Abstract
Seven new briarane diterpenoids, gemmacolides AZ–BF (1–7), were isolated together with eight known analogues (8–15) from the South China gorgonian Dichotella gemmacea. Their structures were elucidated based on detailed spectroscopic analysis and a comparison with reported data. In an in vitro bioassay, these compounds exhibited different levels of growth inhibition activity against A549 and MG63 cells, giving continuous evidences about the biological contribution of functional groups at C-2, C-12, C-13, and C-16. These compounds were also evaluated for their antibacterial and antifungal activities. Compound 8 exhibited a potential antibacterial activity against both Gram-positive bacterium Bacillus megaterium and Gram-negative bacterium Escherichia coli.
1. Introduction
Briarane diterpenoids are a family of structurally intriguing metabolites that are mainly obtained from gorgonians [1]. These metabolites are reported to display a wide spectrum of bioactivities, including cytotoxic, anti-inflammatory, antiviral, antifouling, insecticidal, and immunomodulatory effects [1,2,3,4]. In the course of our searching for novel and bioactive secondary metabolites from the South China Sea invertebrates [5,6,7,8,9,10,11,12], the cluster of metabolites attracted our attention due to their significant tumor tell growth inhibitory activity [7,8,9,10,11,12]. Considering the structural complexity of the metabolites, specific research regarding the determination of absolute configuration was carried out using a solution TDDEFT/ECD approach [12]. Successive investigation on several collections of the gorgonian Dichotella gemmacea and Junceella gemmacea led to the isolation and structure elucidation of 48 new briaranes and 29 known analogues. On the basis of in vitro screening, a structure–activity relationship analysis of the metabolites was performed [7,8,9,10,11,12]. Three new briaranes, gemmacolides J, V, and Y, showed significant growth inhibitory activity towards A549 cells, being more active than the positive control adriamycin [10,11]. The initial research encourages continuous investigations of this class of metabolites, leading to the isolation and structure elucidation of seven new briaranes, namely gemmacolides AZ–BF (1–7), together with eight known analogues (8–15) [13,14,15,16,17,18,19] (Chart 1) from the gorgonian D. gemmacea. The structures of these compounds were elucidated on the basis of detailed spectroscopic analysis (Figures S1–S47) and a comparison with reported data. The isolates were tested in vitro for their antimicrobial and tumor cell growth inhibition activities. We herein report on the isolation, structure elucidation, and bioactivities of these compounds.
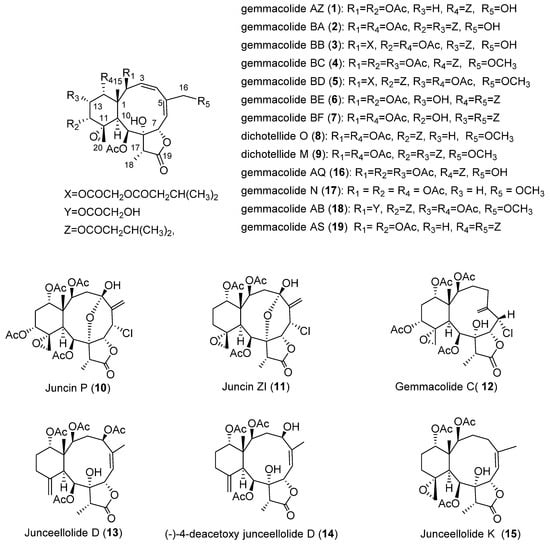
Chart 1.
Structures of compounds 1–19.
2. Results
Freshly collected specimens of D. gemmacea were immediately frozen to −20 °C and stored at this temperature before extraction. The usual workup for the extraction and isolation of briarane diterpenoids [8,9,10,11,12] yielded 15 pure compounds (1–15). The known compounds dichotelllides O and M (8, 9) were once reported from the gorgonian D. gemmacea [13], while gemmacolide C (12) [18] was previously isolated from the gorgonians J. gemmacea. Juncins P (10) [14] and ZI (11) [16] were firstly isolated from the gorgonian J. juncea, and then repeatedly obtained from the gorgonians D. gemmacea [13,20,21], D. fragilis [22], J. gemmacea [7], and J. fragilis [23]. Junceellolide D (13) [17], (−)-4-deacetoxy junceellolide D (14) [15], and junceellolide K (15) [19] were firstly isolated from the gorgonians J. fragilis and then re-isolated from many gorgonian corals, including Verrucella umbraculum [24], J. fragilis [23,25,26,27], D. gemmacea [8,13,20], J. gemmacea [7], Menella sp. [28], and J. juncea [14]. These metabolites displayed antifouling, anti-inflammatory, and cytotoxic activities in the in vitro bioscreening [13,16,21,22,29].
