Abstract
Red alga dulse possesses a unique xylan, which is composed of a linear β-(1→3)/β-(1→4)-xylosyl linkage. We previously prepared characteristic xylooligosaccharide (DX3, (β-(1→3)-xylosyl-xylobiose)) from dulse. In this study, we evaluated the prebiotic effect of DX3 on enteric bacterium. Although DX3 was utilized by Bacteroides sp. and Bifidobacterium adolescentis, Bacteroides Ksp. grew slowly as compared with β-(1→4)-xylotriose (X3) but B. adolescentis grew similar to X3. Therefore, we aimed to find the key DX3 hydrolysis enzymes in B. adolescentis. From bioinformatics analysis, two enzymes from the glycoside hydrolase family 43 (BAD0423: subfamily 12 and BAD0428: subfamily 11) were selected and expressed in Escherichia coli. BAD0423 hydrolyzed β-(1→3)-xylosyl linkage in DX3 with the specific activity of 2988 mU/mg producing xylose (X1) and xylobiose (X2), and showed low activity on X2 and X3. BAD0428 showed high activity on X2 and X3 producing X1, and the activity of BAD0428 on DX3 was 1298 mU/mg producing X1. Cooperative hydrolysis of DX3 was found in the combination of BAD0423 and BAD0428 producing X1 as the main product. From enzymatic character, hydrolysis of X3 was completed by one enzyme BAD0428, whereas hydrolysis of DX3 needed more than two enzymes.
Keywords:
red alga; dulse; xylooligosaccharide; β-(1→3)/β-(1→4)-xylan; Bifidobacterium; GH43; β-xylosidase 1. Introduction
Terrestrial xylan is a β-(1→4)-xylosyl polymer possessing arabinofuranose or glucuronic acid side chains at C2 or C3 positions [1]. These oligosaccharides (XOS) are nondigestible ingredients and have the advantage of stability at a low pH and heat resistance for food industry usage as compared with other types of oligosaccharides [2,3]. XOS are utilized in human gut microbiota and show various beneficial effects on human health [4,5]. XOS as a dietary supplementation improv the blood sugar and lipids in type-2 diabetes mellitus [6]. XOS from birchwood xylan show a favorable effect on human intestinal flora [7]. Namely, in vitro analysis has shown that Bifidobacteria utilizes XOS, whereas Escherichia coli and Clostridium spp. does not utilize XOS. In addition, in vivo analysis has shown that the growth of Bifidobacteria was promoted by 5 g/day. Therefore, XOS have been used as a functional food material, for example, as dietary sweeteners for low-calorie foods [8,9].
Marine algae contain polysaccharides with no lignin and less cellulose [10,11]. Most red algae possess sulfated galactan such as agar and carrageenan [12,13], and those oligosaccharides show a prebiotic effect [14]. Among red algae, dulse (Palmaria palmata) is known to contain a mix-linked β-(1→3)/β-(1→4)-xylan in the cell wall [15,16,17,18]. These studies have focused on the elucidation of their structure, whereas our previous study showed the method of dulse XOS production [19]. The xylotriose (DX3) possesses β-(1→3)-xylosyl linkage at the nonreducing end.
Terrestrial β-(1→4)-XOS has been hydrolyzed by β-xylosidase (EC 3.2.1.37), which showed the bifunctional activities of α-l-arabinofuranosidase due to their spatial similarities between D-xylopyranose and L-arabinofuranose. On the one hand, the enzyme character has been well characterized in gut microbiota such as Bifidobacterium and Bacteroides [20]. Most enzymes in these bacteria were intracellular enzymes [21]. The oligosaccharide uptake transporter has been characterized in Bifidobacterium [22]. On the other hand, the enzyme which hydrolyzed β-(1→3)-xylosyl linkage was not reported from gut microbiota. β-(1→3)-Xylosidase was reported from Streptomyces sp. SWU10 and Vibrio sp. XY-214 [23,24]. These enzymes were classified into the glycoside hydrolase family (GH) 43 and subfamily 11 (Vibrio sp. XY-214) and 12 (Streptomyces sp. SWU10) [25]. The characterized enzyme activity of GH43_11 and GH43_12 was β-(1→4)-xylosidase and α-l-arabinofuranosidase, respectively, indicating that a key enzyme for hydrolysis of β-(1→3)-xylosyl linkage in gut microbiota was not clear.
A previous study has shown that the dulse in Japan is abundant in proteins (approximately 40 g/100 g dried dulse) and the major component is phycoerythrin (PE) [26]. The peptides produced from thermolysin hydrolysis showed the inhibitory activity of the angiotensin-I-converting enzyme (ACE) which was found in soluble proteins from many red algae [27,28,29,30,31]. The chromophores from PE showed antioxidant activity [32]. To obtain more functionality of dulse, we prepared xylooligosaccharide from dulse, which possessed the unique structure of β-(1→3)/β-(1→4)-xylotriose (DX3) [19]. In this study, we investigated the prebiotic effect of DX3 on enteric bacteria, showing that Bifidobacterium adolescentis grew quickly among the tested bacteria. We then attempted to find the key enzyme for DX3 hydrolysis from B. adolescentis and to discuss the metabolic pathway of DX3.
2. Results
2.1. In Vitro Utilization of XOS by Entric Bacteria
DX3 is a xylotriose from red alga dulse, which possesses β-(1→3)-xylosyl linkage at the nonreducing end. This structure differs in β-(1→4)-xylotriose (X3) from terrestrial plant, which is composed of only β-(1→4)-xylosyl linkage. To evaluate the effect of DX3 on entric bacterial growth, ΔOD600 and ΔpH were measured after incubation for 96 h using 10 bacteria (Figure 1 and Table S1). The data were obtained by subtraction of the sample without carbohydrates. Glucose (G1) and xylose (X1) were used as positive controls. In the case of Bacteroides thetaiotaomicron, the growth rate of X3 and DX3 was slow as compared with the medium in G1 and X1. X3 was a more suitable carbon source for the growth of Bacteroides sp. than DX3. Bifidobacterium sp. was classified into three types. The growth of B. longum subsp. infantis increased in only G1, and the growth of B. longum subsp. longum increased in G1 and X1 but not in X3 and DX3. The growth of B. adolescentis increased in X3 and DX3 similar to G1. The rest of the species, which did not metabolize xylose, could also not utilize X3 and DX3. The growth of all bacteria tested in this study was accompanied by a decrease in pH, due to the production of short chain fatty acid resulting from the bacterial fermentation [33,34]. Among the tested bacteria, the utilization of XOS between Bacteroides sp. and B. adolescentis differed. Then, we compared the time course growth rate between them.
