Aurasperone A Inhibits SARS CoV-2 In Vitro: An Integrated In Vitro and In Silico Study
Abstract
:1. Introduction
2. Results and Discussion
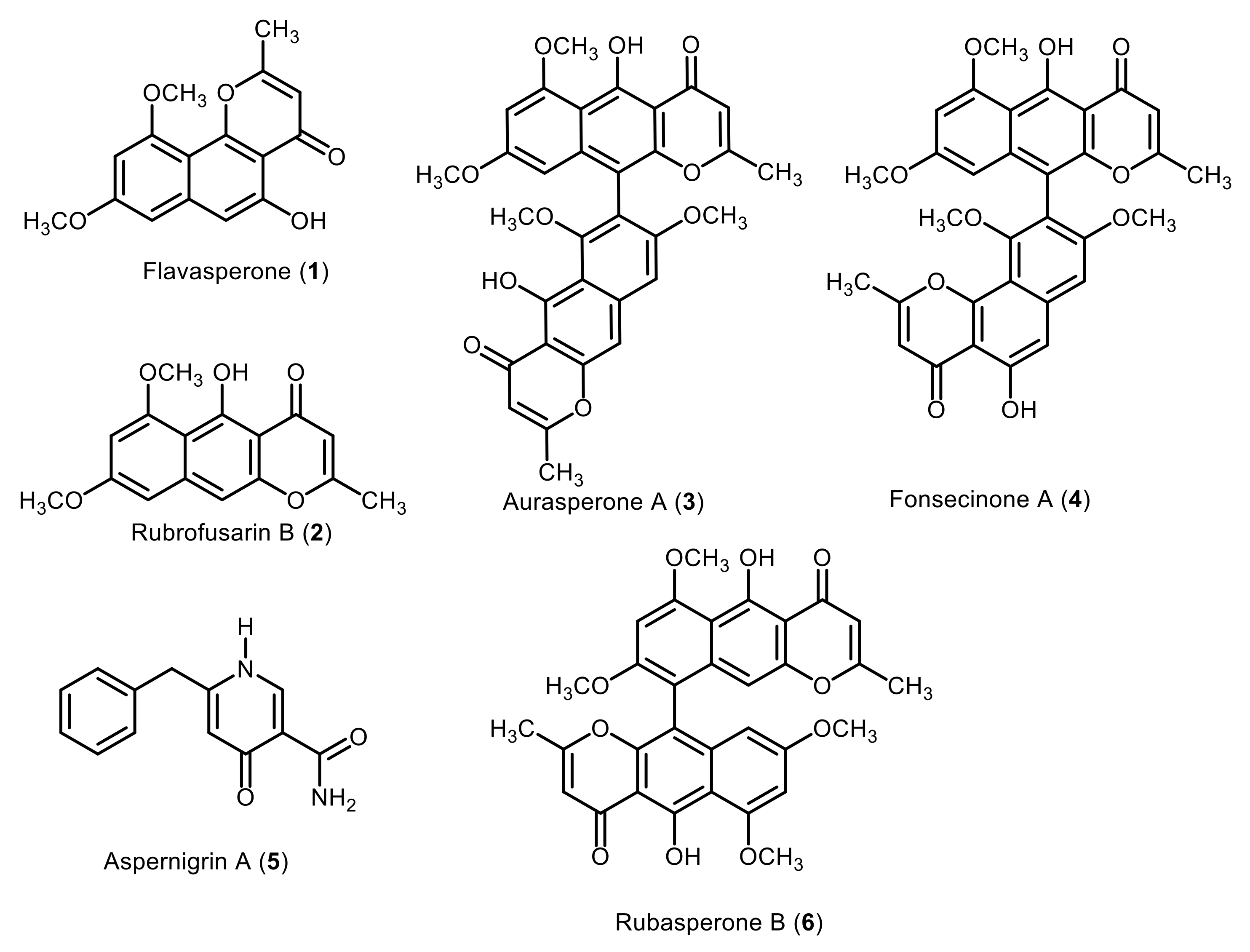
2.1. Purification and Characterization of Compounds 1–6
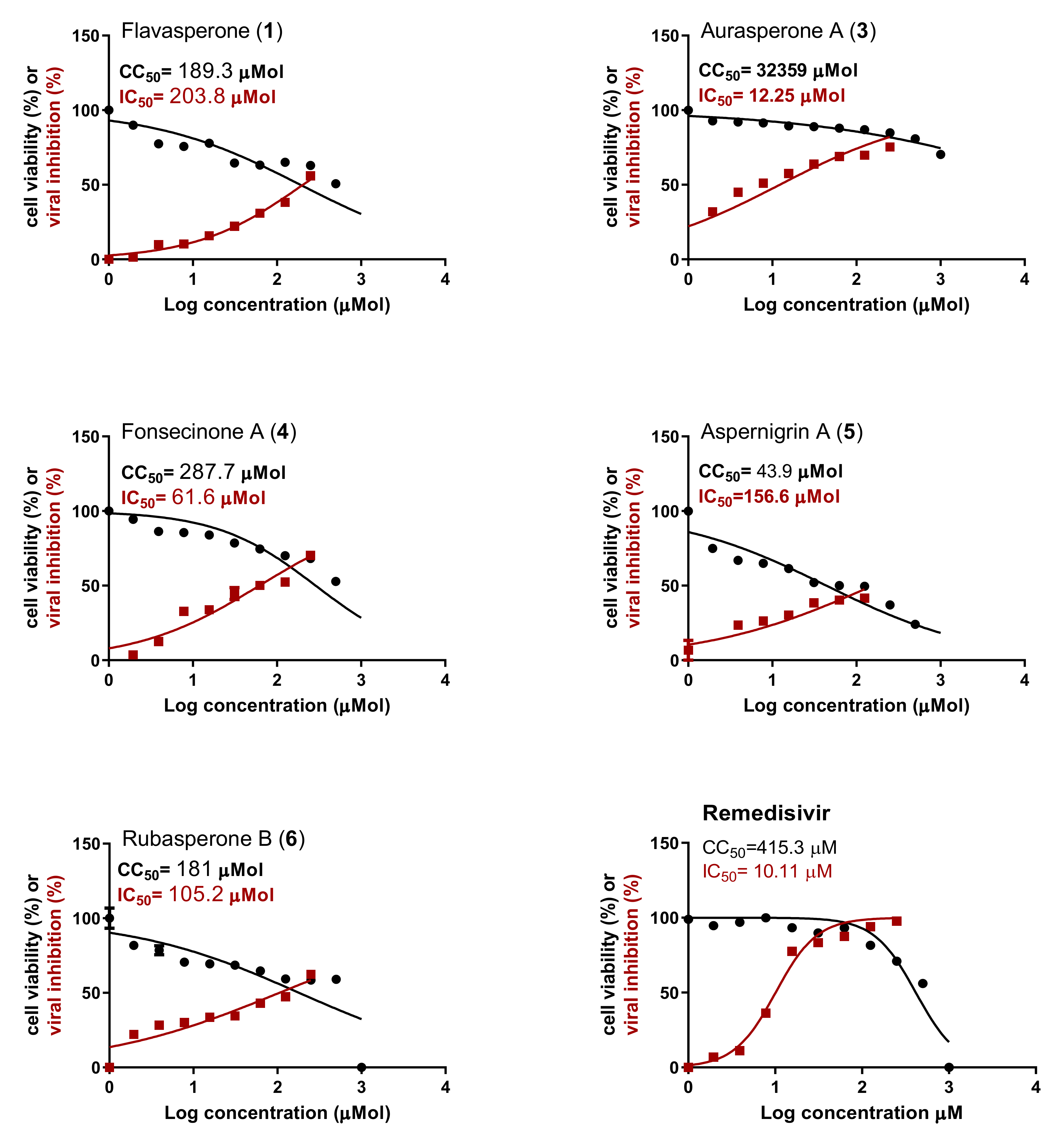
2.2. In Vitro Antiviral Activity
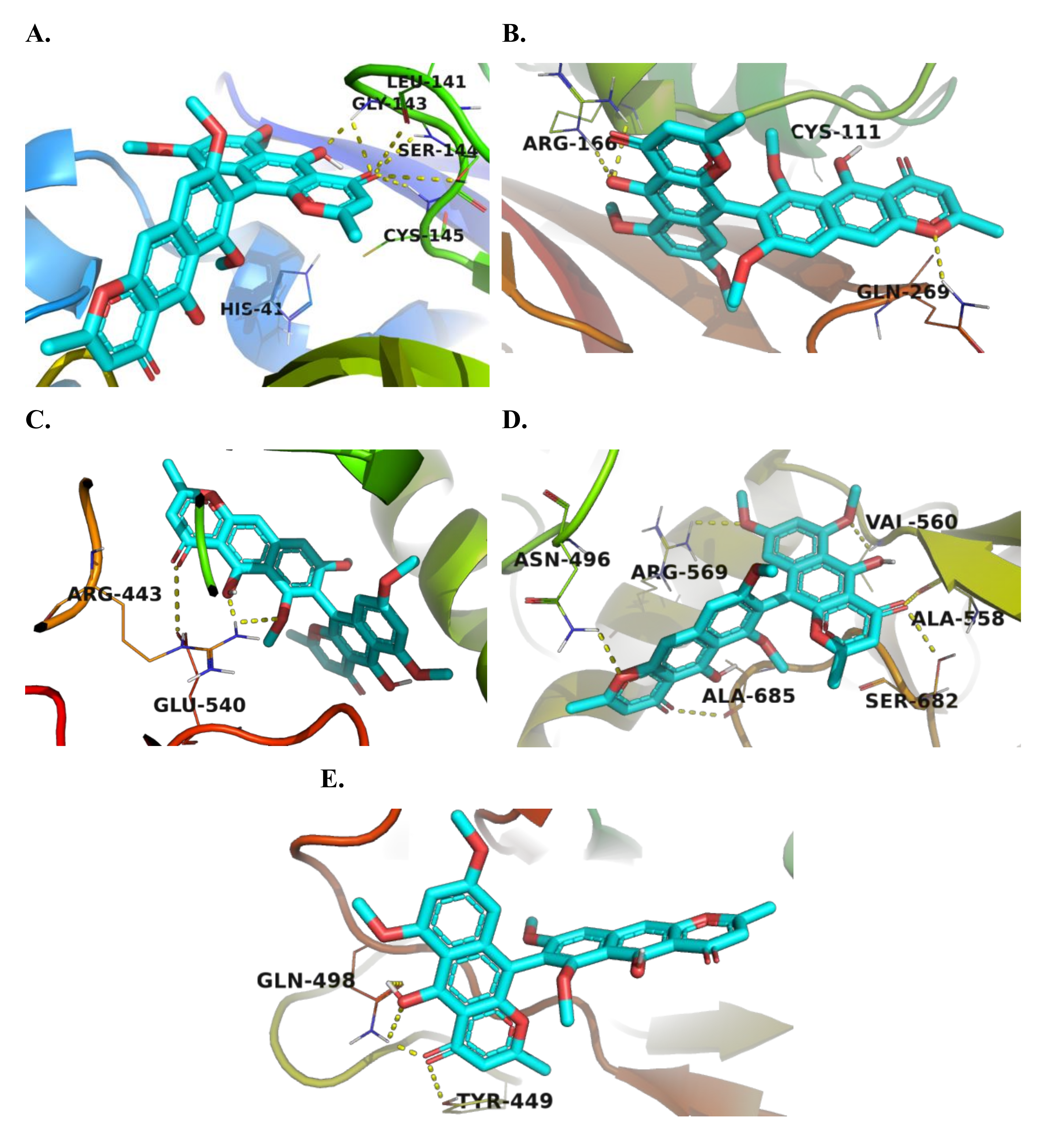
2.3. Docking Study
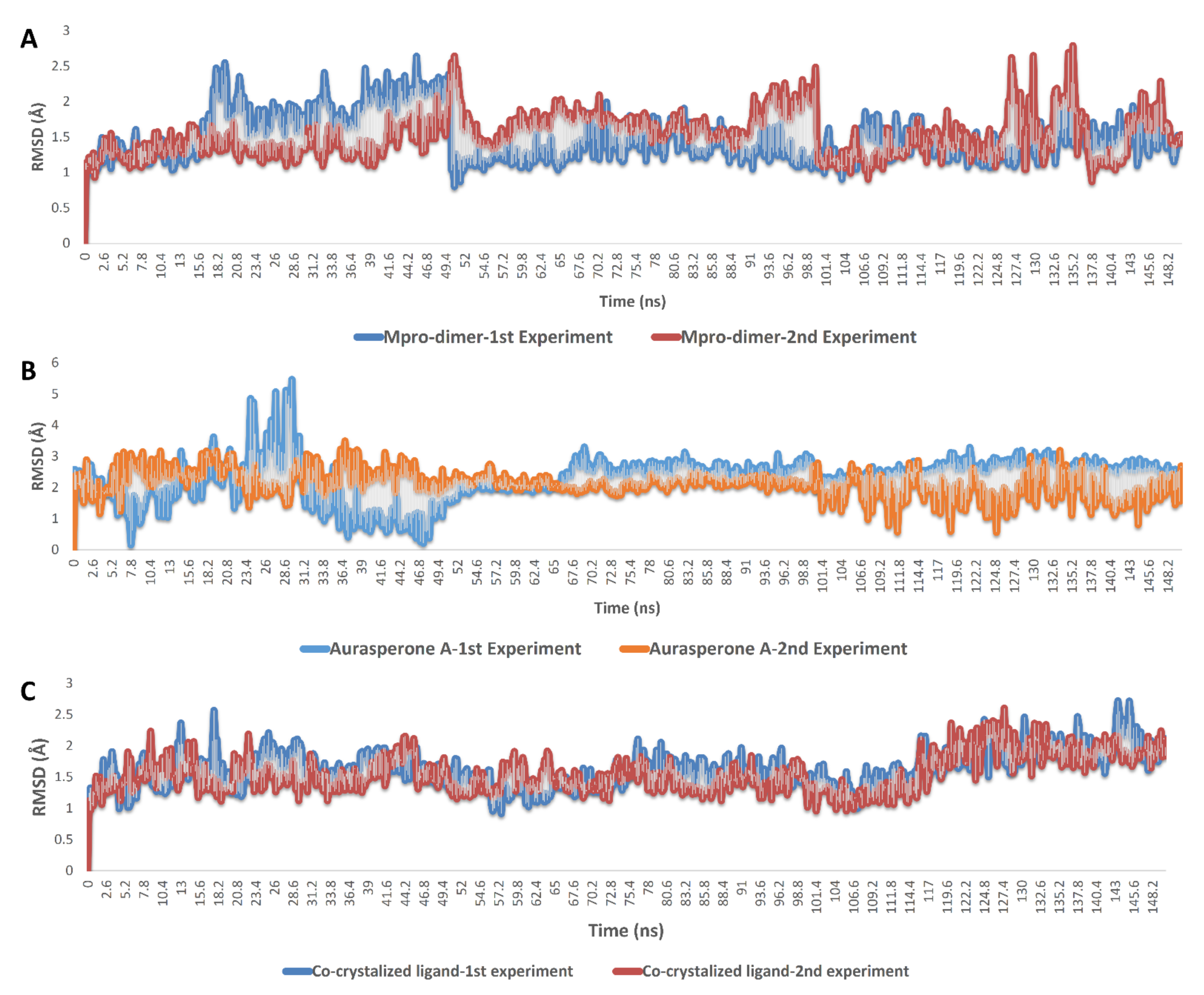
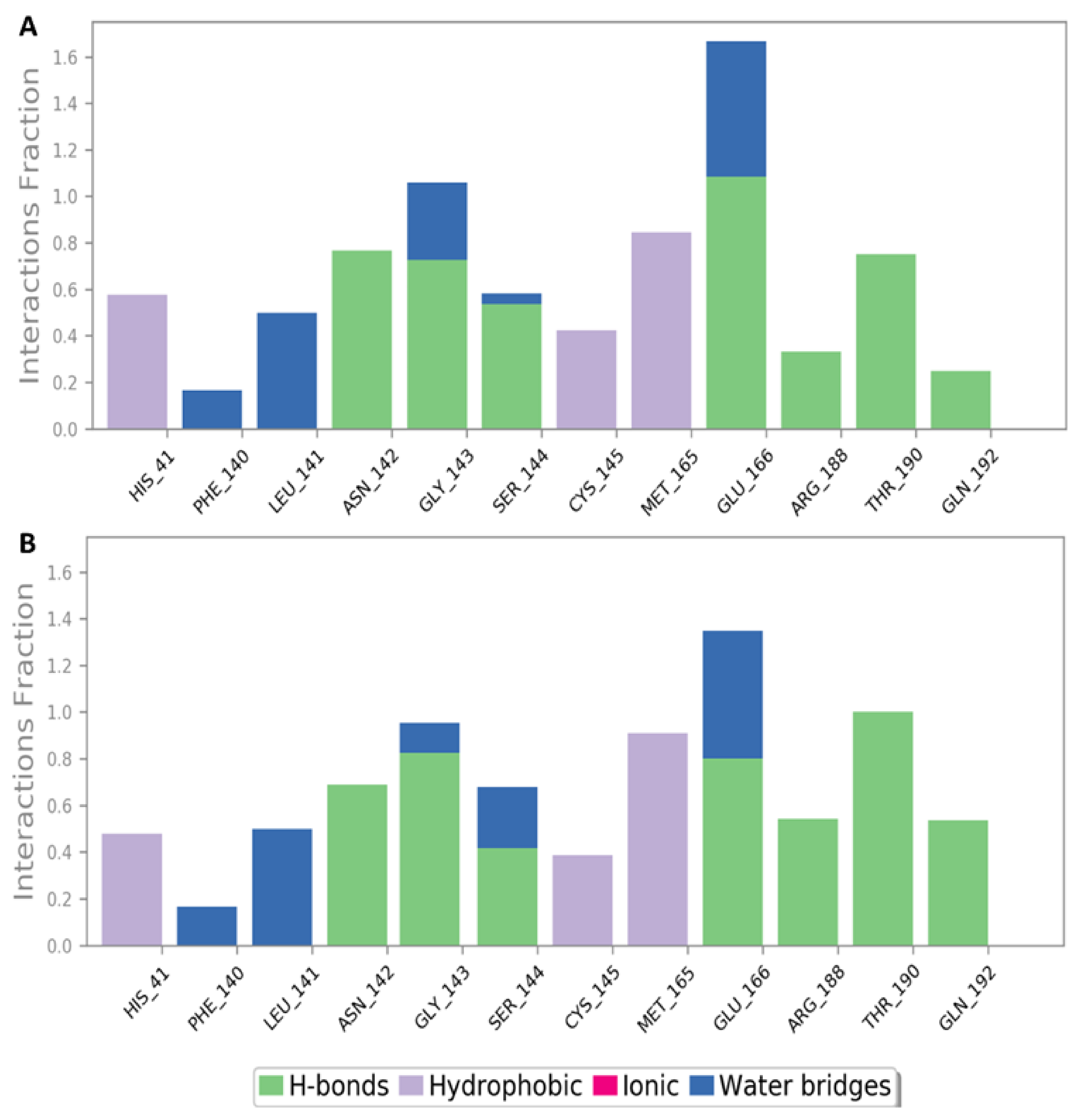
2.4. Molecular Dynamics Simulation
3. Materials and Methods
3.1. General Experimental Procedures
3.2. The Used Fungal Isolate, Fermentation, and Extract Preparation
3.3. Compounds Isolation and Characterization
Isolation and Purification of Compound 6
3.4. Antiviral Activity
3.4.1. MTT Cytotoxicity Assay
3.4.2. Inhibitory Concentration 50 (IC50) Determination
3.5. Docking Study
3.6. Molecular Dynamic Simulation and Binding Free Energy Calculation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feehan, J.; Apostolopoulos, V. Is COVID-19 the worst pandemic? Maturitas 2021, 149, 56–58. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.P.; Das, S.S.; Yadav, S.; Khan, W.; Afzal, M.; Alarifi, A.; Ansari, M.T.; Hasnain, M.S.; Nayak, A.K. Global impacts of pre-and post-COVID-19 pandemic: Focus on socio-economic consequences. Sens. Int. 2020, 1, 100042. [Google Scholar] [CrossRef] [PubMed]
