Abstract
The mechanism for how fucoxanthin (FX) suppressed adipose accumulation is unclear. We aim to investigate the effects of FX on metabolic rate and expressions of genes related to thermogenesis, mitochondria biogenesis and homeostasis. Using a 2 × 2 factorial design, four groups of mice were respectively fed a high sucrose (50% sucrose) or a high-fat diet (23% butter + 7% soybean oil) supplemented with or without 0.2% FX. FX significantly increased oxygen consumption and carbon dioxide production and reduced white adipose tissue (WAT) mass. The mRNA expressions of peroxisome proliferator-activated receptor (PPAR) γ coactivator-1α (PGC-1α), cell death-inducing DFFA-like effecter a (CIDEA), PPARα, PPARγ, estrogen-related receptor α (ERRα), β3-adrenergic receptor (β3-AR) and deiodinase 2 (Dio2) were significantly upregulated in inguinal WAT (iWAT) and epididymal WAT (eWAT) by FX. Mitochondrial biogenic genes, nuclear respiratory factor 1 (NRF1) and NRF2, were increased in eWAT by FX. Noticeably, FX upregulated genes of mitochondrial fusion, mitofusin 1 (Mfn1), Mfn2 and optic atrophy 1 (OPA1), but not mitochondrial fission, Fission 1, in both iWAT and eWAT. In conclusion, dietary FX enhanced the metabolic rate and lowered adipose mass irrespective of the diet. These were associated with upregulated genes of the PGC-1α network and mitochondrial fusion in eWAT and iWAT.
1. Introduction
Obesity, defined as excess accumulation of adipose, is a worldwide endemic health problem. Obesity and its related disorders are associated with increased morbidity, mortality and healthcare costs [1]. Mitochondria play an important role in adipose biology [2]. Mitochondria dysfunction is linked to obesity [3] and type 2 diabetes [4,5]. Mitochondria biogenesis is thus considered a potential target for the intervention of insulin resistance in obesity and diabetes [6,7,8]. Mitochondrion is a dynamic organelle that is continuously remodeled by fusion and fission [9], which is important for bioenergetic adaptation to metabolic demand. Cells exposed to a nutrient-rich environment tend to maintain their mitochondria in a separated state, while cells under starvation tend to have mitochondria in a connected state [10]. Reduction in the mitochondrial network, but unaltered mitochondrial mass have been reported in skeletal muscle of obese Zucker rats [3] and type 2 diabetic patients [11]. Moreover, mRNA and the protein, Mfn2, an important protein located in the mitochondrial outer membrane and mediating mitochondrial fusion, were reduced in skeletal muscle of obese subjects compared to lean subjects [12].
Peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) is a strong promoter of mitochondrial biogenesis and oxidative metabolism through nuclear respiratory factor, NRF1 and NRF2, ERRα, PPARγ and PPARα [13,14,15]. ERRα further activated the transcriptional activity of the Mfn2 promoter, and the effects were synergic with those of PGC-1α [16]. In addition, NRF1 and NRF2 themselves are able to stimulate mitochondrial transcription factor A (TFAM), a mitochondrial matrix protein essential for the replication and transcription of mitochondria DNA [17,18]. PGC-1α mRNA expression is reduced in subcutaneous fat of morbidly obese patients [19]. Ectopic expression of PGC-1α in white adipose tissue (WAT) increased brown adipocyte specific genes, including uncoupling protein 1 (UCP1) and mitochondrial activity [20].
Fucoxanthin (FX) is a major carotenoid in brown algae and has an unusual allenic structure [21]. FX has a suppressive effect on adipose accumulation in genetically diabetic KK-Ay mice [22,23,24], Wistar rats [25,26] and diet-induced obese C57BL/6J mice [27,28,29]. FX feeding significantly increased fecal triglyceride and cholesterol excretion and β-oxidation in liver and epididymal WAT (eWAT) and also reduced fatty acid, cholesterol synthesis-related enzyme activity, serum and hepatic lipid accumulation [28,30,31]. Previous studies also showed that FX increased UCP1 mRNA expression in abdominal WAT of mice with reduced adipose mass [25,28,31]. PGC-1α expression in the skeletal muscles was upregulated in KK-Ay mice fed FX [32].