Gemmacolide AZ (1) was isolated as a white amorphous powder. The molecular formula C31H42O13 was established by the HRESIMS. The IR spectrum showed strong absorption bands of hydroxyl (3468 cm−1), γ-lactone (1775 cm−1), and ester (1738 cm−1) functionalities. This observation was in agreement with the signals in the 13C NMR and DEPT spectra (Table 1) for 9 sp2 carbon atoms (5 × OC = O, CH = CH, CH = C) at lower field and 22 sp3 carbon atoms at higher field (1 × C, 3 × CH, 2 × CH2, 7 × CH3, 2 × OC, 5 × OCH, 2 × OCH2), accounting for seven double bond equivalents. The remaining double bond equivalents were due to the presence of four rings in the molecule.

Table 1.
13C NMR data for gemmacolides AZ–BF (1–7) a.
The 1H and 13C NMR spectra data (Table 1 and Table 2) of 1 showed great similarity to those of gemmacolide AQ (16) [9], except for the absence of an acetoxy group. The missing group was readily assigned at C-13 due to the proton sequence of H-12/H2-13/H-14, established by 1H–1H COSY experiment (Figure 1). The assignment was fully supported by the 1H–1H COSY and HMBC spectra as shown in Figure 1. The relative configuration of 1 at chiral centers was proven the same as that of 16 by a NOESY experiment (Figure 2), showing a β configuration of H-7, H-12, H-14, Me-15, H-17, and CH2-20, and an α configuration of H-2, H-9, H-10, and Me-18. The geometry of the Δ3 double bond was assigned as (Z) based on the proton coupling constant between H-3 and H-4 (J = 10.6 Hz), while Δ5,6 was determined as (E) due to the NOESY correlation between H-6 and H2-16. The NMR assignments of 1 were further supported by a comparison with the reported data of gemmacolide N (17), an analogue obtained from the same species of animals with its absolute stereochemistry being determined by a detailed TDDFT/ECD analysis [12]. Compound 17 differs from 1 in the functional groups at C-14 and C-16, having acetoxy and primary hydroxyl groups instead of isovaleryl and methoxy groups, respectively. As compound 1 contained the same lactone and diene chromophores as gemmacolide N [12], and they differed only in the nature of the ester groups and R5, the ECD spectrum of gemmacolide N could be used as ECD reference for the configurational assignment. The absolute configuration of 1 is therefore determined as (1R,2S,7S,8S,9S,10S,11R,12R,14S,17R).

Table 2.
1H NMR data for gemmacolides AZ–BC (1–4) a.
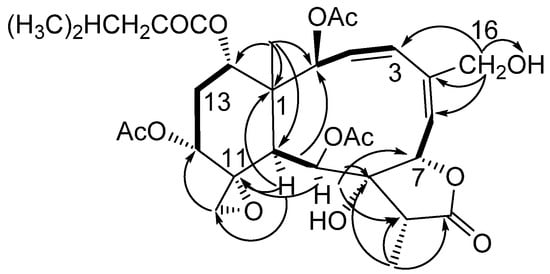
Figure 1.
Key HMBC (arrow H → C) and COSY (bond) spin coupling systems for compound 1.
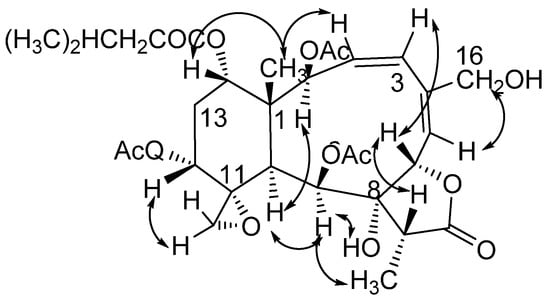
Figure 2.
Key NOESY correlations for compound 1.
Gemmacolide BA (2), a white amorphous powder, had the molecular formula of C36H50O15 on the basis of its HRESIMS. Its 1H and 13C NMR spectra data (Table 1 and Table 2) were similar to those of gemmacolide AQ (16) [9]. However, an acetyl group in 16 was replaced by an isovaleryl group in 2. The three acetyl groups were assigned at C-2, C-9, and C-14 due to the obvious HMBC correlations from the secondary alcohol protons to the respective ester carbonyl groups. The location of two isovaleryl groups at C-12 and C-13 was supported by the obvious HMBC correlations from H-12 and H-13 to the respective carbonyl carbon of the isovaleryl groups. The relative configuration and the absolute configuration of 2 were proven to be the same as that of 16 on the basis of NOESY experiment and their similar ECD spectra.