- Telenti, A.; Arvin, A.; Corey, L.; Corti, D.; Diamond, M.S.; García-Sastre, A.; Garry, R.F.; Holmes, E.C.; Pang, P.S.; Virgin, H.W. After the pandemic: Perspectives on the future trajectory of COVID-19. Nature 2021, 596, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Riccio, G.; Ruocco, N.; Mutalipassi, M.; Costantini, M.; Zupo, V.; Coppola, D.; de Pascale, D.; Lauritano, C. Ten-year research update review: Antiviral activities from marine organisms. Biomolecules 2020, 10, 1007. [Google Scholar] [CrossRef]
- Moghadamtousi, S.Z.; Nikzad, S.; Kadir, H.A.; Abubakar, S.; Zandi, K. Potential antiviral agents from marine fungi: An overview. Mar. Drugs 2015, 13, 4520–4538. [Google Scholar] [CrossRef]
- Takahashi, J.A.; Barbosa, B.V.R.; Lima, M.T.N.S.; Cardoso, P.G.; Contigli, C.; Pimenta, L.P.S. Antiviral fungal metabolites and some insights into their contribution to the current COVID-19 pandemic. Bioorganic Med. Chem. 2021, 46, 116366. [Google Scholar] [CrossRef]
- Zahran, E.M.; Sayed, A.M.; Abdelwahab, M.F.; Albohy, A.; Abdulrazik, B.S.; Ibrahim, A.M.; Bringmann, G.; Abdelmohsen, U.R. Identifying the specific-targeted marine cerebrosides against SARS-CoV-2: An integrated computational approach. RSC Adv. 2021, 11, 36042–36059. [Google Scholar] [CrossRef]
- Yu, R.; Liu, J.; Wang, Y.; Wang, H.; Zhang, H. Aspergillus niger as a secondary metabolite factory. Front. Chem. 2021, 9, 701022. [Google Scholar] [CrossRef]