UCP1 is a thermogenic mitochondria protein that was thought to express exclusively in brown adipose tissue (BAT). However, its expression in WAT has been confirmed in most recent studies, and these UCP1 expressing adipocytes in WAT were named “Beige/Brite” or “recruited (in contrast to the traditional “constituted”) brown adipocytes [33,34]. Increases in the recruited brown adipocytes can enhance thermogenic activity and reduce adiposity in rodents [33,34] and humans [35]. As a dominant regulator of mitochondrial biogenesis and oxidative metabolism [15], PGC-1α regulates thermogenic activity by inducing the expression of UCP1 and key enzymes of the mitochondrial respiratory chain in both constituted and recruited brown adipocytes [33]. Mitochondrial biogenesis and dynamics regulate mitochondrial function, respiratory capacity, apoptosis and oxidative phosphorylation and, thus, impact energy balance [3,12,36]. Whether FX increased expressions of genes in the PGC-1α regulated pathways in WAT and metabolic rate has not been reported.
Here, we examined the suppressive effect of adipose accumulation of FX under a high sucrose or a high saturated fat diet using a 2 × 2 factorial design. Whole body O2 consumption and CO2 production were measured and expressions of PGC-1α regulated genes in WAT analyzed.
2. Results and Discussion
2.1. FX Decreased White Adipose Weight without Altering Food Intake
Four groups of mice were respectively fed the 4 test diets shown in Table 1. The results of two-way ANOVA on the four groups (Table 2) of mice showed that neither FX nor diet affected final body weight, body weight gain and energy efficiency (p > 0.05). The high fat diet, but not FX, decreased food and energy intake (p < 0.05) (Table 2). FX significantly decreased relative weight of both abdominal, eWAT and retroperitoneal WAT (rWAT), and subcutaneous iWAT (p < 0.001). High fat diet decreased, but FX increased, the relative weight of BAT, (p < 0.05) (Table 3). Liver, kidney, spleen and heart relative weights were significantly increased by FX (Supplementary Table S1).

Table 1.
Composition of the 4 test diets: HS, high sucrose diet; HS + F, high sucrose diet supplemented with fucoxanthin; HF, high fat diet; HF + F, high fat diet supplemented with fucoxanthin. The composition of vitamin mix and mineral mix are in accordance with AIN-93 and AIN-93G, respectively [37].
| Ingredients of Diet (g/kg) | HS | HS + F | HF | HF + F |
|---|---|---|---|---|
| Corn starch | 129.5 | 129.5 | 209.35 | 209.35 |
| Sucrose | 500 | 500 | 100 | 100 |
| Butter | − | − | 230 | 228 |
| Soybean oil | 70 | 68 | 70 | 70 |
| Casein | 200 | 200 | 260 | 260 |
| Cellulose | 50 | 50 | 65 | 65 |
| AIN-93 vitamin mix | 10 | 10 | 13 | 13 |
| AIN-93G mineral mix | 35 | 35 | 45.5 | 45.5 |
| l-Cystine | 3 | 3 | 3.9 | 3.9 |
| Choline bitartrate | 2.5 | 2.5 | 3.25 | 3.25 |
| Fucoxanthin | − | 2 | − | 2 |

Table 2.
Initial body weight, final body weight, body weight gain, food/energy intake and energy efficiency of C57BL/6J male mice fed test diets for five weeks. Values are the means ± SD (n = 4). * denotes significant influence by either dietary factor at p < 0.05 analyzed by two-way ANOVA. Energy efficiency = grams of body weight gain/1000 kcal energy intake. FX, fucoxanthin.