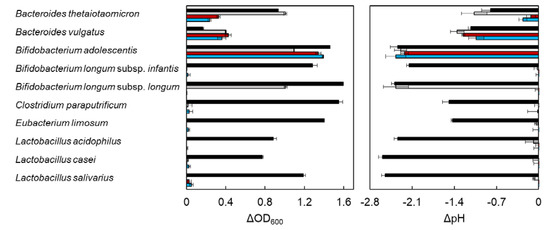
Figure 1.
Effect of carbohydrate on bacterial growth and pH. The bacterial strains were cultured at 37 °C for 96 h in PYF (Peptone−Yeast extract−Fildes) medium containing 0.5% carbohydrate samples under anaerobic conditions. The data were obtained as ΔOD600 and ΔpH by the subtraction of each bacterial growth without carbohydrate samples. Error bars indicate SD (n = 3). Growth data of C. paraputrificum and E. limosum for β-(1→4)-xylotriose (X3) were not determined. Symbols: black bar, glucose; gray bar, xylose; red bar, X3; and blue bar, β-(1→3)/β-(1→4)-xylotriose (DX3).
2.2. Time Course Growth in B. vulgatus and B. adolescentis
Growth rate of bacteria in PYF medium containing carbohydrates (G1, X1, X3 and DX3) was monitored at 24, 28, and 32 h (Figure 2). B. vulgatus grew quickly in G1 and X1. On the one hand, growth of B. vulgatus among XOS was the same up to 24 h, but growth in X3 was fast at 24 to 32 h. On the other hand, B. adolescentis grew rapidly in G1, and the growth rate was almost the same in X1, X3 and DX3. This result suggested that B. adolescentis utilized DX3 similar to X3, and the utilization of DX3 in B. vulgatus was slow as compared with X3. The difference of DX3 and X3 was the linkage at the nonreducing end. Exo hydrolase enzymes generally act on the nonreducing end of carbohydrates [35,36]. This means that B. adolescentis would possess the key enzyme for DX3 hydrolysis. Therefore, we attempted to determine the enzymes by comparing the related genes from CAZy database.
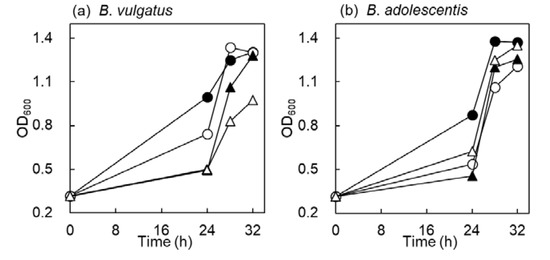
Figure 2.
Time course growth rate of B. vulgatus (a) and B. adolescentis (b). Bacteria grow in PYF medium containing 0.5% carbohydrates at 37 °C in anaerobic condition. Symbols: ●, glucose (G1); ◯, xylose (X1); ▲, X3; △, DX3. Mean ± standard deviation of three replicate determinations and the error bars were within the symbols.
2.3. Comparison of β-Xylosidase Genes in the Tested Bacterium
GH was classified into their primary sequences. Therefore, each family possessed enzymes having different substrate specificity. β-Xylosidase (EC 3.2.1.37) was classified into 13 families, and genes of the tested bacteria belonged to GH 1, 2, 3, 30, 43, 51, and 120 (Figure 3). We attempted to find the specific DX3 hydrolysis enzymes from B. adolescentis. B. adolescentis possessed putative 22 β-xylosidase genes. Among them, Blast analysis revealed that two GH1 and six GH3 enzymes were β-glucosidase, and three GH2 enzymes were β-galactosidase. The GH30 enzyme was subdivided into nine subfamilies, and only the subfamily 2 consisted of β-xylosidase. The B. adolescentis GH30 enzyme was subfamily 2, but two B. longum subsp. longum GH30 genes were subfamily 5. Within two Bacteroides sp., B. vulgatus possessed two subfamily 2 enzymes. One GH30 from the B. vulgatus enzyme showed 60% identity with the B. adolescentis GH30 enzyme. In addition to the low identity between them, B. thetaiotaomicron did not possess the enzyme, indicating that this is not a key enzyme for DX3 hydrolysis. The activity of GH51 enzyme in the tested bacterium was reported as α-l-arabinofuranosidase. Bacteria, which can or cannot utilize X3 and DX3, also possessed GH51, indicating that this family was not the key enzyme for DX3 hydrolysis. The GH120 enzyme is classified into β-xylosidase, and only found in B. adolescentis in the tested bacterium. Previous studies have reported that the enzyme preferred longer XOS larger than X4 [33,37,38]. The target XOS in this study is X3. Therefore, we excluded this enzyme, and a more detailed explanation is included in the Discussion section. GH43 is a large family containing 37 subfamilies, and many studies have been performed with the understanding of the relationship between Bifidobacterium sp. and XOS [39,40,41,42,43,44,45]. To find the key enzymes for DX3 hydrolysis, the enzymes in GH43 were evaluated.
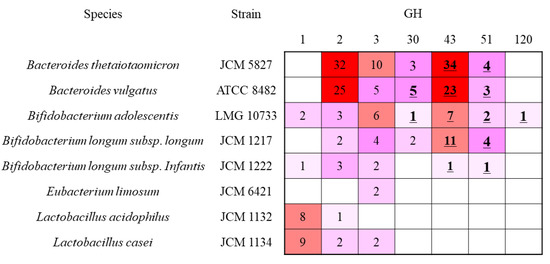
Figure 3.
Relationship between bacteria and the number of enzymes containing β-xylosidase (EC 3.2.1.37) in glycoside hydrolase families (GHs). The colors are related to the number of enzymes from pink (low) to red (high). The bold and underlined number means that GHs contain β-xylosidase.
2.4. β-Xylosidase in GH43 Subfamily
Among 37 subfamilies in GH43, 21 subfamilies were characterized as β-xylosidase or α-l-arabinofuranosidase (Figure 4). Six of seven B. adolescentis enzymes showed the enzyme activity, except for subfamily 26. A comparison of the GH43 subfamilies of B. adolescentis (DX3 metabolic bacterium) to B. longum sp. (DX3 non-metabolic bacteria), showed that subfamilies 11 and 12 were only found in B. adolescentis. Although B. vulgatus also possessed GH43 subfamily 12 (GH43_12), the bacterium also utilized X3 and DX3. From these characteristics, GH43_11 and _12 were the candidates for DX3 hydrolysis. GH43_11 was characterized as β-xylosidase (BAD0428 and BAD1203) and GH43_12 as α-l-arabinofuranosidase (BAD0423). In addition, information about the activity of β-(1→3)-xylosidase from Streptomyces sp. (GH43_12) and Vibrio sp. (GH43_11) would be a clue for finding β-(1→3)-xylosidase from gut microbiota. GH43_22 (BAD1527) β-xylosidase was also employed for the enzymatic analysis.