- Lima, M.A.S.; de Oliveira, M.d.C.; Pimenta, A.T.; Uchôa, P.K. Aspergillus niger: A hundred years of contribution to the natural products chemistry. J. Braz. Chem. Soc. 2019, 30, 2029–2059. [Google Scholar] [CrossRef]
- Zhang, X.; Elliot, M.A. Unlocking the trove of metabolic treasures: Activating silent biosynthetic gene clusters in bacteria and fungi. Curr. Opin. Microbiol. 2019, 51, 9–15. [Google Scholar] [CrossRef]
- Xu, Y.; Vinas, M.; Alsarrag, A.; Su, L.; Pfohl, K.; Rohlfs, M.; Schäfer, W.; Chen, W.; Karlovsky, P. Bis-naphthopyrone pigments protect filamentous ascomycetes from a wide range of predators. Nat. Commun. 2019, 10, 3579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; Tian, J.; Sun, W.; Meng, J.; Wang, X.; Fu, X.; Wang, A.; Lai, D.; Liu, Y.; Zhou, L. Bis-naphtho-γ-pyrones from fungi and their bioactivities. Molecules 2014, 19, 7169–7188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Fang, W.; Tan, S.; Lin, X.; Xun, T.; Yang, B.; Liu, S.; Liu, Y. Aspernigrins with anti-HIV-1 activities from the marine-derived fungus Aspergillus niger SCSIO Jcsw6F30. Bioorganic Med. Chem. Lett. 2016, 26, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Hiort, J.; Maksimenka, K.; Reichert, M.; Perović-Ottstadt, S.; Lin, W.; Wray, V.; Steube, K.; Schaumann, K.; Weber, H.; Proksch, P. New Natural Products from the Sponge-Derived Fungus Aspergillus niger. J. Nat. Prod. 2004, 67, 1532–1543. [Google Scholar] [CrossRef] [PubMed]
