Abstract
The marine microorganisms thraustochytrids have been explored for their potential in the production of various bioactive compounds, such as DHA, carotenoids, and squalene. Squalene is a secondary metabolite of the triterpenoid class and is known for its importance in various industrial applications. The bioinformatic analysis for squalene synthase (SQS) gene (the first key enzyme in the tri-terpenoid synthesis pathway), that is prevailing among thraustochytrids, is poorly investigated. In-silico studies combining sequence alignments and bioinformatic tools helped in the preliminary characterization of squalene synthases found in Aurantiochytrium limacinum. The sequence contained highly conserved regions for SQS found among different species indicated the enzyme had all the regions for its functionality. The signal peptide sequence and transmembrane regions were absent, indicating an important aspect of the subcellular localization. Secondary and 3-D models generated using appropriate templates demonstrated the similarities with SQS of the other species. The 3-D model also provided important insights into possible active, binding, phosphorylation, and glycosylation sites.
1. Introduction
Squalene (a triterpene), a potential antioxidant, is a bio-product in high demand due to its application in various industries, such as cosmetics, pharmaceuticals, and food industries [1,2,3]. Many deep-sea sharks have been hunted for a long period for the extraction of squalene [4,5]. Apart from other sources, such as plants and chemical synthesis, researchers are focusing on the development of microbial factories for the commercial production of squalene [6]. Various microorganisms, such as yeast and microalgae, have been explored [7,8]. Among all, marine heterotrophic microorganisms, such as thraustochytrids, are promising sources of squalene [2,3,9]. Aurantiochytrium species among many thraustochytrids has been the center of interest and promising for the production of squalene at commercial level (up to 30% of cell dry weight) [3,10,11].
There are two major pathways for terpenoid synthesis: (i) The MVA (mevalonate) pathway and (ii) the MEP (2-C-methyl-d-erythritol-4-phosphate) pathway [12,13]. Squalene synthesis in thraustochytrids is believed to follow the MVA pathway [14]. Squalene synthase (EC 2.5.1.21; SQS) is the first rate-limiting enzyme of the reaction of the squalene synthesis in thraustochytrids. Two molecules of farnesyl pyrophosphate (FPP) resulting from the acetyl-CoA via subsequent multi-step reaction are utilized to produce squalene via the catalytic action of squalene synthase [14]. The mechanism of SQS has been well characterized as in two steps (1) condensation: Pre-squalene diphosphate (PSPP) generation from two molecules of farnesyl diphosphate (FPP) and (2) squalene production from PSPP in the presence of NADPH and Mg2+ [15]. The effect on squalene and other secondary metabolite production by overexpression and disruption of squalene synthase in various organisms has been extensively studied [2,14]. Thus, the bioinformatic analysis for the sequences available from genomes of thraustochytrids will aid in designing future enhanced squalene or carotenoids synthesis.
The physicochemical prediction, detailed bioinformatics analysis, and 3-D structure of the SQS enzyme in any thraustochytrid species have not been fully reported yet [2]. The genome sequences obtained for Aurantiochytrium limacinum reveal that SQS is very important for overall terpenoid synthesis. The key features and information of the SQS like potential active sites, phosphorylation sites, etc., will be helpful in rational designing of the genetic engineering experiment in the future to improve squalene production [2]. In the present studies, for the first time, in-silico analysis for the SQS gene found in thraustochytrid Aurantiochytrium limacinum species was performed. A detailed analysis of the conserved regions and important domains responsible for binding substrate and ligands was studied. The secondary and 3D structure for the SQS in the selected species was designed using appropriate known structures of the enzyme (Human SQS) as a template.