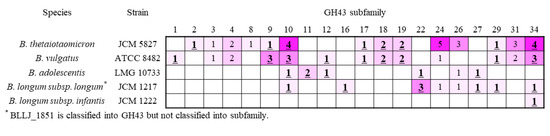
Figure 4.
Relationship between bacteria and the number of enzymes in the GH43 subfamily. The colors are related to the number of enzymes from pink (low) to purple (high). The bold and underlined number shows subfamilies contain β-xylosidase.
2.5. Enzymatic Character of GH43 Enzymes
Although we selected four GH43 genes to evaluate the DX3 hydrolysis, the BAD1203 gene did not amplify by PCR. Therefore, we used three GH43 enzymes (BAD0423, BAD0428, and BAD1527). These enzymes were expressed as bacterial recombinant enzymes. The p-nitrophenyl-β-d-xylopyranoside (pNP-X) activity of BAD0423, BAD0428, and BAD1527 were 6, 77,035, and 10 mU/mg, respectively, showing that the characters of the enzymes were the same as previous reports [21,45,46,47]. Using these enzymes, hydrolysis products of X2, X3, and DX3 were evaluated (Figure 5). BAD0423 showed low activities on X2 and X3 but high activity on DX3. The hydrolysis ratio reached 52% for 1 h hydrolysis, and the hydrolysis products were X1 and X2. BAD0428 hydrolyzed XOS producing X1 as the main product. The activity on DX3 was weak (19%) as compared with X2 and X3. The low activity of BAD0428 on DX3 was due to the low hydrolysis activity on β-(1→3)-linkage at the nonreducing end since the hydrolysis products of DX3 was X1, meaning that the produced X2 was immediately hydrolyzed. BAD1527 weakly hydrolyzed XOS. The specific activity of BAD0423, BAD0428, and BAD1527 on DX3 was determined by HPLC as 2988, 1440, and 84 mU/mg, respectively. The hydrolysis products of BAD0423 on DX3 remained X2, indicating that the cooperative hydrolysis was necessary for complete DX3 hydrolysis. Therefore, BAD0423 and BAD0428 were used for DX3 hydrolysis, resulting in the hydrolysis ratio of 77%, which was slightly high hydrolysis by each enzyme (71%), and that of X1 was 90%. From these data, BAD0423 was the key enzyme for DX3, and the hydrolysis products contained X2, therefore, the cooperative hydrolysis with highly active β-xylosidase (BAD0428) were needed (Figure 6).
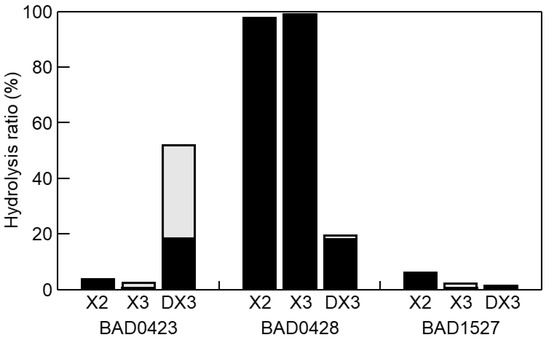
Figure 5.
Hydrolysis products of oligosaccharides (XOS). Ten mM XOS was hydrolyzed by 50 μg/mL BAD0423, BAD0428, and BAD1527 at pH 6.5 and 37 °C for 1 h. The products were analyzed by HPLC. Black and gray indicate the amount of X1 and X2, respectively.
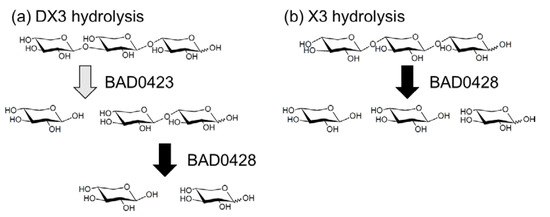
Figure 6.
The putative hydrolysis mechanism of DX3 and X3. (a) DX3; (b) X3.
3. Discussion
Red alga dulse possesses a unique xylan in the cell wall, and we have previously developed the preparation method of DX3 (β-(1→3)-xylosyl-xylobiose) [19]. In this study, we evaluated the prebiotic effect of DX3 on bacteria. Among the tested bacteria, Bifidobacterium adolescentis effectively metabolized DX3, indicating that the bacterium possessed the enzyme having β-(1→3)-xylosidase activity. Two candidate enzymes (BAD0423: GH43_12 and BAD0428: GH43_11) were obtained from bioinformatics analysis. We characterized these enzymes, revealing that BAD0423 was the key enzyme for the hydrolysis of β-(1→3)-xylosyl linkage, and BAD0428 was necessary for the complete hydrolysis of DX3 for X1. These genes were members of the xylooligosaccharide utilization cluster composed of xylose isomerase (BAD0422), GH43_12 arabinofuranosidase (BAD0423), LacI transcriptional regulators (BAD0424), solute binding protein (BAD0425), two ABC transporter (BAD0426 and BAD0427), GH43_11 β-xylosidase (BAD0428), two esterase (BAD0429 and BAD0430), and xylulose kinase (BAD0431) [48]. It has been reported that the solute binding protein from Bifidobacterium animals subsp. lactis Bl-04 captures XOS and arabino-xylooligosaccharides with the specificity for tri- and tetrasaccharides, which show 69% identity with BAD0425 [22]. The structure of arabinoxylobiose was α-(1→3)-arabinofuranosyl xylobiose, which was quite similar to DX3, suggesting that DX3 would also be incorporated into B. adolescentis by the same solute binding protein (BAD0425). Bifidobacterium sp., which did not increase in XOS containing medium in this test, did not possess the cluster, indicating that the enzymes containing this cluster are necessary for DX3 hydrolysis.
The activities of BAD0423 (α-l-arabinofuranosidase) on pNP-Ara and pNP-X have been reported as 249 and 4 mU/mg, respectively [46], and we also obtained the same results. In addition, the DX3 activity of BAD0423 was 2988 mU/mg, which was 10 and 500 times higher than that of pNP-Ara and pNP-X, respectively. The arabinose releasing activity against arabinoxylan of AXH-m23 from B. adolescentis DSM 20083 (BAD0423) has been reported as 80,000 mU/mg [45], meaning that the favorite substrate at subsite-1 of BAD0423 would be α-(1→3)-arabinofuranose residues. The activity of the GH43_12 enzyme from Streptomyces sp. SWU10 showed β-(1→3)-xylosidase activity with the specific activity of 1330 mU/mg toward β-(1→3)-xylobiose, which was 440-fold higher than that of X2 [24]. The identity of this enzyme and BAD0423 was low (20%), suggesting that there are many enzymes having β-(1→3)-xylosidase activity in GH43_12.
