, the origins of occasional cases of diarrhea along the Black Sea coast, typically occurring during the summer season, are associated with food contamination. There is no evidence that the Black Sea or Dam of Iskar waters could be a direct source of diarrhea infections associated to salmonellosis [13]. The detected concentrations are below the EU and Bulgarian standards for total coliforms; 900 cfu/100 mL, for recreational waters. In four additional samples there were no detectable amounts of Cryptosporidium in two samples from the Black Sea and two samples from the Dam of Iskar using immunomagnetic separation and fluorescence microscopy (Black Sea: <0.01 oocysts/10 L; Dam of Iskar: <0.01 oocysts/10 L) [14]. Cryptosporidiosis is a zoonotic disease, with C. parvum and C. hominis being associated most with human infection. Although industrial animal farming is not permitted around the Dam of Iskar, wildlife still provides a source of pathogens. Mass spectrometry analysis on the same four samples did not identify the presence of cyanobacterial toxins [15].
Routine monitoring of water is necessary for prevention of human and animal health. Pathogenic microorganisms occur in relatively low concentrations in surface waters. Often the contamination episode is not detected until the public shows symptoms of infection. Detection of indicator bacteria of known pathogens in the water indicates potential presence of contamination. E. coli has been chosen as biological indicator of water safety, and is part of drinking water regulations (EU Council directive 98/83/EC on the quality of water intended for human consumption). E. coli is also used as a fecal pollution indicator for recreational bathing waters in Europe (Directive 2006/7/EC concerning the management of bathing water quality and repealing Directive 76/160/EEC). The applied ultrafiltration approach allowed us to concentrate 50 L samples and decrease the filtration duration. The ultrafiltration system showed effective simultaneous collection of several microorganism types (viruses, bacteria, parasites, algae) from large volumes of water without clogging.

Table 1.
Occurrence of targeted pathogens from environmental water obtained from the Black Sea and the Dam of Iskar during the two year monitoring campaign. The results are based on qPCR and regular PCR.
| Target microorganism | Black Sea (n = 14) | Dam of Iskar (n = 24) |
|---|---|---|
| Mycobacterium spp. | 0 | 0 |
| Vibirio spp. | 6 | 0 |
| Listeria monocytogenes | 0 | 0 |
| Campilobacter spp. | 0 | 0 |
| Pseudomonas spp. | 1 | 0 |
| Shigella spp. | 0 | 0 |
| Salmonella spp. | 0 | 0 |
| Legionella spp. | 0 | 1 |
| Yersinia enterocolitica | 0 | 0 |
| Clostridium perfringens | 0 | 0 |
| Clostridium botulinum | 0 | 0 |
| E. coli EHEC stx1 and stx2 | 3 | 4 |
| Staphylococcus aureus | 0 | 0 |
| Aeromonas spp. | 0 | 0 |
| Rotavirus A | 0 | 4 |
The Black Sea basin has unique ecological characteristics [16]. The sea is almost closed and water exchange with the Mediterranean Sea is through the Bosphorus (Bosfor) Strait. The Black Sea is meromictic, characterized by an aerobic surface layer with a depth up to 100 m comprising only 13% of the total volume of the basin (salinity is 17–18‰), and a deep, anaerobic, more saline (22‰) layer up to 2245 m deep. The surface layer is supplied from rivers. Biodiversity is appreciably lower than seen in comparable North Atlantic waters [17]. A monitoring study [18] in 2007 at 23 stations of the bathing waters in Varna’s Black Sea coastal area confirmed that microbiological parameters including ‘total coliforms’, ‘faecal coliforms’ and ‘faecal streptococci’ were in compliance with the current EC Bathing Water Directive (76/EC/160) and exhibited good water quality characteristics. Similar results of generally good microbial status of the Georgian coastal zone were also reported [17]. Continuous monitoring of water samples for presence of potential pathogens, such as Mycobacterium, Vibrio, Listeria and Clostridium species, is essential especially during warm seasons. For many of the opportunistic pathogens targeted in this study, no internationally acceptable levels of the pathogens in fresh or marine water have been established.
4. Conclusions
During a two-year period in 2012–2014, we evaluated the water quality of the Black Sea near Bulgaria and the Dam of Iskar by applying molecular monitoring tests. Enteropathogenic Vibrio, E. coli and Rotavirus species have been detected at low concentration in seasonal summer water samples from the Dam of Iskar. The studied sampling sites along the Black Sea coast were not yet seriously threatened by these pathogenic microorganisms, in spite of the rapid development of recreation areas during the last years. No Mycobacterium spp. were detected in either the Black Sea or Dam of Iskar water samples during the two years. The sampling sites, during the two year period, were considered free of mycobacteria and safe for baths and industrial production of drinking tap water based on the presence of these pathogens. The application of rapid molecular tools for the detection of pathogens in water samples represents a valuable tool for monitoring to aid control and prevention of spread.