2. Results and Discussions
2.1. Conserved Domain Search and Multiple Sequence Analysis
Conserved domain (CD) search performed for the selected protein sequence confirmed the identity of the protein as squalene synthase belonging to Isoprenoid_Biosyn_C1 superfamily (cl00210; https://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?ascbin=8&maxaln=10&seltype=2&uid=cl00210/, accessed on 29 November 2021), as shown in Figure 1. CD-search revealed information about the enzyme, such as substrate binding pocket, Substrate-Mg2+ binding site, aspartate rich regions (1 and 2) indicating the deduced amino acid sequence can code for a functional enzyme. The enzymes in the isoprenoid biosynthesis class 1 superfamily contain similar ‘isoprenoid synthase fold’. The SQS enzyme generally requires NAD(P)H and Mg2+ for its functioning. SQS gene belonging to Aurantiochytrium sp. KRS101, with a 99.2% similarity of the selected gene for the study, was cloned and expressed in E. coli BL21. Recombinant SQS obtained was able to convert FPP to squalene in the presence of NADPH and Mg2+, and the product was further confirmed by GC-MS analysis [15]. The Multiple sequence alignment (MSA) analysis was performed by clustal omega using SQS sequences belonging to a different group of organisms (obtained from NCBI), such as humans, yeast, algae, plant, and fungi. Figure 2 shows the presence of five different highly conserved domains (I–V). Similarities were found in the conserved domains detected with the previous report for SQS from different microorganisms [15,16,17]. Conserved domain I was detected in the sequence for the amino acids (AA) 9 to 29 (IRLAVGIFYIVLRALDTVEDD). The aspartate-rich region 1 as found in domain I, DTVED (AA 24 to 28) was found to have similarities with human squalene synthase (AA 80 to 84) and was predicted for substrate binding [15,18,19]. Conserved domain II was detected in the sequence for the amino acids 145 to 162 (YCHYVAGTVGDGLTRIFA). Motif search tool with hidden Markov model revealed the information about the presence of Pfam id as SQS_PSY motif (position AA 08 to 303 of protein sequence, I-E-Value: 9.2 × 10−25). Signature motif YCHYVAGTVGDGLTRI for squalene synthase on conserved region II was found at a position of AA 145 to 160 in the sequence. Conserved domain II was also identified as an active site for SQS reported in the previous literature [16]. AA sequence from position 180 to 197 was identified as conserved domain III, which was previously recognized as a catalytic region [16]. From AA position 192 to 200 (RDYLEDLVD) in the selected SQS AA, the sequence was found to be aspartate-rich region 2. The role of these regions (Aspartate rich 1 and 2) is well characterized as a binding site for phosphate groups of the prenyl acceptor via a magnesium salt bridge [20]. The second aspartate-rich region containing DYLED (AA 193 to 200), which matches the human squalene synthase (AA 219 to 223) is the aspartate-rich region. Any mutation in the region AA 219 to 223 led to the loss of function of the enzyme. Thus, it is mainly responsible for the inactivation of the enzyme activity [17,18]. Conserved domain IV region PQVMAIATL (AA 277 to 281) was also found in the selected sequence, which was earlier predicted to be the sequence for NAD(P)H binding and catalyzing the second reaction of the squalene synthesis [17,21]. VKIRK (AA 299 to 303) conserved region V was also found similar to the human squalene synthase that is important for binding to NAD(P)H [18]. A region rich in the hydrophobic amino acids, which may contribute to transmembrane activity, was missing in the selected SQS sequence, in contrast to what was observed in many other species, which was also supported by TMHMM analysis (Section 2.3) [22]. The absence of the transmembrane (TM) region in the selected SQS may suggest improved solubility and activity [23]. This was verified by deleting the TM region (containing the hydrophobic amino acids) of Magnolia officinalis squalene synthase by genetic engineering resulting in improvement of the solubility and expression [23].
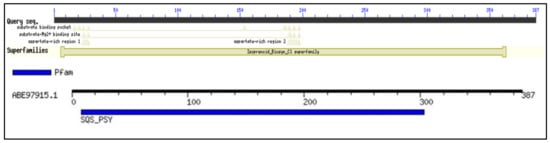
Figure 1.
Conserved domains and motifs for the SQS genes of Aurantiochytrium limacinum: Showing the conserved motifs, superfamily, substrate binding, and aspartate rich regions for the selected sequence.

Figure 2.
Multiple sequence alignment analysis for SQS belonging to different species: the eight sequences used in the analysis are of squalene synthase; Aurantiochytrium limacinum SR21 (ABE97915.1), Homo sapiens (AAA36645.1), Auxenochlorella protothecoides (KFM22694.1), Zea mays (NP_001104839.1), Arabidopsis thaliana (AAD00296.1), Withania somnifera (ADC95435.1), Saccharomyces cerevisiae (GFP68539.1), Aspergillus niger (PYH59865.1). The identical and similar amino acids are mentioned using dots and asterisk signs.