The growth rate of B. adolescentis in X3 and DX3 was almost the same, indicating that the utilization of X3 and DX3 corresponded to the enzyme activities. This means that the high amount of BAD0423 in the bacterium was needed as compared with BAD0428. A study on transcriptional analysis and proteome analysis in Bifidobacterium animalis sp. showed that the expression level of β-xylosidase (a homolog of BAD0428) was high [48,49,50], indicating that additional β-(1→3)-xylosidases were required. Although there are fewer reports on β-(1→3)-xylosidase activity, our results suggested that the enzyme having α-(1→3)-arabinofuranosidase activity also showed β-(1→3)-xylosidase activity. Therefore, we attempted to find the enzyme that corresponded with the Bacteroides sp. The common enzyme families having α-(1→3)-arabinofuranosidase were found in GH30, -43, and -51.
In GH30, only subfamily 2 possessed β-xylosidase activity. Two enzymes from B. vulgatus showed 60% identity, but these showed a low identity to the B. adolescentis enzyme (27% and 30%). In addition, B. thetaiotaomicron, which showed the same growth rate with B. vulgatus, did not possess these subfamily enzymes. The GH30 enzyme from Bifidobacterium breve K-110 showed pNP-X activity but not pNP-Ara [51], indicating that high β-(1→3)-xylosidase activity was not expected in this family.
The activities for DX3, on seven of the three GH43 enzymes, were characterized in this study. GH43_10 (BAD0301) has been reported as the double substituted xylan α-1,3-l-specific arabinofuranosidase [45,46]. GH43_27 from B. longum sp. (BLLJ_1852) has shown an arabinan-degrading exo-1,2-1,3-α-l-arabinofuranosidase [40] and the study showed that the activity of arabinose releasing from arabinoxylan of BLLJ_1852 was 0.62-fold of pNP-Ara, suggesting that the character did not agree with the high DX3 hydrolysis activity. In addition, they also mentioned that the subfamily 27 possessed various substrate specific enzymes. The identity of GH43_27 BAD0149 with BLLJ_1852 was 40%. However, two Bacteroides sp. did not possess this subfamily enzymes, suggesting a low possibility of high DX3 hydrolysis activity. GH43_26 BAD0152 was classified into exo-α-1,5-l-arabinofuranosidase, suggesting that this was not suitable. Although the main activity of GH43_11 BAD0428 was β-(1→4)-xylosidase activity, the enzyme also showed β-(1→3)-xylosidase activity. The activity of the GH43_11 enzyme from Vibrio sp. XY-214 has shown high β-(1→3)-xylosidase activity [23]. The identity of these enzymes was 34%. We attempted to express GH43_11 BAD1203 enzymes resulting in failure of characterization. The primary sequence identities of BAD1203 to BAD0423 and Vibrio sp. enzyme were 32% and 33%, respectively, remaining the potential for DX3 hydrolase. Although the activity of GH43_22 BAD1527 on the tested substrate was low, the high activity was reported on arabinogalactan from B. longum subsp. longum showing 20% identity [44]. Among GH43, the subfamily 11 (BAD1203) had potential for β-(1→3)-xylosidase activity, however only one Bacteroides sp. in the tested possessed this subfamily enzyme, indicating that alternative enzymes could exist.
In GH51, BAD1205 and BAD1524, which was designated as α-l-arabinofuranosidase, showed low identity (13%). The two enzymes also showed low identities with four GH51 from B. longum subsp. longum JCM 1217 (11% to 18%). α-l-Arabinofuranosidase from B. longum B667, which possess 100% identity with BLLJ0445, showed high activity on the arabinose side chain of arabinan as compared with that of arabinoxylan [52]. The GH51 enzymes within Bacteroides sp. showed high identities as follows: BT0368 and BVU1001 (74%), BT3657 and BVU0496 (57%), and BT0438 and BVU4054 (77%). BAD1205 showed 27% and 29% identities with BT0348 and BVU4054, respectively, and BAD1524 showed 30% and 32% identities with BT3657 and BVU0496, respectively, suggesting the potential for DX3 hydrolysis. Studies on GH51 have shown the high activity on arabinan side chain than arabinoxylan [53,54,55], and pNP-Ara activity of GH51 was lower than that of GH43 [53]. Therefore, the activity of DX3 could be low.
The activity of GH120 β-xylosidase has been reported as pNP-Ara and pNP-X [47]. When the enzyme hydrolyzed AXOS, only X1 was released but not Ara, suggesting that the possibility of DX3 hydrolysis activity of this enzyme was low.
Bacteroides vulgatus also possesses XOS utilization locus, containing four GH43 enzymes [55]. The study reported that two GH43 enzymes (BVU_0039 and BVU_0040, subfamily 1) were sufficient for XOS utilization. B. adolescentis possesses the same subfamily 12 with BVU_0039, and the rest of subfamily BVU_0040 (subfamily 1) would work like BAD0428, a β-xylosidase.
4. Materials and Methods
4.1. Materials
Dulse (Palmaria palmata in Japan) was harvested near Usujiri, Hokkaido, Japan in February 2017 and stored at −30 °C until use [30,31]. Xylose, β-(1→4)-xylobiose (X2) and β-(1→4)-xylotriose (X3) were purchased from Wako Pure Chemical Industries (Osaka, Japan). β-(1→4)-Xylotetraose (X4) and β-(1→4)-xylopentaose (X5) were purchased from Megazyme (Bray, Ireland). All other reagents were purchased from Wako Pure Chemical Industries (Osaka, Japan).
4.2. Preparation of DX3 from Red Alga Dulse
DX3 (23-β-d-xylosyl-xylobiose) (99.0%>) was prepared as previously in [19]. The frozen dulse thalli were lyophilized and homogenized by a Wonder Blender WB-1 (OSAKA CHEMIKAL CO., Osaka, Japan) into powder. The dulse powder was suspended in 40 volumes (v/w) of distilled water, and the suspension was autoclaved at 121 °C for 20 min. Then, the solution was centrifuged at 15,000× g for 10 min. The supernatant was dialyzed against distilled water with a dialysis tube (molecular weight cut off, approximately 14 kDa, EIDIA Co., Ltd., Tokyo, Japan). The dialysis solution was centrifuged at 15,000× g for 10 min to remove small amount of insoluble materials. The supernatant (10 mg/ml) was hydrolyzed for 6 h by 2 wt% (5 U) of SucraseX (Mitsubishi-Chemical Foods Corporation, Japan) at pH 6.0 and 50 °C. The enzyme reactions were stopped by heating at 100 °C for 10 min. The products were applied to an activated carbon column (φ 3 × 35 cm) pre-equivalated with distilled water. The carbohydrate was eluted by a stepwise system consisting of 0%, 10%, 15%, 20%, and 25% ethanol. The DX3 fractions were detected by HPLC equipped with a Sugar-D column and refractive index detector (RI). The DX3 fractions were pooled, evaporated, and lyophilized as DX3 powder. The purity of DX3 was evaluated by HPLC.