Acknowledgments
This work was supported by an European FP7 project µAQUA—Universal microarrays for the evaluation of fresh-water quality based on detection of pathogens and their toxins THEME [KBBE.2010.3.2-04] [Innovative aquatic biosensors—Call: FP7-KBBE-2010-4] Grant agreement no: 265409.
Author Contributions
Stefan Panaiotov, Ivan Simeonovski, Viktoria Levterova conceived and designed the experiment, Ivan Simeonovski, Viktoria Levterova, Ventzislav Karamfilov, Nadia Brankova, Kristin Tankkova, Katrina Campbell, Pauline Jacob, Karim Helmi, Bas Boots, Emilio D’Ugo, Stefania Marcheggiani, Laura Mancini, Ulrich Breitenbach, Erik Mielke performed the experiments; Todor Kantardjiev contributed reagents/materials/analysis tools; Stefan Panaiotov wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix

Table A1.
List of water samples collected from the Black Sea and the Dam of Iskar between 26.06.2012–07.02.2014. Sampling coordinates and results of detected pathogens. NA—result not available; Black Sea—Rezovo village 41o58’57.76” N 28o1’54.00”E; Black Sea—Ahtopol 42o5’57.51” N27o56’38.54”E; Black Sea—Sozopol 42o25’05.50” N 27o41’18.47”E; Dam of Iskar—Shtarkelovo gnezdo 42o27’28.79”N 23o33’31.71”E; Dam of Iskar—Zlo kuche 42o23’35.27”N 23o33’10.51.
| №/Date | Location/GPS coordinates | Yersinia enterocolitica | Vibirio spp | Listeria monocytogenes | Mycobactrium spp. | Salmonella spp | Shigella spp. | Campilobacter spp. | E. coli EHEC stx1 and stx2 | Legionella spp. | Clostridium perfringens | Clostridium botulinum | S. aureus | Aeromonas spp | P.aeruginosa | Rotavirus A |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. 16.01.2013 | Dam of Iskar—Zlo kuche | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA | |
| 2. 09.06.2013 | Dam of Iskar—Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | |
| 3. 16.06.2013 | Dam of Iskar—Zlokuche | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | |
| 4. 22.06.2013 | Dam of Iskar—Zlokuche | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | ||
| 5. 29.06.2013 | Dam of Iskar—Zlokuche | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NEG |
| 6. 14.07.2013 | Dam of Iskar—Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA |
| 7. 21.07.2013 | Dam of Iskar—Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NEG |
| 8. 28.07.2013 | Dam of Iskar—Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA |
| 9. 04.08.2013 | Dam of Iskar—Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA |
| 10. 18.08.2013 | Dam of Iskar–Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA |
| 11. 15.09.2013 | Dam of Iskar—Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA |
| 12. 23.09.2013 | Dam of Iskar—Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA | |
| 13. 29.09.2013 | Dam of Iskar—Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA | |
| 14. 04.10.2013 | Dam of Iskar—Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NEG | |
| 15. 13.10.2013 | Dam of Iskar—Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NEG |
| 16. 20.10.2013 | Dam of Iskar—Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA |
| 17. 27.10.2013 | Dam of Iskar—Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA |
| 18. 03.11.2013 | Dam of Iskar—Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA |
| 19. 06.11.2013 | Dam of Iskar—Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA |
| 20. 10.11.2013 | Dam of Iskar—Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA |
| 21. 17.11.2013 | Dam of Iskar—Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA |
| 22. 24.11.2013 | Dam of Iskar—Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA |
| 23. 12.05.14 | Dam of Iskar—Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | |
| 24. 30.06.14 | Dam of Iskar—Shtarkelovo gnezdo | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | ||
| 25. 09.09.2012 | Black Sea—Rezovo village | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA | |
| 26. 16.09.2012 | Black Sea—Ahtopol | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | ||
| 27. 23.09.2012 | Black Sea—Rezovo village | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NEG | |
| 28. 12.01.2013 | Black Sea—Sozopol | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA | |
| 29. 01.05.2013 | Black Sea – Ahtopol | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA | |
| 30. 23.06.2013 | Black Sea—Rezovo village | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA |
| 31. 20.07.2013 | Black Sea—Rezovo village | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA | |
| 32. 31.08.2013 | Black Sea—Rezovo village | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA |
| 33. 01.09.2013 | Black Sea—Rezovo village | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NEG | |
| 34. 10.10.2013 | Black Sea—Rezovo village | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NA |
| 35. 14.04.2014 | Black Sea—Rezovo village | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NEG | |
| 36. 25.07.2014 | Black Sea—Rezovo village | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NEG |
| 37. 25.08.2014. | Black Sea—Rezovo village | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NEG |
| 38. 05.09.2014 | Black Sea—Rezovo village | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NA | NEG |
References
- Panaiotov, S.; Ciccozzi, M.; Brankova, N.B.; Levterova, V.S.; Mitova-Tiholova, M.; Amicosante, M.; Rezza, G.; Kantardjiev, T.V. An outbreak of Q fever in Bulgaria. Ann. Ist. Super Sanita 2009, 45, 83–86. [Google Scholar] [PubMed]
- Kantardjiev, T.; Ivanov, I.; Velinov, T.; Padeshki, P.; Popov, B.; Nenova, R.; Mincheff, M. Tularemia outbreak, Bulgaria, 1997–2005. Emerg. Infect. Dis. 2006, 12, 678–680. [Google Scholar] [CrossRef] [PubMed]
- Christova, I.; Velinov, T.; Kantardjiev, T.; Galev, A. Tularemia outbreak in Bulgaria. Scand. J. Infect. Dis. 2004, 38, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, P.M.; Hedberg, K.; Saulson, A.; McNelly, E.; Winthrop, K.L. Nontuberculous mycobacterial disease prevalence and risk factors: A changing epidemiology. Clin. Infect. Dis. 2009, 49. [Google Scholar] [CrossRef] [PubMed]
- Falkinham, J.O. Surrounded by mycobacteria: Nontuberculous mycobacteria in the human environment. J. Appl. Microbiol. 2009, 107, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Bachiyska, E. Handbook for Microbiological Diagnosis of Tuberculosis, 1st ed.Publisher Ministry of Health: Sofia, Bulgaria, 2005; pp. 1–95.