2.2. Physicochemical Characteristics In-Silico Analysis
Physicochemical characteristics for the selected gene were predicted using the ProtParam analysis tool, and the calculated values were as shown in Table 1. The SQS sequence selected for analysis consists of ORF (Open-Reading Frame) of 1161 nucleotides in length encoding 387 amino acids. The gene sequence for SQS in Aurantiochytrium sp. KRS101 was identified to be of 1551 bp, with two exons and an intron (1164 bp encoding 387 amino acids) [15]. The estimated molecular weight of SQS was 43.28 kDa that was falling within the range of SQS from different species, which is 40–50 kDa with certain exceptions [17]. Isoelectric point (pI) is the pH value at which the overall net charge on the protein is zero (Zwitter-ion form). The theoretical value of pI was predicted to be 5.17. The value of pI has a very large range for SQS originating from different sources and is predicted to be as high as 8.20 in some cases [24] and as low as 6.64 [17]. The values of negatively charged and positively charged amino acid residues were 56 and 43, respectively. This information is helpful during enzyme purification as well as to understand the surface topology of the protein [25]. The total number of atoms predicted in the protein was 6029. The protein extinction co-efficient at 280 nm in water was predicted to be 1.028, and it provides information regarding the estimation methods using spectrophotometry. The value of the half-life for the selected protein was 30 h, which means it will take 30 h to vanish half the proteins after its synthesis. The half-life predicted was similar to those SQS sequences found in the plant species previously [26]. The protein instability index was 41.82, showing that the protein was not stable (value higher than 40). The aliphatic index was 86.72, which consists of a basic indication of aliphatic side chain amino acids (leucine, valine, alanine, and isoleucine). The GRAVY score was −0.256, indicating that the protein is nearly non-polar in nature (value close to 0) [27]. There were 28 phosphorylation sites (15 Serine, 10 Threonine, and 4 Tyrosine residues) predicted using netphos 3.1 (https://services.healthtech.dtu.dk/service.php?NetPhos-3.1, accessed on 8 December 2021, Supplementary Materials Figure S1). The phosphorylation sites predicted are very important as they are related to the active state of the enzyme upon phosphorylation using different kinases [28]. The analysis for prediction of glycosylation sites using NetNGlyc 1.0 showed two sites for possible glycosylation (Asp 31 and As134) based on Asn-Xaa-Ser/Thr sequon presence (Supplementary Materials Figure S2). However, the analysis further revealed that although it contained the sequon, it may not be exposed to the N-glycosylation site as there is no signal peptide sequence detected in the selected sequence (Section 2.4). In general, all this information will help for designing future experiments for cloning or other modification of the SQS found in the A. limacinum species for overproduction of squalene.

Table 1.
Physicochemical characteristics of the squalene synthase of Aurantiochytrium limacinum.
2.3. Transmembrane Domain and Hydropathy Analysis
The Transmembrane (TM) helices prediction performed using TMHMM 2.0 (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0, accessed on 6 December 2021) revealed that the squalene synthase from A. limacinum did not contain any TM helices (Figure 3A), while two TM helices were reported in most of the squalene synthase that was analyzed for other species, such as plants. The absence of TM region, particularly in the SQS of Aurantiochytrium species, indicates the location and solubility of the enzyme in the thraustochytrids may differ from most of the SQS reported from other groups [29]. In-vitro studies about the effects of an absence of TM region on the expression and enzyme activities need to be further investigated. The hydropathy analysis performed using protscale was in the range between −2.733 and +2.678 and window size 9, as shown in Figure 3B.
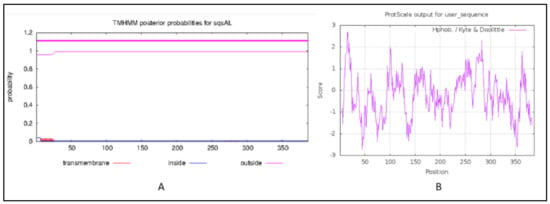
Figure 3.
(A) Transmembrane helices prediction, (B) hydropathy analysis: The red lines on the X-axis indicates the probability of TM regions. Y-axis indicates the probability of occurrence.
2.4. Signal Peptide and Peroxisome Targeting Signal Prediction
The signal peptide prediction was performed using SignalP 5.0 (https://services.healthtech.dtu.dk/service.php?SignalP-5.0, accessed on 28 November 2021). The output from the analysis, as shown in Figure 4, shows different scores for amino acid sequences. As shown in Figure 4, no signal peptide was predicted with an above-threshold value in the selected sequences, which were similar to previous studies [27]. The peroxisome targeting signal 1 (PTS1; SKL AA position 369 to 371) and 2 (PTS2; AA position 327, 330, 344, and 348) were observed at the C-terminal domain as previously identified for SQS from Aurantiochytrium sp. KRS101 as a possible anchor to the membrane [15].
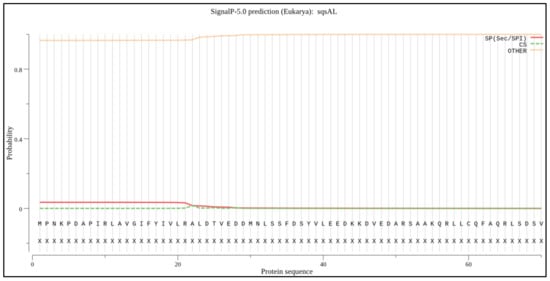
Figure 4.
Signal peptide prediction: The X-axis shows the position of the amino acid sequence, the Y-axis indicates the probability of occurrence.