4.3. Bacterial Growth
The bacterial strains used in this study (Bacteroides thetaiotaomicron JCM 5827, Bacteroides vulgatus JCM 5826, Bifidobacterium adolescentis JCM 7046, Bifidobacterium longum subsp. infantis JCM 1222, Bifidobacterium longum subsp. longum JCM 1217, Clostridium paraputrificum JCM 1293, Eubacterium limosum JCM 6421, Lactobacillus acidophilus JCM 1132, and Lactobacillus casei JCM 1134) were provided by Japan Collection of Microorganisms, RIKEN BRC which is participating in the National BioResource Project of the MEXT, Japan. The defibrinated blood was purchased from Cosmo Bio CO., LTD.
PYF medium consisted of 10 g Trypticase Peptone (BBL, Becton Dickinson & Company, Cockeysville, MD, USA), 5 g yeast extract (Difco Lab, Detroit, MI, USA), 0.5 g L-cysteine hydrochloride, 40 mL Fildes solution, and 40 mL salt solution per 1 L. Fildes solution contained 0.85% NaCl, 6 mL HCl, 50 mL horse defibrinated blood, and pepsin 1:10,000 (Fujifilm Wako Pure Chemical Industries, Ltd., Osaka, Japan). The salt solution contained 0.2 g CaCl2, 0.2 g MgSO4 (7H2O), 1.0 g KH2PO4, 1.0 g K2HPO4, 10 g NaHCO3, and 2.0 g NaCl in 1 L deionized water. Before the fermentation trials, bacterial strains were grown on GAM plate (Nissui Pharmaceutical, Japan) and then precultured in PYF broth containing 1% glucose.
Precultured bacterial suspensions were prepared at a concentration of 1 × 107 CFU/tube in the PFY medium containing 0.5% carbohydrates (glucose, xylose, X3, and DX3), and then incubated at 37 °C for 96 h in anaerobiosis condition (Anaeropack Anaero, Mitsubishi Gas Chemical Co., Inc., Germany). The growth of bacteria in each carbohydrate was evaluated as ΔpH and ΔOD600 by the lowering of pH and the increase in OD600 by subtraction of the values without carbohydrate.
Time course growth of B. adolescentis and B. vulgatus was monitored up to 32 h and the OD600 was measured at 24, 28, and 32 h. All the growth assays were performed in triplicate.
4.4. Bioinformatics
The genes related to carbohydrate-active enzymes in bacteria were collected from the CAZy database (http://www.cazy.org/). Among them, the glycoside hydrolase families containing β-xylosidase genes (EC 3.2.1.37) were selected. The function of genes that classified into GH containing several enzymes was manually confirmed by Blast search (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Sequences in the same family were confirmed by CLUSTALW (https://www.genome.jp/tools-bin/clustalw). The bacterial genomic sequences used in this study were as follows: B. thetaiotaomicron JCM 5827 (GenBank AE015928), B. vulgatus JCM 5826 (GenBank CP000139), B. adolescentis JCM 7046 (GenBank LNKM00000000), B. longum subsp. infantis JCM 1222 (GenBank AP010889), B. longum subsp. longum JCM 1217 (GenBank AP010888), C. paraputrificum JCM 1293 (GenBank UAWH00000000), E. limosum JCM 6421 (GenBank CP019962), L. acidophilus JCM 1132 (GenBank AZCS00000000), and L. casei JCM 1134 (GenBank AP012544).
4.5. Cloning, Expression, and Purification of Enzymes
Genomic DNA from B. adolescentis JCM 7046 was extracted using an innu PREP Bacteria DNA Kit (Analytik Jena, Jena, Germany). We prepared the full length of xylosidase constructs (BAD0423, BAD0428, and BAD1527) having the His-tag at N-terminus, since these did not possess signal peptide (http://www.cbs.dtu.dk/services/SignalP-4.1/). The xylosidase genes were amplified from the genomic DNA by PCR using KOD FX new DNA polymerase (Toyobo, Japan) with the sets of primers (Table S2). The PCR products were cloned into the pBluescript II SK(+) vector and sequenced. The fragments were digested by restriction enzymes (Table S1) and ligated into pET28a vector with the digestion of the sets of restriction enzymes. Escherichia coli BL21 (DE3) was transformed with the expression plasmid. Protein expression and purification were performed as previously [56]. The protein expression was induced by 0.1 mM isopropyl β-d-thiogalactopyranoside. The cells were collected by centrifugation and disrupted by sonication. The recombinants were purified by TALON metal affinity resin (Talon, Clontech, Japan) according to the manufacturer’s protocol. The active fractions were dialyzed with a dialysis tube (molecular weight cutoff about 14 kDa, EIDIA Co., Ltd., Tokyo, Japan) against 20 mM sodium phosphate buffer (pH 6.5). The purity of recombinants was confirmed as a single band by SDS-PAGE (Figure S1). The protein concentrations were determined by the absorption at 280 nm using molecular absorptivity of each protein.
4.6. Activity Assay
β-d-Xylosidase activity was measured based on the release of p-nitrophenyl (pNP) from pNP-β-d-Xylopyranoside (pNP-X). The reaction mixture containing 18.4 mM pNP-X, an appropriate amount of purified enzyme, and 10 mM sodium phosphate buffer (pH 6.5) was incubated at 37 °C for 30 min. The reaction was terminated by adding 1.5 volumes of 1.0 M Na2CO3, and the released pNP was measured at 400 nm. One unit was defined as the amount of enzyme that released 1.0 μmol pNP per minute.
4.7. HPLC Analysis
Hydrolysis products of XOS were determined by HPLC using a RI detector. The enzyme reaction was performed as follows: 10 mM XOS (X2, X3, and DX3) were hydrolyzed by 50 μg/mL or enzymes in 20 mM sodium phosphate buffer (pH 6.5) at 37 °C for 1 h. The reaction was terminated by heating at 95 °C for 5 min. The products were applied to HPLC equipped with a Sugar-D column (4.6 × 250 mm, Nakalai Tesque, Kyoto, Japan) with the column oven temperature at 40 °C. The products were eluted with an isocratic elution system of acetonitrile/water (4:1, v/v) at a flow rate of 1.0 ml/min. The enzyme activities were determined by the amount of hydrolysis products. Xylose and XOS were used as standards.