| Dietary groups | Initial body weight | Final body weight | Body weight gain | Food intake | Energy intake | Energy efficiency |
|---|---|---|---|---|---|---|
| g | g | g | g/day | kcal/day | ||
| HS | 19.27 ± 1.06 | 24.19 ± 1.75 | 4.92 ± 0.76 | 3.12 ± 0.39 | 12.37 ± 1.53 | 11.42 ± 1.66 |
| HS + F | 19.24 ± 0.58 | 24.63 ± 1.03 | 5.39 ± 0.59 | 3.16 ± 0.20 | 12.45 ± 0.78 | 12.35 ± 0.92 |
| HF | 19.28 ± 0.69 | 24.27 ± 1.71 | 4.99 ± 1.31 | 2.29 ± 0.14 | 11.44 ± 0.72 | 12.45 ± 3.01 |
| HF + F | 19.21 ± 0.61 | 24.84 ± 1.26 | 5.63 ± 0.98 | 2.27 ± 0.06 | 11.28 ± 0.30 | 14.28 ± 2.67 |
| p values | ||||||
| Diet | 0.9769 | 0.8469 | 0.7474 | <0001 * | 0.0460 * | 0.2085 |
| FX | 0.9001 | 0.5050 | 0.2656 | 0.9331 | 0.9391 | 0.2381 |
| Diet * FX | 0.9717 | 0.9257 | 0.8628 | 0.7604 | 0.8075 | 0.6943 |

Table 3.
Relative tissue weights (percent of body weight) of C57BL/6J male mice fed test diets for five weeks. Values are the means ± SD (n = 4). * denotes significant influence by either dietary factor at p < 0.05 analyzed by two-way ANOVA. iWAT, inguinal white adipose tissue (WAT); eWAT, epididymal WAT; rWAT, retroperitoneal WAT; BAT, brown adipose tissue.
| Group | HS | HS+F | HF | HF + F | p-values | ||
|---|---|---|---|---|---|---|---|
| Diet | FX | Diet * FX | |||||
| iWAT | 0.56 ± 0.13 | 0.39 ± 0.03 | 0.63 ± 0.08 | 0.34 ± 0.08 | 0.8149 | 0.0002 * | 0.1599 |
| eWAT | 1.77 ± 0.48 | 1.10 ± 0.12 | 1.90 ± 0.42 | 1.21 ± 0.21 | 0.3944 | 0.0006 * | 0.9527 |
| rWAT | 0.40 ± 0.18 | 0.13 ± 0.04 | 0.44 ± 0.24 | 0.14 ± 0.06 | 0.7321 | 0.0003 * | 0.9982 |
| BAT | 0.29 ± 0.05 | 0.37 ± 0.04 | 0.26 ± 0.03 | 0.30 ± 0.02 | 0.0198 * | 0.0066 * | 0.3840 |
2.2. FX Enhanced Metabolic Rate
Mice were placed in respiratory chambers at week 3 for six days to continuously monitor O2 consumption (VO2), CO2 production (VCO2). These and calculated respiratory quotients (RQ) values throughout a day and respective area under the curve (AUC) values were shown in Figure 1. At seven out of 24 hour time points, high sucrose diet supplemented with fucoxanthin (HS + F) or high-fat diet (HF + F) groups had significantly higher VO2 and VCO2, respectively, compared to HS and HF groups (Figure 1A,B). Diet did not change the AUC of VO2 (Figure 1A), but the high fat diet decreased that of the VCO2 (Figure 1B). FX increased the AUC of both VO2 and VCO2 through the dark phase and all day without an interaction with the diet. Mice fed high fat diets had significantly lower AUC of RQ (p < 0.05), indicating that these mice used more fat as their energy source. FX supplementation did not change RQ (Figure 1C), implying that FX might have enhanced the common aerobic metabolic pathway of glucose and fat metabolism. Indeed, FX was shown to enhance the utilization of glucose in skeletal muscle [32] and liver [29], as well as fatty acid β-oxidation in WAT [28] and liver [30].