2.5. Secondary Structure Prediction and Analysis
Secondary structure prediction was performed using the SOPMA-NPSA method (https://npsa-prabi.ibcp.fr/cgi-bin/secpred_sopma.pl, accessed on 29 November 2021). It revealed that the SQS from the A. limacinum species is mainly composed of α-helix (66.14%) followed by random coils (25.84%), extended strands (4.39%), and β-Sheets (3.36%) (Figure 5A). Our findings were matching in the same range with a slight variation with the secondary structure predicted previously for the SQS of different species, such as Withania somnifera species, Gienseng species and species belonging to Fabaceae family [16,26,30,31]. These data were supported along with information about the Psi-phi dihedral angles and surface accessibility using NetsurfP 2.0 (https://services.healthtech.dtu.dk/service.php?NetSurfP-2.0/, accessed on 8 December 2021). The prediction regarding the relative surface accessibility and disorder residues was as shown in Figure 5B.
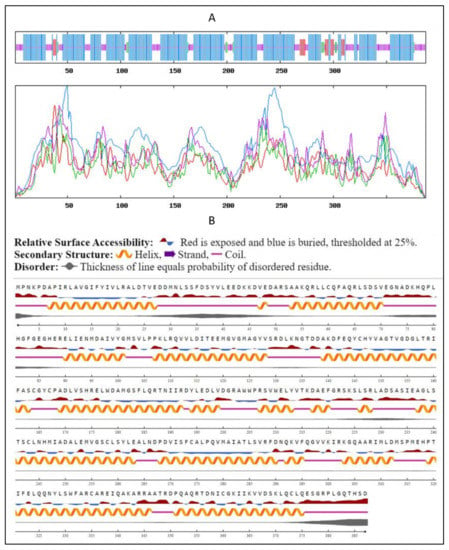
Figure 5.
Secondary structure prediction using (A) SOPMA-NPSA and (B) NetsurfP 2.0 method: Predicted secondary structure of sqs, where blue lines indicate α-helix, the purple lines indicate random coils, the red lines indicate extended strands, and the green lines indicate β-strands.
2.6. 3-D Structure Modeling and Analysis
The 3-D structure modeling was performed using appropriate templates. Nine different templates (3vj9.1.A, 3wce.1.B, 3v66.1.A, 3vja.1.A, 3wca.1.A, 3vj8.1.A, 3vja.1.A, 3vja.1.A, and 3weg.1.A) with lower e-value, higher identity, and availability of X-ray crystal structure were selected for the model prediction on SWISS-MODEL platform (http://swissmodel.expasy.org/, accessed on 16 December 2021). One model out of nine was finalized based on the Q-Mean score (−4.18) predicted based on the template structure (3vj9.1.A; crystal structure of human squalene synthase). The 3-D structure predicted was observed as shown in Figure 6A. The 3-D model generated SQS of selected species was predicted to majorly contain α- helixes and was made of a monomer with large space in the center as a binding site. The active site at the center provides space to interact with NAD(P)H and Mg2+ via hydrogen bonding [32]. The 3-D structure was similar to many predicted models reported earlier for the SQS from different species [16,17,26,30]. The Ramachandran plot, as shown in Figure 6B, showed that 93.37% of the model structure was in favored regions, and the protein was stereo-chemically stable. The dark green in the graph indicates the allowed regions. The stereochemical specificity and dihedral angles [ψ, ϕ] were indicated in Figure 6B. The modeled structure was further assessed by the ProSa-web server by analyzing the protein folding energy. The Z score for the SQS structure modeled was found to be −7.93. Z-score is the estimation based on the energy calculation for each of the amino acids and the comparison with the known template structure. The two different colors, as shown in Figure 7A, indicate the groups of structures explained by X-ray or NMR (Nuclear Magnetic Resonance). The value of the Z-score of modeled structure for SQS (A. limacinum species) was in the acceptable range (+10 to −10, a preferably negative value is good) [33]. Figure 7B shows the energy calculation for each amino acid, with all values predicted to be negative. Amino acid residues resulting in positive values were absent in the predicted SQS model [26]. H-bond energy using the VADAR analysis showed the value of −1.5 against the expected value of −2.0.
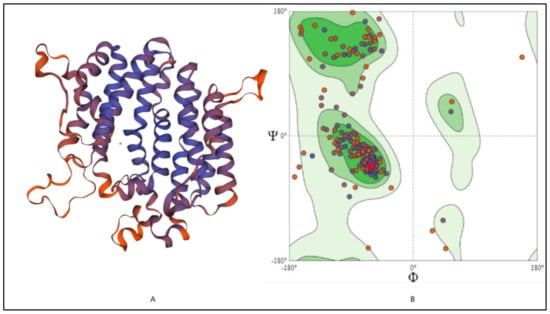
Figure 6.
(A) 3-D structure modeling and (B) Ramachandran plot for the SQS in A. limacinum sp.: Showing the 3D structure of SQS predicted by SWISS-MODEL using appropriate template 3vj9.1.A (crystal structure of human SQS) and prediction of amino acids in favored regions using Ramachandran plot.