5. Conclusions
Bifidobacterium adolescentis utilized DX3 from red alga dulse similar to X3. Therefore, key enzymes for β-(1→3)-xylosidase activity were investigated, with the result that BAD0423 (GH43_12) and BAD0428 (GH43_11) were required for DX3 hydrolysis. Although xylose was released from DX3 by the above two enzymes, the rate of xylose release from X3 was faster than DX3, which did not coincide with the growth rate of B. adolescentis. In search of enzymes, GH43_11 and GH51 enzymes occurred as candidates. However, the putative activity for DX3 could be low. Therefore, cooperative hydrolysis of a repertoire of various α-l-arabinofuranosidases are needed.
Supplementary Materials
The following are available online at https://www.mdpi.com/1660-3397/18/3/174/s1, Table S1: Effect of carbohydrate on bacterial growth and pH, Table S2: Primers used in this study, Figure S1: SDS-PAGE of recombinant proteins.
Author Contributions
H.K. and H.Y. conceived and designed the research; H.K. and H.Y. contributed to sample collection; M.K., Y.Y., and Y.K. performed the experiments and analyzed the data; M.K., Y.K., and H.K. contributed to writing and editing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported in part by MEXT/JSPS KAKENHI (grant number 18K05810).
Acknowledgments
We gratefully acknowledge the sampling assistant of Palmaria palmata in Japan at Usujiri by Hiroyuki Munehara and Atsuya Miyajima. The authors wish to thank Koji Yamazaki for his technical assistance with bacterial growth.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ebringerová, A.; Hromádková, Z.; Heinze, T. Hemicellulose. In Polysaccharides I: Structure, Characterization and Use; Heinze, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 1–67. [Google Scholar]
- Carvalho, A.F.A.; de Oliva Neto, P.; da Silva, D.F.; Pastore, G.M. Xylo-oligosaccharides from lignocellulosic materials: Chemical structure, health benefits and production by chemical and enzymatic hydrolysis. Food Res. Int. 2013, 51, 75–85. [Google Scholar] [CrossRef]
- Singh, R.D.; Banerjee, J.; Arora, A. Prebiotic potential of oligosaccharides: A focus on xylan derived oligosaccharides. Bioact. Carbohydrates Diet. Fibre 2015, 5, 19–30. [Google Scholar] [CrossRef]
- Manisseri, C.; Gudipati, M. Prebiotic activity of purified xylobiose obtained from Ragi (Eleusine coracana, Indaf-15) bran. Indian J. Microbiol. 2012, 52, 251–257. [Google Scholar] [CrossRef]
- Zhu, Z.-Y.; Zhao, L.; Ge, X.-R.; Tang, Y.-L.; Chen, L.-J.; Pang, W.; Zhang, Y. Preparation, characterization and bioactivity of xylobiose and xylotriose from corncob xylan by xylanase. Eur. Food Res. Technol. 2015, 241, 27–35. [Google Scholar] [CrossRef]
- Sheu, W.H.-H.; Lee, I.-T.; Chen, W.; Chan, Y.-C. Effects of Xylooligosaccharides in type 2 diabetes mellitus. J. Nutr. Sci. Vitaminol. 2008, 54, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, M.; Fujikawa, S.; Matsumoto, N. Effect of xylooligosaccharide on the growth of Bifidobacteria. Bifidobact. Microflora 1990, 9, 77–86. [Google Scholar] [CrossRef]
- Aachary, A.A.; Prapulla, S.G. Xylooligosaccharides (XOS) as an emerging prebiotic: Microbial synthesis, utilization, structural characterization, bioactive properties, and applications. Compr. Rev. Food Sci. Food Saf. 2011, 10, 2–16. [Google Scholar] [CrossRef]
- Akpinar, O.; Erdogan, K.; Bostanci, S. Production of xylooligosaccharides by controlled acid hydrolysis of lignocellulosic materials. Carbohydr. Res. 2009, 344, 660–666. [Google Scholar] [CrossRef]
- Martone, P.T.; Estevez, J.M.; Lu, F.; Ruel, K.; Denny, M.W.; Somerville, C.; Ralph, J. Discovery of lignin in seaweed reveals convergent evolution of cell-wall architecture. Curr. Biol. 2009, 19, 169–175. [Google Scholar] [CrossRef]
- Hong, I.K.; Jeon, H.; Lee, S.B. Comparison of red, brown and green seaweeds on enzymatic saccharification process. J. Ind. Eng. Chem. 2014, 20, 2687–2691. [Google Scholar] [CrossRef]
- McCandless, E.L.; Craigie, J.S. Sulfated polysaccharides in red and brown algae. Ann. Rev. Plant Physiol. 1979, 30, 41–53. [Google Scholar] [CrossRef]
- Pereira, M.G.; Benevides, N.M.B.; Melo, M.R.S.; Valente, A.P.; Melo, F.R.; Mourão, P.A.S. Structure and anticoagulant activity of a sulfated galactan from the red alga, Gelidium crinale. Is there a specific structural requirement for the anticoagulant action? Carbohydr. Res. 2005, 340, 2015–2023. [Google Scholar] [CrossRef] [PubMed]
- Cherry, P.; Yadav, S.; Strain, C.R.; Allsopp, P.J.; McSorley, E.M.; Ross, R.P.; Stanton, C. Prebiotics from Seaweeds: An Ocean of Opportunity? Mar. Drugs 2019, 17, 327. [Google Scholar] [CrossRef] [PubMed]
- Lahaye, M.; Rondeau-Mouro, C.; Deniaud, E.; Buléon, A. Solid-state 13C NMR spectroscopy studies of xylans in the cell wall of Palmaria palmata (L. Kuntze, Rhodophyta). Carbohydr. Res. 2003, 338, 1559–1569. [Google Scholar] [CrossRef]
- Morgan, K.C.; Wright, J.L.C.; Simpson, F.J. Review of chemical constituents of the red alga Palmaria palmata (dulse). Econ. Bot. 1980, 34, 27–50. [Google Scholar] [CrossRef]
- Lahaye, M.; Michel, C.; Barry, J.L. Chemical, physicochemical and in-vitro fermentation characteristics of dietary fibres from Palmaria palmata (L.) Kuntze. Food Chem. 1993, 47, 29–36. [Google Scholar] [CrossRef]