Gemmacolide BB (3) was obtained as a white amorphous powder with the molecular formula of C38H52O17 being established by HRESIMS. The 1H and 13C NMR spectra of 3 showed similarity to those of compound 2 (Table 1 and Table 2). The structure of 3 differed from that of 2 with the presence of an isovaleric acetyl group instead of an acetyl group at C-2 and an acetyl group replace of the isovaleryl group at C-12 (Table 1 and Table 2). The conclusion was fully supported by the distinct HMBC correlations from H-2 to the carbonyl carbons of the isovaleric acetyl group at δ 167.8, and from H-12 to the acetyl ester carbon at δ 169.8, respectively. Its absolute configuration was proven to be the same as that of 2 on the basis of their similar ECD spectra.
Gemmacolide BC (4) was obtained as a white amorphous powder. Its HRESIMS demonstrated the same molecular formula as C34H46O15 by HRESIMS. Comparison of overall 1H and 13C NMR spectra data (Table 1 and Table 2) of 4 with those of gemmacolide AQ (16) [9] revealed great similarity with the only difference of an appearance of an oxygenated methyl group at C-16 (δH 3.44, s; δC 58.5, CH3). The result was supported by the obvious HMBC correlations from the methoxyl protons to C-16. The absolute structure of 4 was proven to be the same as that of 16 by the analysis of NOESY and ECD spectra.
Gemmacolide BD (5) was obtained as a white amorphous powder and exhibited a molecular formula of C39H54O17 as deduced from its HRESIMS. 1H and 13C NMR spectra of 5 (Table 1 and Table 3) were very similar to those of compound gemmacolide AB (18) [9]. The primary alcohol of the hydroxyacetyl group, however, was further isovalerianated. This conclusion was proved by the long-range correlation from secondary hydrogens of both the isovaleryl group and the hydroxyacetyl group to the carbonyl carbon of the isovaleryl group (δ 172.3), and from both H-2 and the secondary hydrogens of the hydroxyacetyl group to the carbonyl carbon of the hydroxyacetyl group (δ 166.6). The same absolute geometry was obtained on the basis of the ECD spectrum.

Table 3.
1H NMR data for gemmacolides BD–BF (5–7) a.
Gemmacolide BE (6) was found to be a white amorphous powder, having the molecular formula of C36H50O15 based on the HRESIMS. 1H and 13C NMR spectra (Table 1 and Table 3) of 6 resembled to those of gemmacolide AS (19) [8] except for a hydrogen at C-13 in 19 was replaced by a hydroxyl group in 6. The assignment was in agreement with its NMR shift values (δH-13 4.07, br s; δC-13 67.4, CH), compared to those of the ester group substitution in gemmacolide P, gemmacolide AC, gemmacolide AD, gemmacolide AF, gemmacolide AG, gemmacolide AR, and gemmacolide AT (δH-13 5.08–5.10, d, J = 2.7–3.5; δC-13 66.3–66.7, CH) [8,9,12], and further supported by the proton sequences from H-12 to H-14 established by the 1H–1H COSY experiment. The hydroxyl group was assigned as an α-orientation due to the NOESY correlation of H-13 with H-15. The two isovaleryl groups were deduced to be attached to C-14 and C-16 based on the 2D NMR (1H–1H COSY, HMBC) analysis and a comparison to those reported data of analogues [7,8,9,10,11,12]. The relative and absolute configuration of 6 was also proven to be the same as those of 19 by the NOESY and ECD experiments.