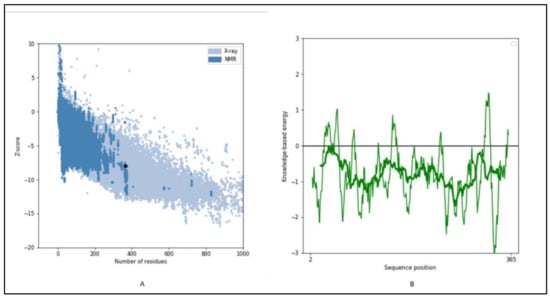
Figure 7.
(A) Prosa analysis for the modeled structure of SQS, (B) energy plot for all AA residues: Showing the Z-score of energy for overall model quality.
2.7. Prediction of Anticipated Binding Sites in SQS
Active and binding sites were predicted using CASTp servers (http://sts.bioe.uic.edu/castp/index.html, accessed on 2 December 2021). The predicted binding sites in the SQS from A. limacinum are shown in Figure 8. The amino acids highlighted in blue color show the AA taking part in the formation of the binding site at the center of the protein. The binding site prediction for the SQS matched a previous report [26]. According to the analysis done with the 3D ligand site, it was observed that four sites were predicted for ligand binding for Mg2+ (Glu27, Asp28, Tyr145, and TRP204), which was also supported by the CD analysis, as mentioned in Section 2.1.
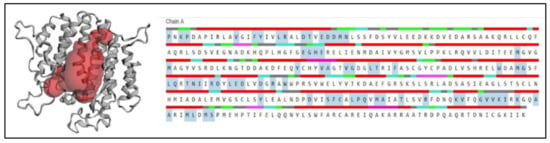
Figure 8.
3-D structure prediction of the binding site using CASTp analysis: Showing the predicted sited of binding using CASTp servers. The amino acids highlighted in blue color indicate the predicted active site residues of SQS.
3. Materials and Methods
3.1. Retrieval of the Sequence Data:
The sequences for SQS gene available for all the thraustochytrids species were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/, accessed on 29 November 2021) and UniProtKB database (https://www.uniprot.org/uniprot/, accessed on 29 November 2021). The respective four sequences were as mentioned in Table 2 with the basic information provided. The sequence for SQS from Aurantiochytrium limacinum SR21 (ATCC-MYA-1381) (previously known as Schizochytrium limacinum) was selected for further analysis (NCBI ID: DQ464066.2, UniProtKB id: ABE97915.1).

Table 2.
List of available squalene synthase genes among thraustochytrids.
3.2. Physicochemical Characteristics Prediction
MOTIF search tool (http://www.genome.jp/tools/motif, accessed on 28 November 2021) using PROSITE database was used for motifs identification. The physicochemical characteristics, such as isoelectric point (pI), half-life, and GRAVY (Grand average of hydropathy) analysis, were obtained using the ProtParam tool of ExPasy (https://web.expasy.org/protparam/, accessed on 28 November 2021) [34]. The selected sequence was analyzed for the presence of transmembrane domains using TMHMM analysis tool (http://www.cbs.dtu.dk/services/TMHMM/, accessed on 6 December 2021) [29]. Analysis of hydrophobic and hydrophilic properties of membrane-spanning regions of the protein was performed by protscale (http://web.expasy.org/cgi-bin/protscale/protscale.pl, accessed on 28 November 2021) [26]. Analysis and identification of signal peptide sequence were performed using SignalP 5.0 (http://www.cbs.dtu.dk/services/SignalP/, accessed on 28 November 2021) [35]. Phosphorylation sites were predicted using netphos 3.1 (https://services.healthtech.dtu.dk/service.php?NetPhos-3.1, accessed on 8 December 2021) [24]. Possible N-glycosylation sites were predicted using NetNglyc-1.0 (https://services.healthtech.dtu.dk/service.php?NetNGlyc-1.0, accessed on 9 December 2021) [24]. The sequence was also checked for the possible presence of Peroxisome targeting signal (PTS) by the presence of PTS1 ((S/A/C)-(K/R/H)-(L/A) amino acids consensus sequence) and PTS2 ((R/K)-(L/V/I)-XXXXX-(H/Q)-(L/A/F) (where X is any amino acid) consensus sequence) motif at C-terminal domain [15]. Multiple sequence alignment (MSA) analysis was done using clustal omega (https://www.ebi.ac.uk/Tools/msa/clustalo/, accessed on 3 December 2021).