- Kremer, K.; Arnold, C.; Cataldi, A.; Gutierrez, M.C.; Haas, W.H.; Panaiotov, S.; Skuce, R.A.; Supply, P.; van der Zanden, A.G.; van Soolingen, D. Discriminatory power and reproducibility of novel DNA typing methods for Mycobacterium tuberculosis complex strains. J. Clin. Microbiol. 2005, 43, 5628–5638. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Park, H.J.; Cho, S.N.; Bai, G.H.; Kim, S.J. Species identification of mycobacteria by PCR-restriction fragment length polymorphism of the rpoB gene. J. Clin. Microbiol. 2000, 38, 2966–2971. [Google Scholar] [PubMed]
- Roth, A.; Reischl, U.; Streubel, A.; Naumann, L.; Kroppenstedt, R.M.; Habicht, M.; Fischer, M.; Mauch, H. Novel diagnostic algorithm for identification of mycobacteria using genus-specific amplification of the 16S–23S rRNA gene spacer and restriction endonucleases. J. Clin. Microbiol. 2000, 38, 1094–1104. [Google Scholar] [PubMed]
- Wilson, S.M.; McNerney, R.; Nye, P.M.; Godfrey-Faussett, P.D.; Stoker, N.G.; Voller, A. Progress towards a simplified polymerase chain reaction and its application to diagnosis of tuberculosis. J. Clin. Microbiol 1993, 31, 776–782. [Google Scholar] [PubMed]
- Xiang, X.; Qiu, D.; Hegele, R.D.; Tan, W.C. Comparison of different methods of total RNA extraction for viral detection in sputum. J. Virol. Methods 2001, 94, 129–135. [Google Scholar] [CrossRef]
- Soller, S.A.; Bartrand, T.; Ashbolt, N.J.; Ravenscroft, J.; Wade, T.J. Estimating the primary etiologic agents in recreational freshwaters impacted by human sources of faecal contamination. Waters Res. 2010, 44, 4736–4747. [Google Scholar] [CrossRef] [PubMed]
- Petrov, P.; Asseva, G.; Parmakova, K.; Kantardjiev, T. Analysis of human salmonelloses in Bulgaria, 2007–2011. European Food Safety Authority: Parma, Italy, 2013; IN-236; p. 20. [Google Scholar]
- Boots, B.; UCD School of Biosystems Engineering, University College Dublin, Dublin, Ireland. Personal communication, 2014.
- Greer, B.; McNamee, S.; Boots, G.; Cimarelli, L.; Guillebault, D.; Helmi, K.; Marcheggiani, S.; Panaiotov, S.; Breitenbach, V.; Akcaalan, R.; Medlin, L.; Elliott, C.; Campbell, K. A survey of the prevalence of freshwater toxins in samples from six European countries using a validated UPLC-MS/MS procedure. Personal communication, 2015. [Google Scholar]
- Kideys, A.E. Fall and rise of the Black Sea ecosystem. Science 2002, 297, 1482–1484. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.; Komakihidze, A.; Osadchaya, T.; Alyomov, S.; Romanov, A.; Tediashvili, M. Evaluating ecological quality in the north-eastern Black Sea coastal zone. Marine Pollu. Bull. 2008, 57, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Simeonova, A.K.; Chuturkova, R.Z.; Bojilova, V.B. Bathing water quality monitoring of Varna Black Sea coastal zone, Bulgaria. Water Res. 2010, 37, 520–527. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).