Gemmacolide BF (7) was a white amorphous powder and had the same molecular formula of C36H50O15 as that of 6 as deduced from its HRESIMS. The structure of 7 differed from that of 6 only in the sequence of substituent groups. The hydroxyl, isovaleryl, and acetoxy groups at C-13, C-14, and C-12 in 6 were instead assigned at C-12, C-13, and C-14 in 7, respectively. The location of hydroxyl at C-12 was supported by 1H and 13C NMR spectra data (δH-12 3.48, br d, J = 4.7; δC-12 75.3, CH), compared to those of ester group substitution (δH-12 4.88–4.93, br d, J = 2.8–3.5; δC-12 72.8–73.3, CH) [8,9,12]. A β-orientation of H-12 was deduced from its NOESY correlation with H-20b. Two isovaleryl groups were attached at C-13 and C-16 due to the HMBC correlations of H-13 and H-16 with the respective carbonyl carbon of the isovaleryl groups. The assignment was supported by the proton sequence of H-12/H-13/H-14, as deduced from the 1H–1H COSY experiment. The established structure of 7 was further supported by a detailed analysis of its 1D NMR and 2D NMR data. Its absolute configuration was determined as (−)-(1S,2S,3Z,5E,7S,8S,9S,10S,11R,12R,13S,14R,17R) based on NOESY and ECD experiments.
The compounds were evaluated for their antimicrobial and tumor cell growth inhibition activities. In the in vitro bioassays, compounds 1–9 exhibited a different level of growth inhibition against cell lines A549 and MG63 (Table 4). The activity of 5 with 2-isovaleric acetyl substitution marked decreased compared with its 2-glycolyl analogue (IC50 for A549 = 19.4 μM, for MG63 = 22.8 μM) [9]. The previous conclusion that 13-isovalerate may decrease the activity [9] was further supported by comparing the activity of 1 and 9 with those of 16 (IC50 = 28.7 and > 100 μM for A549 and MG63, respectively) [9] and 8, respectively. Comparing the activity of 2 and 8 with respect to those of gemmacolides AT (IC50 > 30.0 μM for both A549 and MG63) [8] and N (IC50 > 50.5 μM for both A549 and MG63) [12] supported the conclusion that 12-O-isovalerate gave a positive contribution to the activity as mentioned previously [9,11,12]. The replacement of an isovaleryl group by an acetyl group at C-14 would increase the activity as observed in 4 and juncenolide D [12]. The 16-substituents displayed an activity order as 16-OH > 16-OMe when the activity of 2 and 3 was compared with those of 9 (IC50 > 30.0 μM for both A549 and MG63) and gemmacolide AL (IC50 > 37.8 μM for both A549 and MG63) [9], respectively.

Table 4.
Cytotoxic assay for compounds 1–9 (IC50 μM).
In antimicrobial test in vitro, compounds 4 and 8 showed potential antibacterial activity against the Gram-negative bacterium Escherichia coli, while 8 exhibited significant antibacterial activity against the Gram-positive bacterium Bacillus megaterium. Compounds 1, 2, 4, 5, and 8 exhibited weak antifungal activity against both Septoria tritici and Microbotryum violaceum, while compound 9 only displayed weak activity against S. tritici (Table 5).

Table 5.
Agar diffusion assays for antibacterial and antifungal activities a,b.
3. Materials and Methods
3.1. General Experimental Procedures
Commercial silica gel (Yantai, China, 200–300; 400–500 mesh) and RP silica gel (Merck, Darmstadt, Germany, 43–60 μm) were used for column chromatography (CC). Precoated silica gel plates (Yantai, China, HSGF-254) and RP silica gel (Macherey-Nagel, Düren, Germany, RP-18 F254) were used for analytical thin-layer chromatography (TLC). Spots were detected on TLC under UV or by heating after spraying with an anisaldehyde-sulphuric acid reagent. The NMR spectra were recorded at 300 K on a Bruker DRX 400 spectrometer (Ettlingen, Germany). Chemical shifts are reported in parts per million (δ), with use of the residual CHCl3 signal (δH 7.26 ppm) as an internal standard for 1H NMR and CDCl3 (δC 77.0 ppm) for 13C NMR; Coupling constants (J) are reported in Hz. 1H NMR and 13C NMR assignments were complemented 1H–1H COSY, HSQC, HMBC, and NOESY experiments. The following abbreviations are used to describe spin multiplicity: s = singlet; d = doublet; t = triplet; q = quartet; m = multiplet; br s = broad singlet; dd = doublet of doublets; ov = overlapped signals. Optical rotations were measured in CHCl3 with an Autopol IV polarimeter at the sodium D line (590 nm). Infrared spectra were recorded in thin polymer films on a Nexus 470 FT-IR spectrophotometer (Nicolet, Madison, WI, USA); peaks are reported in cm−1. UV absorption spectra were recorded on a Varian Cary 100 UV-Vis spectrophotometer (Palo Alto, CA, USA); peaks wavelengths are reported in nm. Circular dichroism spectra were recorded with a JASCO J-715 circular dichroism spectropolarimeter (Mary’s Court Easton, MD, USA). The MS and HRMS were performed on a Q-TOF Micro mass spectrometer (Bruker, Bremen, Germany), resolution 5000. An isopropyl alcohol solution of sodium iodide (2 mg/mL) was used as a reference compound. Semi-preparative RP-HPLC was performed on an Agilent 1100 system (Santa Clara, CA, USA) equipped with a refractive index detector using an YMC Pack ODS-A column (Kyoto, Japan, particle size 5 μm, 250 mm × 10 mm).