3.3. Secondary and 3-D Structure Modelling
Secondary structure prediction was performed using NPSA-SOPMA (Self-optimized Prediction Method with Alignment) (https://npsa-prabi.ibcp.fr/cgi-bin/secpred_sopma.pl, accessed on 29 November 2021) [16,36]. Psi-phi dihedral angles and surface accessibility analysis were performed using NetSurfP-2.0 (https://services.healthtech.dtu.dk/service.php?NetSurfP-2.0/, accessed on 8 December 2021) [37]. The 3-D structure modelling for the selected sequence was performed by SWISS-MODEL using appropriate template selection (http://swissmodel.expasy.org/, accessed on 16 December 2021). Protein folding energy scores for the modeled structures were predicted using prosa web servers by uploading the PDB files obtained from the SWISS-MODEL prediction (https://prosa.services.came.sbg.ac.at/prosa.php, accessed on 16 December 2021) [38]. Tertiary protein analysis was done by VADAR tools (http://vadar.wishartlab.com/, accessed on 16 December 2021).
3.4. Binding Site and Ligand Binding Prediction
Appropriate binding sites were predicted using CASTp servers (http://sts.bioe.uic.edu/castp/index.html?2 accessed on 2 December 2021) [39]. The ligand binding sites were also predicted using 3D ligand site (https://www.wass-michaelislab.org/3dlig/, accessed on 2 December 2021) [40].
4. Conclusions
In the present study, theoretical modeling and bioinformatics analysis for the squalene synthase found in the thraustochytrid species A. limacinum was performed. The sequence homology revealed that selected SQS could have all the important conserved domains for its regular functions and binding to substrate and co-factors. The physicochemical properties predicted will be helpful during enzyme purification as well as to understand the surface topology of the protein. Secondary and 3-D structure models predicted will be useful in future enzyme-substrate interactions using docking and computational tools. Experimental studies, such as cloning of putative gene sequence, are further required to confirm the functioning of the SQS. Detailed analysis of genes, such as SQS, could help in rational designing of possible future biotechnological interventions for a beneficial purpose, such as improved squalene production using thraustochytrids.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/md20030180/s1. Figure S1: Predicted possible phosphorylation sites in the SQS; Figure S2: Predicted possible N-glycosylation sites in the SQS.
Author Contributions
S.V.: conceptualization, methodology, data curation, writing—original draft preparation; M.B.: writing—review & editing, visualization, supervision; U.R.: writing—review & editing, visualization, supervision; P.C.: formal analysis, visualization, supervision; L.M.: validation, visualization, writing—review & editing; A.P.: funding acquisition, validation, visualization, writing —review & editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Swedish Research Council (FORMAS) and Kempestiftelserna, Sweden for supporting this work as part of the projects ‘Green and sustainable approach to valorise high saline and oily fish processing effluents for the production of nutraceuticals’ (INVENTION; 2020-01028) and Boosting the squalene content in thraustochytrids by genetic engineering using CRISPR–Cas9 System to replace the shark-based squalene as an adjuvant for COVID 2019 vaccine (JCK-2115 L3).
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not Applicable.
Data Availability Statement
Not Applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Patel, A.; Karageorgou, D.; Katapodis, P.; Sharma, A.; Rova, U.; Christakopoulos, P.; Matsakas, L. Bioprospecting of Thraustochytrids for Omega-3 Fatty Acids: A Sustainable Approach to Reduce Dependency on Animal Sources. Trends Food Sci. Technol. 2021, 115, 433–444. [Google Scholar] [CrossRef]
- Xie, Y.; Sen, B.; Wang, G. Mining Terpenoids Production and Biosynthetic Pathway in Thraustochytrids. Bioresour. Technol. 2017, 244, 1269–1280. [Google Scholar] [CrossRef] [PubMed]