- Deniaud, E.; Quemener, B.; Fleurence, J.; Lahaye, M. Structural studies of the mix-linked β-(1→3)/β-(1→4)-d-xylans from the cell wall of Palmaria palmata (Rhodophyta). Int. J. Biol. Macromol. 2003, 33, 9–18. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Kishimura, H.; Kinoshita, Y.; Saburi, W.; Kumagai, Y.; Yasui, H.; Ojima, T. Enzymatic production of xylooligosaccharides from red alga dulse (Palmaria sp.) wasted in Japan. Process Biochem. 2019, 82, 117–122. [Google Scholar] [CrossRef]
- van den Broek, L.A.; Hinz, S.W.; Beldman, G.; Vincken, J.P.; Voragen, A.G. Bifidobacterium carbohydrases-their role in breakdown and synthesis of (potential) prebiotics. Mol. Nutr. Food Res. 2008, 52, 146–163. [Google Scholar] [CrossRef]
- Amaretti, A.; Bernardi, T.; Leonardi, A.; Raimondi, S.; Zanoni, S.; Rossi, M. Fermentation of xylo-oligosaccharides by Bifidobacterium adolescentis DSMZ 18350: Kinetics, metabolism, and β-xylosidase activities. Appl. Microbiol. Biotechnol. 2013, 97, 3109–3117. [Google Scholar] [CrossRef]
- Ejby, M.; Fredslund, F.; Vujicic-Zagar, A.; Svensson, B.; Slotboom, D.J.; Abou Hachem, M. Structural basis for arabinoxylo-oligosaccharide capture by the probiotic Bifidobacterium animalis subsp. lactis Bl-04. Mol. Microbiol. 2013, 90, 1100–1112. [Google Scholar] [CrossRef] [PubMed]
- Umemoto, Y.; Onishi, R.; Araki, T. Cloning of a novel gene encoding β-1,3-xylosidase from a marine bacterium, Vibrio sp. strain XY-214, and characterization of the gene product. Appl. Environ. Microbiol. 2008, 74, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Phuengmaung, P.; Fujiwara, D.; Sukhumsirichart, W.; Sakamoto, T. Identification and characterization of the first β-1,3-D-xylosidase from a gram-positive bacterium, Streptomyces sp. SWU10. Enzyme Microb. Technol. 2018, 112, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Mewis, K.; Lenfant, N.; Lombard, V.; Henrissat, B. Dividing the large glycoside hydrolase family 43 into subfamilies: A motivation for detailed enzyme characterization. Appl. Environ. Microbiol. 2016, 82, 1686–1692. [Google Scholar] [CrossRef]
- Miyabe, Y.; Furuta, T.; Takeda, T.; Kanno, G.; Shimizu, T.; Tanaka, Y.; Gai, Z.; Yasui, H.; Kishimura, H. Structural properties of phycoerythrin from dulse Palmaria palmata. J. Food Biochem. 2017, 41, e12301. [Google Scholar] [CrossRef]
- Kitade, Y.; Miyabe, Y.; Yamamoto, Y.; Takeda, H.; Shimizu, T.; Yasui, H.; Kishimura, H. Structural characteristics of phycobiliproteins from red alga Mazzaella japonica. J. Food Biochem. 2018, 42, e12436. [Google Scholar] [CrossRef]
- Furuta, T.; Miyabe, Y.; Yasui, H.; Kinoshita, Y.; Kishimura, H. Angiotensin I converting enzyme inhibitory peptides derived from phycobiliproteins of dulse Palmaria palmata. Mar. Drugs 2016, 14, 32. [Google Scholar] [CrossRef]
- Fitzgerald, C.; Mora-Soler, L.; Gallagher, E.; O’Connor, P.; Prieto, J.; Soler-Vila, A.; Hayes, M. Isolation and characterization of bioactive pro-peptides with in vitro renin inhibitory activities from the macroalga Palmaria palmata. J. Agric. Food Chem. 2012, 60, 7421–7427. [Google Scholar] [CrossRef]
- Kumagai, Y.; Miyabe, Y.; Takeda, T.; Adachi, K.; Yasui, H.; Kishimura, H. In silico analysis of relationship between proteins from plastid genome of red alga Palmaria sp. (Japan) and angiotensin I converting enzyme inhibitory peptides. Mar. Drugs 2019, 17, 190. [Google Scholar] [CrossRef]
- Kumagai, Y.; Ryota, T.; Miyabe, Y.; Takeda, T.; Adachi, K.; Yasui, H.; Kishimura, H. Complete sequence of mitochondrial DNA of red alga dulse Palmaria palmata (Linnaeus) Weber & Mohr in Japan. Mitochondrial DNA Part B Resour. 2019, 4, 3177–3178. [Google Scholar]
- Sato, N.; Furuta, T.; Takeda, T.; Miyabe, Y.; Ura, K.; Takagi, Y.; Yasui, H.; Kumagai, Y.; Kishimura, H. Antioxidant activity of proteins extracted from red alga dulse harvested in Japan. J. Food Biochem. 2019, 43, e12709. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, B.; Cao, Y.; Wang, C. In vitro fermentation of xylooligosaccharides from wheat bran insoluble dietary fiber by Bifidobacteria. Carbohydr. Polym. 2010, 82, 419–423. [Google Scholar] [CrossRef]
- van Zanten, G.C.; Knudsen, A.; Röytiö, H.; Forssten, S.; Lawther, M.; Blennow, A.; Lahtinen, S.J.; Jakobsen, M.; Svensson, B.; Jespersen, L. The effect of selected synbiotics on microbial composition and short-chain fatty acid production in a model system of the Human colon. PLoS ONE 2012, 7, e47212. [Google Scholar] [CrossRef] [PubMed]
- Taylor, E.J.; Smith, N.L.; Turkenburg, J.P.; D’Souza, S.; Gilbert, H.J.; Davies, G.J. Structural insight into the ligand specificity of a thermostable family 51 arabinofuranosidase, Araf51, from Clostridium thermocellum. Biochem. J. 2006, 395, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Hövel, K.; Shallom, D.; Niefind, K.; Belakhov, V.; Shoham, G.; Baasov, T.; Shoham, Y.; Schomburg, D. Crystal structure and snapshots along the reaction pathway of a family 51 α-l-arabinofuranosidase. EMBO J. 2003, 22, 4922–4932. [Google Scholar] [CrossRef]
- Yamada, T.; Akiyama, T.; Hatano, H.; Kimura, K. In Vitro assessment of oligosaccharides assimilation by intestinal anaerobic bacteria. Milk Sci. 2015, 64, 87–98. [Google Scholar]
- Moura, P.; Barata, R.; Carvalheiro, F.; Gírio, F.; Loureiro-Dias, M.C.; Esteves, M.P. In vitro fermentation of xylo-oligosaccharides from corn cobs autohydrolysis by Bifidobacterium and Lactobacillus strains. LWT Food Sci. Technol. 2007, 40, 963–972. [Google Scholar] [CrossRef]