- Abdelwahab, G.M.; Mira, A.; Cheng, Y.-B.; Abdelaziz, T.A.; Lahloub, M.F.I.; Khalil, A.T. Acetylcholine esterase inhibitory activity of green synthesized nanosilver by naphthopyrones isolated from marine-derived Aspergillus niger. PLoS ONE 2021, 16, e0257071. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, U.R.; Albohy, A.; Abdulrazik, B.S.; Bayoumi, S.A.; Malak, L.G.; Khallaf, I.S.; Bringmann, G.; Farag, S.F. Natural coumarins as potential anti-SARS-CoV-2 agents supported by docking analysis. RSC Adv. 2021, 11, 16970–16979. [Google Scholar] [CrossRef]
- Owis, A.I.; El-Hawary, M.S.; El Amir, D.; Aly, O.M.; Abdelmohsen, U.R.; Kamel, M.S. Molecular docking reveals the potential of Salvadora persica flavonoids to inhibit COVID-19 virus main protease. RSC Adv. 2020, 10, 19570–19575. [Google Scholar] [CrossRef]
- Singh, R.; Bhardwaj, V.K.; Sharma, J.; Kumar, D.; Purohit, R. Identification of potential plant bioactive as SARS-CoV-2 Spike protein and human ACE2 fusion inhibitors. Comput. Biol. Med. 2021, 136, 104631. [Google Scholar] [CrossRef]
- Dömling, A.; Gao, L. Chemistry and biology of SARS-CoV-2. Chem 2020, 6, 1283–1295. [Google Scholar] [CrossRef]
- Luan, B.; Huynh, T.; Cheng, X.; Lan, G.; Wang, H.-R. Targeting proteases for treating COVID-19. J. Proteome Res. 2020, 19, 4316–4326. [Google Scholar] [CrossRef]