- Aasen, I.M.; Ertesvåg, H.; Heggeset, T.M.B.; Liu, B.; Brautaset, T.; Vadstein, O.; Ellingsen, T.E. Thraustochytrids as Production Organisms for Docosahexaenoic Acid (DHA), Squalene, and Carotenoids. Appl. Microbiol. Biotechnol. 2016, 100, 4309–4321. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.; Mu, L.; Shi, Y.; Rova, U.; Christakopoulos, P.; Matsakas, L. Novel Biorefinery Approach Aimed at Vegetarians Reduces the Dependency on Marine Fish Stocks for Obtaining Squalene and Docosahexaenoic Acid. ACS Sustain. Chem. Eng. 2020, 8, 8803–8813. [Google Scholar] [CrossRef]
- Gohil, N.; Bhattacharjee, G.; Khambhati, K.; Braddick, D.; Singh, V. Engineering Strategies in Microorganisms for the Enhanced Production of Squalene: Advances, Challenges and Opportunities. Front. Bioeng. Biotechnol. 2019, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- Pyne, M.E.; Narcross, L.; Martin, V.J.J. Engineering Plant Secondary Metabolism in Microbial Systems. Plant Physiology 2019, 179, 844–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, L.J.; Kwak, S.; Liu, J.J.; Lane, S.; Hua, Q.; Kweon, D.H.; Jin, Y.S. Improved Squalene Production through Increasing Lipid Contents in Saccharomyces Cerevisiae. Biotechnol. Bioeng. 2018, 115, 1793–1800. [Google Scholar] [CrossRef]
- Potijun, S.; Jaingam, S.; Sanevas, N.; Vajrodaya, S.; Sirikhachornkit, A. Green Microalgae Strain Improvement for the Production of Sterols and Squalene. Plants 2021, 10, 1673. [Google Scholar] [CrossRef]
- Morabito, C.; Bournaud, C.; Maës, C.; Schuler, M.; Aiese Cigliano, R.; Dellero, Y.; Maréchal, E.; Amato, A.; Rébeillé, F. The Lipid Metabolism in Thraustochytrids. Prog. Lipid Res. 2019, 76, 101007. [Google Scholar] [CrossRef]
- Li, Q.; Chen, G.Q.; Fan, K.W.; Lu, F.U.P.; Aki, T.; Jiang, Y. Screening and Characterization of Squalene-Producing Thraustochytrids from Hong Kong Mangroves. J. Agric. Food Chem. 2009, 57, 4267–4272. [Google Scholar] [CrossRef]
- Patel, A.; Rova, U.; Christakopoulos, P.; Matsakas, L. Simultaneous Production of DHA and Squalene from Aurantiochytrium sp. Grown on Forest Biomass Hydrolysates. Biotechnol. Biofuels 2019, 12, 255. [Google Scholar] [CrossRef] [PubMed]
- Varela, J.C.; Pereira, H.; Vila, M.; León, R. Production of Carotenoids by Microalgae: Achievements and Challenges. Photosynth. Res. 2015, 125, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Ma, X.; Wang, Y. Production of Squalene by Microbes: An Update. World J. Microbiol. Biotechnol. 2016, 32, 195. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Wang, Y.Z.; Xu, Y.S.; Shi, T.Q.; Liu, W.Z.; Sun, X.M.; Huang, H. Biotechnological Production of Lipid and Terpenoid from Thraustochytrids. Biotechnol. Adv. 2021, 48, 107725. [Google Scholar] [CrossRef]
- Hong, W.K.; Heo, S.Y.; Park, H.M.; Kim, C.H.; Sohn, J.H.; Kondo, A.; Seo, J.W. Characterization of a Squalene Synthase from the Thraustochytrid Microalga Aurantiochytrium Sp. KRS101. J. Microbiol. Biotechnol. 2013, 23, 759–765. [Google Scholar] [CrossRef] [Green Version]
- Jiang, D.; Rong, Q.; Chen, Y.; Yuan, Q.; Shen, Y.; Guo, J.; Yang, Y.; Zha, L.; Wu, H.; Huang, L.; et al. Molecular Cloning and Functional Analysis of Squalene Synthase (SS) in Panax Notoginseng. Int. J. Biol. Macromol. 2017, 95, 759–765. [Google Scholar] [CrossRef]
- Kalra, S.; Kumar, S.; Lakhanpal, N.; Kaur, J.; Singh, K. Characterization of Squalene Synthase Gene from Chlorophytum Borivilianum (Sant. and Fernand.). Mol. Biotechnol. 2013, 54, 944–953. [Google Scholar] [CrossRef]
- Pandit, J.; Danley, D.E.; Schulte, G.K.; Mazzalupo, S.; Pauly, T.A.; Hayward, C.M.; Hamanaka, E.S.; Thompson, J.F.; Harwood, H.J. Crystal Structure of Human Squalene Synthase. J. Biol. Chem. 2000, 275, 30610–30617. [Google Scholar] [CrossRef] [Green Version]
- Zhan, D.; Zhang, Y.; Song, Y.; Sun, H.; Li, Z.; Han, W.; Liu, J. Computational Studies of Squalene Synthase from Panax Ginseng: Homology Modeling, Docking Study and Virtual Screening for a New Inhibitor. J. Theor. Comput. Chem. 2012, 11, 1101–1120. [Google Scholar] [CrossRef]
- Gu, P.; Ishii, Y.; Spencer, T.A.; Shechter, I. Function-Structure Studies and Identification of Three Enzyme Domains Involved in the Catalytic Activity in Rat Hepatic Squalene Synthase. J. Biol. Chem. 1998, 273, 12515–12525. [Google Scholar] [CrossRef] [Green Version]