3.2. Animal Material
The South China Sea gorgonian coral Dichotella gemmacea (3.5 kg, wet weight) was collected from the South China Sea in August 2007 and identified by Xiu-Bao Li, South China Sea Institute of Oceanology, Chinese Academy of Sciences. A voucher specimen (ZS-3) was deposited in the Second Military Medical University.
3.3. Extraction and Isolation
The frozen specimen was extracted ultrasonically three times with acetone and MeOH, respectively. The combined residue was partitioned between H2O and EtOAc to afford 16.1 g of an EtOAc extract. The EtOAc extract was further partitioned between MeOH and hexane, affording 11.2 g of MeOH soluble residue. The MeOH extract was subjected to column chromatography (CC) on silica gel to give 16 fractions, using hexane and acetone (from 100:0 to 0:100) as an eluent. Fraction 5 was repeatedly subjected to normal phase silica gel column chromatography, Sephadex LH-20, and RP-silical gel column chromatography, then purified via HPLC (eluent MeOH/H2O, 75:25, 1.5 mL/min), yielding 5 (3.4 mg) at 29.6 min. Fraction 6 was chromatographed over Sephadex LH-20 to give four sub-fractions (A–D) eluted with CHCl3 and MeOH (1:1). Sub-fractions A was purified via HPLC (eluent MeOH/H2O, 70:30, 1.5 mL/min), yielding 2 (2.0 mg) at 34.0 min. Sub-fractions B was purified via HPLC (eluent MeOH/H2O, 70:30, 1.5 mL/min), yielding 6 (1.4 mg) and 7 (1.6 mg) at 64.3 and 67.9 min. Sub-fractions C was purified via HPLC (eluent MeOH/H2O, 70:30, 1.5 mL/min), yielding 3 (1.0 mg) at 62.3 min. Fraction 10 was repeatedly chromatographed over normal phase silica gel column and Sephadex LH-20 and purified via HPLC (eluent MeOH/H2O, 70:30, 1.5 mL/min), yielding 4 (3.4 mg) at 51.7 min. Fraction 13 was further fractionated by RP-silical gel column chromatography (gradient elution from MeOH/H2O, 2:7 to 2:1, in 5% increments) to afford 6 sub-fractions (A–F), and sub-fraction E was purified via HPLC (eluent MeOH/H2O, 65:35, 1.5 mL/min), yielding 1 (2.3 mg) at 29.7 min.
Gemmacolide AZ (1): white amorphous powder; −1.3 (c 0.07, CHCl3); UV (MeOH) λmax(log ε) 205 (1.71) nm; CD (CH3CN, c 2.0 × 10−4) λmax (Δε) positive below 190 nm, 209 (−5.50) nm; IR (film) νmax 3468, 1775, 1738 cm−1; 1H NMR spectroscopic data, see Table 2; 13C NMR spectroscopic data, see Table 1; ESIMS m/z 645 [M + Na]+; HRESIMS m/z 645.2527 [M + Na]+ (calcd. for C31H42O13 Na, 645.2523).
Gemmacolide BA (2): white amorphous powder; −43.0 (c 0.07, CHCl3); UV (MeOH) λmax(log ε) 208 (2.26) nm; CD (CH3CN, c 4.2 × 10−4) λmax (Δε) positive below 190 nm, 209 (−4.77) nm; IR (film) νmax 3469, 1775, 1744 cm−1; 1H NMR spectroscopic data, see Table 2; 13C NMR spectroscopic data, see Table 1; ESIMS m/z 745 [M + Na]+; HRESIMS m/z 745.3041 [M + Na]+ (calcd. for C36H50O15Na, 745.3047).
Gemmacolide BB (3): white amorphous powder; −28 (c 0.03, CHCl3); UV (MeOH) λmax(log ε) 204(1.47) nm; CD (CH3CN, c 1.6 × 10−4) λmax (Δε) positive below 190 nm, 205 (−5.36) nm; IR (film) νmax 3469, 1772, 1744 cm−1; 1H NMR spectroscopic data, see Table 2; 13C NMR spectroscopic data, see Table 1; ESIMS m/z 803 [M + Na]+; HRESIMS m/z 803.3107 [M + Na]+ (calcd. for C38H52O17Na, 803.3102).