- Mahmoudvand, S.; Shokri, S. Interactions between SARS coronavirus 2 papain-like protease and immune system: A potential drug target for the treatment of COVID-19. Scand. J. Immunol. 2021, 94, e13044. [Google Scholar] [CrossRef] [PubMed]
- Spratt, A.N.; Gallazzi, F.; Quinn, T.P.; Lorson, C.L.; Sönnerborg, A.; Singh, K. Coronavirus helicases: Attractive and unique targets of antiviral drug-development and therapeutic patents. Expert Opin. Ther. Pat. 2021, 31, 339–350. [Google Scholar] [CrossRef] [PubMed]
- White, M.A.; Lin, W.; Cheng, X. Discovery of COVID-19 inhibitors targeting the SARS-CoV-2 Nsp13 helicase. J. Phys. Chem. Lett. 2020, 11, 9144–9151. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Rathore, A.S. RNA dependent RNA polymerase (RdRp) as a drug target for SARS-CoV2. J. Biomol. Struct. Dyn. 2021. [Google Scholar] [CrossRef]
- Pandey, P.; Rane, J.S.; Chatterjee, A.; Kumar, A.; Khan, R.; Prakash, A.; Ray, S. Targeting SARS-CoV-2 spike protein of COVID-19 with naturally occurring phytochemicals: An in-silico study for drug development. J. Biomol. Struct. Dyn. 2021, 39, 6306–6316. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Bhardwaj, V.K.; Purohit, R. Potential of turmeric-derived compounds against RNA-dependent RNA polymerase of SARS-CoV-2: An in-silico approach. Comput. Biol. Med. 2021, 139, 104965. [Google Scholar] [CrossRef]
- Bhardwaj, V.K.; Singh, R.; Sharma, J.; Rajendran, V.; Purohit, R.; Kumar, S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2021, 39, 3449–3458. [Google Scholar] [CrossRef]