- Robinson, G.W.; Tsay, Y.H.; Kienzle, B.K.; Smith-Monroy, C.A.; Bishop, R.W. Conservation between Human and Fungal Squalene Synthetases: Similarities in Structure, Function, and Regulation. Mol. Cell. Biol. 1993, 13, 2706–2717. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Liu, Y.; Chen, M.; Feng, J.; Ma, Z.; Zhang, X.; Zhu, C. Cloning, Expression Analysis and Functional Characterization of Squalene Synthase (SQS) from Tripterygium Wilfordii. Molecules 2018, 23, 269. [Google Scholar] [CrossRef] [Green Version]
- Zha, L.; Liu, S.; Su, P.; Yuan, Y.; Huang, L. Cloning, Prokaryotic Expression and Functional Analysis of Squalene Synthase (SQS) in Magnolia Officinalis. Protein Expr. Purif. 2016, 120, 28–34. [Google Scholar] [CrossRef]
- Gao, R.; Yu, D.; Chen, L.; Wang, W.; Sun, L.; Chang, Y. Cloning and Functional Analysis of Squalene Synthase Gene from Dryopteris Fragrans (L.) Schott. Protein Expr. Purif. 2019, 155, 95–103. [Google Scholar] [CrossRef]
- Boyd, D.; Beckwith, J. Positively Charged Amino Acid Residues Can Act as Topogenic Determinants in Membrane Proteins. Proc. Natl. Acad. Sci. USA 1989, 86, 9446–9450. [Google Scholar] [CrossRef] [Green Version]
- Aminfar, Z.; Tohidfar, M. In Silico Analysis of Squalene Synthase in Fabaceae Family Using Bioinformatics Tools. J. Genet. Eng. Biotechnol. 2018, 16, 739–747. [Google Scholar] [CrossRef]
- Zhao, H.; Tang, Q.; Mo, C.; Bai, L.; Tu, D.; Ma, X. Cloning and Characterization of Squalene Synthase and Cycloartenol Synthase from Siraitia Grosvenorii. Acta Pharm. Sin. B 2017, 7, 215–222. [Google Scholar] [CrossRef] [Green Version]
- Krebs, E.G.; Beavo, J.A. Phosphorylation-Dephosphorylation of Enzymes. Annu. Rev. Biochem. 1979, 48, 923–959. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete Genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [Green Version]
- Sanchita; Singh, G.; Sharma, A. In Silico Study of Binding Motifs in Squalene Synthase Enzyme of Secondary Metabolic Pathway Solanaceae Family. Mol. Biol. Rep. 2014, 41, 7201–7208. [Google Scholar] [CrossRef]
- Ding, C.; Zhao, C.L.; Chen, Z.J.; Wei, F.G.; Song, D.G.; Miao, K.R. Bioinformatics Analysis of the Squalene Synthase Gene and the Amino Acid Sequence in Ginseng Species. Int. J. Clin. Exp. Med. 2015, 8, 12818–12825. [Google Scholar] [PubMed]
- Kang, J.; Zhang, Q.; Jiang, X.; Zhang, T.; Long, R.; Yang, Q.; Wang, Z. Molecular Cloning and Functional Identification of a Squalene Synthase Encoding Gene from Alfalfa (Medicago Sativa L.). Int. J. Mol. Sci. 2019, 20, 4499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kharwar, S.; Bhattacharjee, S.; Mishra, A.K. Bioinformatics Analysis of Enzymes Involved in Cysteine Biosynthesis: First Evidence for the Formation of Cysteine Synthase Complex in Cyanobacteria. 3 Biotech 2021, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein Identification and Analysis Tools in the ExPASy Server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Stothard, P. The Sequence Manipulation Suite: JavaScript Programs for Analyzing and Formatting Protein and DNA Sequences. BioTechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef] [Green Version]
- Geourjon, C.; Deléage, G. Sopma: Significant Improvements in Protein Secondary Structure Prediction by Consensus Prediction from Multiple Alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Klausen, M.S.; Jespersen, M.C.; Nielsen, H.; Jensen, K.K.; Jurtz, V.I.; Sønderby, C.K.; Sommer, M.O.A.; Winther, O.; Nielsen, M.; Petersen, B.; et al. NetSurfP-2.0: Improved Prediction of Protein Structural Features by Integrated Deep Learning. Proteins Struct. Funct. Bioinform. 2019, 87, 520–527. [Google Scholar] [CrossRef] [Green Version]
- Wiederstein, M.; Sippl, M.J. ProSA-Web: Interactive Web Service for the Recognition of Errors in Three-Dimensional Structures of Proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [Green Version]
- Dundas, J.; Ouyang, Z.; Tseng, J.; Binkowski, A.; Turpaz, Y.; Liang, J. CASTp: Computed Atlas of Surface Topography of Proteins with Structural and Topographical Mapping of Functionally Annotated Residues. Nucleic Acids Res. 2006, 34, W116–W118. [Google Scholar] [CrossRef] [Green Version]
- Wass, M.N.; Kelley, L.A.; Sternberg, M.J.E. 3DLigandSite: Predicting Ligand-Binding Sites Using Similar Structures. Nucleic Acids Res. 2010, 38, W469–W473. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).