Gemmacolide BC (4): white amorphous powder; −44.0 (c 0.24, CHCl3); UV (MeOH) λmax(log ε) 205 (1.55) nm; CD (CH3CN, c 4.1 × 10−4) λmax (Δε) positive below 190 nm, 201.5 (−4.62) nm; IR (film) νmax 3479, 1776, 1743 cm−1; 1H NMR spectroscopic data, see Table 2; 13C NMR spectroscopic data, see Table 1; ESIMS m/z 717 [M + Na]+; HRESIMS m/z 717.2730 [M + Na]+ (calcd. for C34H46O15Na, 717.2734).
Gemmacolide BD (5): white amorphous powder; −36.0 (c 0.12, CHCl3); UV (MeOH) λmax(log ε) 205 (1.61) nm; CD (CH3CN, c 2.0 × 10−4) λmax (Δε) positive below 190 nm, 216 (−7.34) nm; IR (film) νmax 3461, 1775, 1744 cm−1; 1H NMR spectroscopic data, see Table 3; 13C NMR spectroscopic data, see Table 1; ESIMS m/z 817 [M + Na]+; HRESIMS m/z 817.3254 [M + Na]+ (calcd. for C39H54O17Na, 817.3259).
Gemmacolide BE (6): white amorphous powder; −22.0 (c 0.05, CHCl3); UV (MeOH) λmax(log ε) 204 (1.41) nm; CD (CH3CN, c 3.5 × 10−4) λmax (Δε) positive below 190 nm, 198.0 (−5.43) nm; IR (film) νmax 3467, 1776, 1741 cm−1; 1H NMR spectroscopic data, see Table 3; 13C NMR spectroscopic data, see Table 1; ESIMS m/z 745 [M + Na]+; HRESIMS m/z 745.3043 [M + Na]+ (calcd. for C36H50O15Na, 745.3047).
Gemmacolide BF (7): white amorphous powder; −30.0 (c 0.05, CHCl3); UV (MeOH) λmax(log ε) 204 (1.53) nm; CD (CH3CN, c 1.7 × 10−4) λmax (Δε) positive below 190 nm, 198.0 (−10.59) nm; IR (film) νmax 3469, 1777, 1742 cm−1; 1H NMR spectroscopic data, see Table 3; 13C NMR spectroscopic data, see Table 1; ESIMS m/z 745 [M + Na]+; HRESIMS m/z 745.3051 [M + Na]+ (calcd. for C36H50O15Na, 745.3051).
3.4. Cytotoxicity Assay
Cytotoxicity was tested against human lung adenocarcinoma (A549) and human osteosarcoma cell (MG63), using a modification of the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] colorimetric method [30]. DMSO was used as a vehicle control, and adriamycin was used as a positive control, IC50 = 2.8 μM for A549 cells and 3.2 μM for MG63 cells.
3.5. Agar Diffusion Test for Biological Activity
Compounds 1–9 were dissolved in acetone at a concentration of 2 mg/mL; 25 μL of the solution (0.05 mg) were pipetted onto a sterile filter disc (Schleicher & Schuell, Dassel, Germany, 9 cm), which was placed onto an appropriate agar growth medium for the respective test organism and subsequently sprayed with a suspension of the test organize. The test organisms were the Gram-negative bacterium Escherichia coli (Coli), the Gram-positive bacterium Bacillus megaterium (Meg) (both grown on NB medium), the fungus Microbotryum violaceum (Vio), and Septoria tritici (Tri) (both grown on MPY medium). Commencing at the outer edge of the filter disc, the diameter of zone of inhibition was measured in mm. Reference substances were ketoconazole (0.05 mg), penicillin (0.05 mg), and streptomycin (0.05 mg), with the diameter (Ф) of zone of inhibition being 18.0, 18.0, 30.0, 25.0; 26.0, 18.0, 14.0, 12.0; and 18.0, 11.0, 16.0, 11.0 mm, respectively.
Supplementary Materials
The following are available online at www.mdpi.com/1660-3397/14/11/201/s1, Figures S1–S47: Spectra of the new compounds 1–7.