- Huang, H.-B.; Feng, X.-J.; Liu, L.; Chen, B.; Lu, Y.-J.; Ma, L.; She, Z.-G.; Lin, Y.-C. Three dimeric naphtho-γ-pyrones from the mangrove endophytic fungus Aspergillus tubingensis isolated from Pongamia pinnata. Planta Med. 2010, 76, 1888–1891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, J.C.; Fadl, S.; Villanueva, A.J.; Rabeh, W.M. Catalytic Dyad Residues His41 and Cys145 Impact the Catalytic Activity and Overall Conformational Fold of the Main SARS-CoV-2 Protease 3-Chymotrypsin-Like Protease. Front. Chem. 2021, 9, 491. [Google Scholar] [CrossRef]
- Bello, M. Elucidation of the inhibitory activity of ivermectin with host nuclear importin α and several SARS-CoV-2 targets. J. Biomol. Struct. Dyn. 2021. [Google Scholar] [CrossRef]
- Yin, W.; Mao, C.; Luan, X.; Shen, D.-D.; Shen, Q.; Su, H.; Wang, X.; Zhou, F.; Zhao, W.; Gao, M. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 2020, 368, 1499–1504. [Google Scholar] [CrossRef] [PubMed]
- Gurung, A.B.; Ali, M.A.; Lee, J.; Farah, M.A.; Al-Anazi, K.M. Identification of potential SARS-CoV-2 entry inhibitors by targeting the interface region between the spike RBD and human ACE2. J. Infect. Public Health 2021, 14, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rut, W.; Lv, Z.; Zmudzinski, M.; Patchett, S.; Nayak, D.; Snipas, S.J.; El Oualid, F.; Huang, T.T.; Bekes, M.; Drag, M. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: A framework for anti–COVID-19 drug design. Sci. Adv. 2020, 6, eabd4596. [Google Scholar] [CrossRef]