- Duranti, S.; Milani, C.; Lugli, G.A.; Mancabelli, L.; Turroni, F.; Ferrario, C.; Mangifesta, M.; Viappiani, A.; Sanchez, B.; Margolles, A.; et al. Evaluation of genetic diversity among strains of the human gut commensal Bifidobacterium adolescentis. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Komeno, M.; Hayamizu, H.; Fujita, K.; Ashida, H. Two novelα-l-arabinofuranosidases from Bifidobacterium longum subsp. longum belonging to glycoside hydrolase family 43 cooperatively degrade arabinan. Appl. Environ. Microbiol. 2019, 85, e02582-18. [Google Scholar]
- Lee, J.H.; Hyun, Y.J.; Kim, D.H. Cloning and characterization of α-l-arabinofuranosidase and bifunctional α-l-arabinopyranosidase/β-d-galactopyranosidase from Bifidobacterium longum H-1. J. Appl. Microbiol. 2011, 111, 1097–1107. [Google Scholar] [CrossRef]
- Milani, C.; Andrea Lugli, G.; Duranti, S.; Turroni, F.; Mancabelli, L.; Ferrario, C.; Mangifesta, M.; Hevia, A.; Viappiani, A.; Scholz, M.; et al. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci. Rep. 2015, 5, 15782. [Google Scholar] [CrossRef] [PubMed]
- Viborg, A.H.; Sørensen, K.I.; Gilad, O.; Steen-Jensen, D.B.; Dilokpimol, A.; Jacobsen, S.; Svensson, B. Biochemical and kinetic characterisation of a novel xylooligosaccharide-upregulated GH43 β-d-xylosidase/α-l-arabinofuranosidase (BXA43) from the probiotic Bifidobacterium animalis subsp. lactis BB-12. AMB Express 2013, 3, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Sakamoto, A.; Kaneko, S.; Kotake, T.; Tsumuraya, Y.; Kitahara, K. Degradative enzymes for type II arabinogalactan side chains in Bifidobacterium longum subsp. longum. Appl. Microbiol. Biotechnol. 2019, 103, 1299–1310. [Google Scholar] [CrossRef] [PubMed]
- Van Laere, K.M.J.; Voragen, C.H.L.; Kroef, T.; Van Den Broek, L.A.M.; Beldman, G.; Voragen, A.G.J. Purification and mode of action of two different arabinoxylan arabinofuranohydrolases from Bifidobacterium adolescentis DSM 20083. Appl. Microbiol. Biotechnol. 1999, 51, 606–613. [Google Scholar] [CrossRef]
- Lagaert, S.; Pollet, A.; Delcour, J.A.; Lavigne, R.; Courtin, C.M.; Volckaert, G. Substrate specificity of three recombinant α-l-arabinofuranosidases from Bifidobacterium adolescentis and their divergent action on arabinoxylan and arabinoxylan oligosaccharides. Biochem. Biophys. Res. Commun. 2010, 402, 644–650. [Google Scholar] [CrossRef] [PubMed]
- Lagaert, S.; Pollet, A.; Delcour, J.A.; Lavigne, R.; Courtin, C.M.; Volckaert, G. Characterization of two β-xylosidases from Bifidobacterium adolescentis and their contribution to the hydrolysis of prebiotic xylooligosaccharides. Appl. Microbiol. Biotechnol. 2011, 92, 1179–1185. [Google Scholar] [CrossRef]
- Andersen, J.M.; Barrangou, R.; Hachem, M.A.; Lahtinen, S.J.; Goh, Y.J.; Svensson, B.; Klaenhammer, T.R. Transcriptional analysis of oligosaccharide utilization by Bifidobacterium lactis Bl-04. BMC Genom. 2013, 14, 312. [Google Scholar] [CrossRef]
- Gilad, O.; Jacobsen, S.; Stuer-Lauridsen, B.; Pedersen, M.B.; Garrigues, C.; Svensson, B. Combined transcriptome and proteome analysis of Bifidobacterium animalis subsp. lactis BB-12 grown on xylo-oligosaccharides and a model of their utilization. Appl. Environ. Microbiol. 2010, 76, 7285–7291. [Google Scholar]
- Gilad, O.; Svensson, B.; Viborg, A.H.; Stuer-Lauridsen, B.; Jacobsen, S. The extracellular proteome of Bifidobacterium animalis subsp. lactis BB-12 reveals proteins with putative roles in probiotic effects. Proteomics 2011, 11, 2503–2514. [Google Scholar]
- Hyun, Y.J.; Kim, B.; Kim, D.H. Cloning and characterization of ginsenoside Ra1-hydrolyzing β-d-xylosidase from Bifidobacterium breve K-110. J. Microbiol. Biotechnol. 2012, 22, 535–540. [Google Scholar] [CrossRef]
- Margolles, A.; de los Reyes-Gavilán, C.G. Purification and Functional Characterization of a Novel α-l-Arabinofuranosidase from Bifidobacterium longum B667. Appl. Environ. Microbiol. 2003, 69, 5096–5103. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, H.R.; Jørgensen, C.T.; Hansen, C.H.; Jørgensen, C.I.; Pedersen, S.; Meyer, A.S. A novel GH43 α-l-arabinofuranosidase from Humicola insolens: Mode of action and synergy with GH51 α-l-arabinofuranosidases on wheat arabinoxylan. Appl. Microbiol. Biotechnol. 2006, 73, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Pastell, H.; Westermann, P.; Meyer, A.S.; Päivi, T.; Tenkanen, M. In vitro fermentation of arabinoxylan-derived carbohydrates by bifidobacteria and mixed fecal microbiota. J. Agric. Food Chem. 2009, 57, 8598–8606. [Google Scholar] [CrossRef] [PubMed]
- Tauzin, A.S.; Laville, E.; Xiao, Y.; Nouaille, S.; Le Bourgeois, P.; Heux, S.; Portais, J.C.; Monsan, P.; Martens, E.C.; Potocki-Veronese, G.; et al. Functional characterization of a gene locus from an uncultured gut Bacteroides conferring xylo-oligosaccharides utilization to Escherichia coli. Mol. Microbiol. 2016, 102, 579–592. [Google Scholar] [CrossRef]
- Kumagai, Y.; Usuki, H.; Yamamoto, Y.; Yamasato, A.; Arima, J.; Mukaihara, T.; Hatanaka, T. Characterization of calcium ion sensitive region for β-Mannanase from Streptomyces thermolilacinus. Biochim. Biophys. Acta Proteins Proteom. 2011, 1814, 1127–1133. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).