Acknowledgments
The research work was financially supported by NSFC (81573342, U1405227, 41576157, 81502978), the Program of Shanghai Subject Chief Scientist (15XD1504600), the Hundred Talents Program of SMCH (XBR2013111), the key project of STCSM (14431902900), and the Youth Program of SMMU (2014QN08).
Author Contributions
Cui Li, Ming-Ping La, and Hua Tang performed the experiments; Peng Sun, Bao-Shu Liu, Chun-Lin Zhuang, and Yang-Hua Yi contributed reagents/materials/analysis tools; Wen Zhang conceived and designed the experiments.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2015, 32, 116–211. [Google Scholar] [CrossRef] [PubMed]
- Berrue, F.; Kerr, R.G. Diterpens from gorgonian corals. Nat. Prod. Rep. 2009, 26, 681–710. [Google Scholar] [CrossRef] [PubMed]
- Sung, P.J.; Su, J.H.; Wang, W.H.; Sheu, J.H.; Fang, L.S.; Wu, Y.C.; Chen, Y.H.; Chung, H.M.; Su, Y.D.; Chang, Y.C. Survey of briarane-type diterpenoids-Part IV. Heterocycles 2011, 83, 1241–1258. [Google Scholar] [CrossRef]
- Zhang, W.; Guo, Y.W.; Gu, Y.C. Secondary metabolites from the South China Sea invertebrates: Chemistry and biological activity. Curr. Med. Chem. 2006, 13, 2041–2090. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Sun, P.; Jiang, N.; Riccio, R.; Lauro, G.; Bifulco, G.; Li, T.J.; Gerwick, W.H.; Zhang, W. New steroids with a rearranged skeleton as (h)P300 inhibitors from the sponge Theonella swinhoei. Org. Lett. 2014, 16, 2224–2227. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Xu, D.X.; Mándi, A.; Kurtán, T.; Li, T.J.; Schulz, B.; Zhang, W. Structure, absolute configuration, and conformational study of 12-membered macrolides from the fungus Dendrodochium sp. associated with sea cucumber Holothuria nobilis Selenka. J. Org. Chem. 2013, 78, 7030–7047. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Li, J.; Hc, E.; Liu, B.S.; Tang, H.; Gerwick, W.; Hua, H.-M.; Zhang, W. Briarane diterpenes from the gorgonian coral Junceella gemmacea. Mar. Drugs 2014, 12, 589–600. [Google Scholar] [CrossRef] [PubMed]
- La, M.P.; Li, J.; Li, C.; Tang, H.; Liu, B.S.; Sun, P.; Zhuang, C.L.; Li, T.J.; Zhang, W. Briarane diterpenoids from the gorgonian Dichotella gemmacea. Mar. Drugs 2014, 12, 6178–6189. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Jiang, M.; La, M.P.; Li, T.J.; Tang, H.; Sun, P.; Liu, B.S.; Yi, Y.H.; Liu, Z.; Zhang, W. Chemistry and tumor cell growth inhibitory activity of 11,20-epoxy-3Z,5(6)E-diene briaranes from the South China Sea gorgonian Dichotella gemmacea. Mar. Drugs 2013, 11, 1565–1582. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; La, M.P.; Tang, H.; Pan, W.H.; Sun, P.; Yi, Y.H.; Zhang, W. Bioactive briaranes from the South China Sea gorgonian Dichotella gemmacea. Bioorg. Med. Chem. Lett. 2012, 5, 4368–4372. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; La, M.P.; Li, L.; Li, X.B.; Tang, H.; Liu, B.S.; Krohn, K.; Sun, P.; Yi, Y.H.; Zhang, W. Bioactive11,20-epoxy-3,5(16)-diene briarane diterpenoids from the South China Sea gorgonian Dichotella gemmacea. J. Nat. Prod. 2011, 74, 1658–1662. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; La, M.P.; Sun, P.; Kurtán, T.; Mándi, A.; Tang, H.; Liu, B.S.; Yi, Y.H.; Li, L.; Zhang, W. Bioactive (3Z,5E)-11,20-epoxybriara-3,5-diene-7,18-olide diterpenoidsfrom the South China Sea gorgonian Dichotella gemmacea. Mar. Drugs 2011, 9, 1403–1418. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.F.; Han, Z.; Zhou, X.F.; Yang, B.; Lin, X.; Liu, J.; Peng, Y.; Yang, X.W.; Liu, Y.H. Antifouling briarane type diterpenoids from South China Sea gorgonians Dichotella gemmacea. Tetrahedron 2013, 69, 871–880. [Google Scholar] [CrossRef]