- Newman, J.A.; Douangamath, A.; Yazdani, S.; Yosaatmadja, Y.; Aimon, A.; Brandao-Neto, J.; Dunnett, L.; Gorrie-Stone, T.; Skyner, R.; Fearon, D. Structure, Mechanism and Crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase. bioRxiv 2021, 12, 4848. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Oshima, H.; Zhang, H.; Kern, N.R.; Re, S.; Lee, J.; Roux, B.; Sugita, Y.; Jiang, W.; Im, W. CHARMM-GUI free energy calculator for absolute and relative ligand solvation and binding free energy simulations. J. Chem. Theory Comput. 2020, 16, 7207–7218. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Alhadrami, H.A.; Sayed, A.M.; Al-Khatabi, H.; Alhakamy, N.A.; Rateb, M.E. Scaffold Hopping of α-Rubromycin Enables Direct Access to FDA-Approved Cromoglicic Acid as a SARS-CoV-2 MPro Inhibitor. Pharmaceuticals 2021, 14, 541. [Google Scholar] [CrossRef]
| Compound | Binding Energy (kcal/mol) | ||||
|---|---|---|---|---|---|
| Mpro | PLpro | Helicase | RdRp | Spike Protein | |
| Flavasperone (1) | −7.1 | −6.4 | −7.6 | −6.9 | −5.8 |
| Rubrofusarin B (2) | −6.9 | −5.9 | −7.0 | −6.8 | −5.8 |
| Aurasperone A (3) | −8.1 | −7.4 | −8.0 | −7.8 | −7.0 |
| Fonsecinone A (4) | −8.0 | −7.1 | −8.1 | −8.2 | −7.1 |
| Aspernigrin A (5) | −6.2 | −6.3 | −6.7 | −6.5 | −5.9 |
| Rubasperone B (6) | −8.5 | −6.8 | −8.0 | −7.9 | −7.0 |
| Reference inhibitor * | −7.5 | −6.7 | −5.6 | −6.6 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
ElNaggar, M.H.; Abdelwahab, G.M.; Kutkat, O.; GabAllah, M.; Ali, M.A.; El-Metwally, M.E.A.; Sayed, A.M.; Abdelmohsen, U.R.; Khalil, A.T. Aurasperone A Inhibits SARS CoV-2 In Vitro: An Integrated In Vitro and In Silico Study. Mar. Drugs 2022, 20, 179. https://doi.org/10.3390/md20030179
ElNaggar MH, Abdelwahab GM, Kutkat O, GabAllah M, Ali MA, El-Metwally MEA, Sayed AM, Abdelmohsen UR, Khalil AT. Aurasperone A Inhibits SARS CoV-2 In Vitro: An Integrated In Vitro and In Silico Study. Marine Drugs. 2022; 20(3):179. https://doi.org/10.3390/md20030179
Chicago/Turabian StyleElNaggar, Mai H., Ghada M. Abdelwahab, Omnia Kutkat, Mohamed GabAllah, Mohamed A. Ali, Mohamed E. A. El-Metwally, Ahmed M. Sayed, Usama Ramadan Abdelmohsen, and Ashraf T. Khalil. 2022. "Aurasperone A Inhibits SARS CoV-2 In Vitro: An Integrated In Vitro and In Silico Study" Marine Drugs 20, no. 3: 179. https://doi.org/10.3390/md20030179
APA StyleElNaggar, M. H., Abdelwahab, G. M., Kutkat, O., GabAllah, M., Ali, M. A., El-Metwally, M. E. A., Sayed, A. M., Abdelmohsen, U. R., & Khalil, A. T. (2022). Aurasperone A Inhibits SARS CoV-2 In Vitro: An Integrated In Vitro and In Silico Study. Marine Drugs, 20(3), 179. https://doi.org/10.3390/md20030179