- Qi, S.H.; Zhang, S.; Huang, H.; Xiao, Z.H.; Huang, J.S.; Li, Q.X. New briaranes from the South China Sea gorgonian Junceella juncea. J. Nat. Prod. 2004, 67, 1907–1910. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.; Rodriguez, J.; Jimenez, C. Absolute structures of new briarane diterpenoids from Junceella fragilis. J. Nat. Prod. 1999, 62, 257–260. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.H.; Zhang, S.; Qian, B.Y.; Xiao, Z.H.; Li, M.Y. Ten new antifouling briarane diterpenoids from the South China Sea gorgonian Junceella juncea. Tetrahedron 2006, 62, 9123–9130. [Google Scholar] [CrossRef]
- Shin, J.; Park, M.; Fenical, W. The junceellolides, new antiinflammatory diterpenoids of the briarane class from the Chinese gorgonian Junceella fragilis. Tetrahedron 1989, 45, 1633–1638. [Google Scholar] [CrossRef]
- He, H.Y.; Faulkner, D.J. New chlorinated diterpenes from the gorgonian Junceella gemmacea. Tetrahedron 1991, 47, 3271–3280. [Google Scholar] [CrossRef]
- Sheu, J.H.; Chen, Y.P.; Hwang, T.L.; Chiang, M.Y.; Fang, L.S.; Sung, P.J. Junceellolides J–L, 11,20-epoxybriaranes from the gorgonian coral Junceella fragilis. J. Nat. Prod. 2006, 69, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.Z.; Liao, X.J.; Liu, F.; Xu, S.Y. Chemical constituents from Dichotella gemmacea. J. Chin. Med. Mater. 2014, 37, 266–269. [Google Scholar]
- Sun, J.F.; Huang, H.; Chai, X.Y.; Yang, X.W.; Meng, L.; Huang, C.G.; Zhou, X.F.; Yang, B.; Hu, J.; Chen, X.Q.; et al. Dichotellides A–E, five new iodine-containing briarane type diterpenoids from Dichotella gemmacea. Tetrahedron 2011, 67, 1245–1250. [Google Scholar] [CrossRef]
- Zhou, Y.M.; Shao, C.L.; Wang, C.Y.; Huang, H.; Xu, Y.; Qian, P.Y. Chemical constituents of the gorgonian Dichotella fragilis (Ridleg) from the South China Sea. Nat. Prod. Commun. 2011, 6, 1239–1242. [Google Scholar] [PubMed]
- Liaw, C.C.; Shen, Y.C.; Lin, Y.S.; Hwang, T.L.; Kuo, Y.H.; Khalil, A.T. Frajunolides E–K, briarane diterpenes from Junceella fragilis. J. Nat. Prod. 2008, 71, 1551–1556. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Wang, B.; Liu, Y. Briarane-type diterpenoids from the gorgonian coral Verrucella umbraculum. Chem. Nat. Compd. 2012, 48, 516–517. [Google Scholar] [CrossRef]
- Shen, Y.C.; Chen, Y.H.; Hwang, T.L.; Guh, J.H.; Khalil, A.T. Four new briarane diterpenoids from the gorgonian coral Junceella fragilis. Helv. Chim. Acta 2007, 90, 1391–1398. [Google Scholar] [CrossRef]
- Wen, Y.M.; Qi, S.H.; Zhang, S. Briarane diterpenes and steroids from the South China Sea gorgonian Junceella fragilis. Nat. Prod. 2006, 18, 234–237. [Google Scholar]
- Zhang, W.; Guo, Y.-W.; Mollo, E.; Cimino, G. Junceellonoids A and B, two new briarane diterpenoids from the Chinese gorgonian Junceella fragilis Ridley. Helv. Chim. Acta 2004, 87, 2341–2345. [Google Scholar] [CrossRef]
- Chai, X.Y.; Sun, J.F.; Tang, L.Y.; Yang, X.W.; Li, Y.Q.; Huang, H.; Zhou, X.F.; Yang, B.; Liu, Y. A novel cyclopentene derivative and a polyhydroxylated steroid from a South China Sea gorgonian Menella sp. Chem. Pharm. Bull. 2010, 58, 1391–1394. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.X.; Wu, H.X.; Xu, Y.; Shao, C.L.; Wang, C.Y.; Qian, P.Y. Antifouling activity of secondary metabolites isolated from Chinese marine organisms. Mar. Biotechnol. 2013, 15, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).