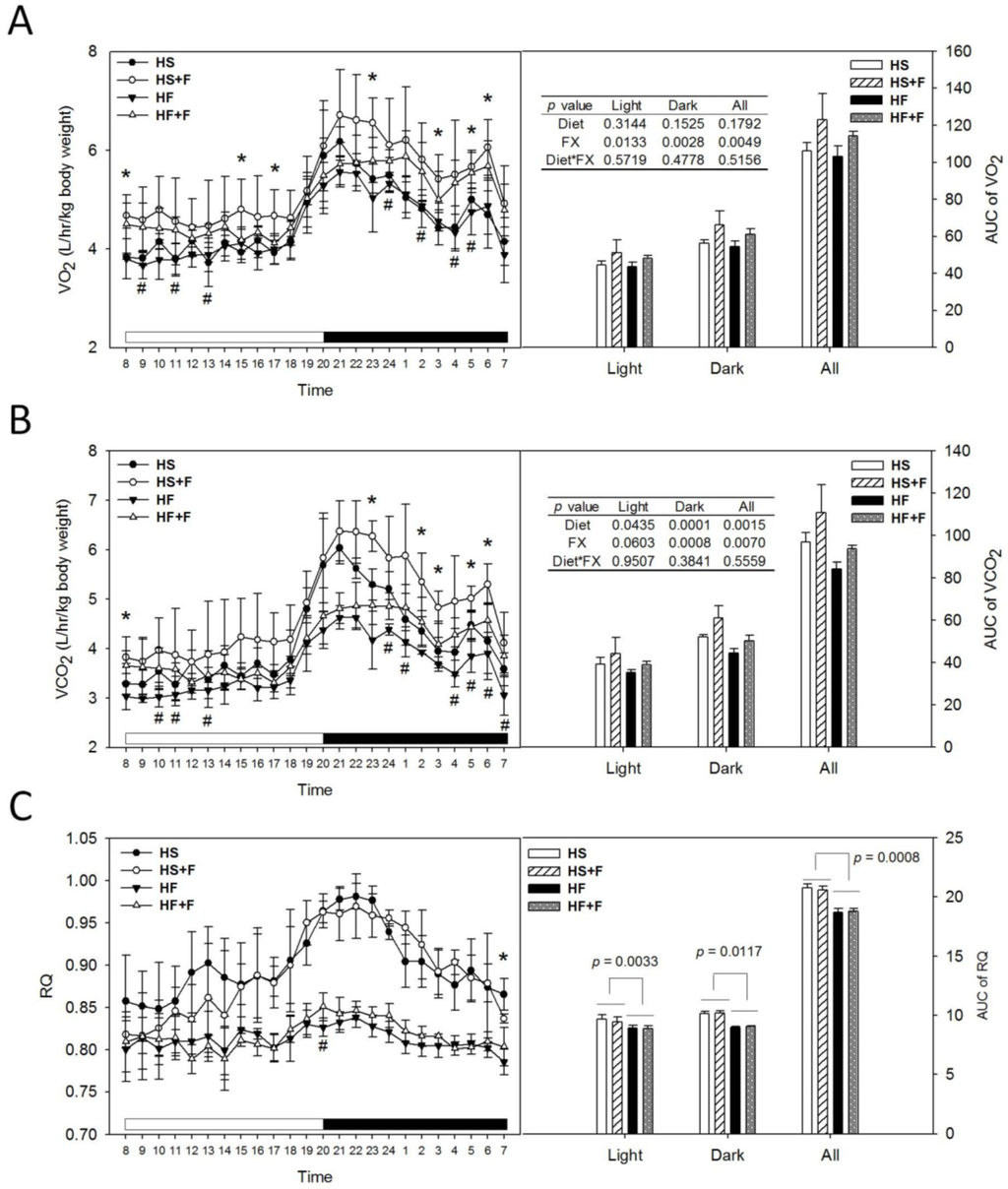
Figure 1.
O2 consumption (A), CO 2 production (B), respiratory quotient (C) and their area under the curve (AUC) of test mice at week 3. Mice were individually placed in metabolic chambers with free access to water and their respective diet and monitored for six days. Values are means and error bars are SD (n = 4). * denotes significant difference between groups HS and HS + F, p < 0.05; # denotes significant difference between groups HF and HF + F, p < 0.05, analyzed by the Student’s t-test. AUCs of O2 consumption (VO2), CO2 production (VCO2) were analyzed by two-way ANOVA. The AUC of RQ was analyzed by the Wilcoxon rank-sum test.
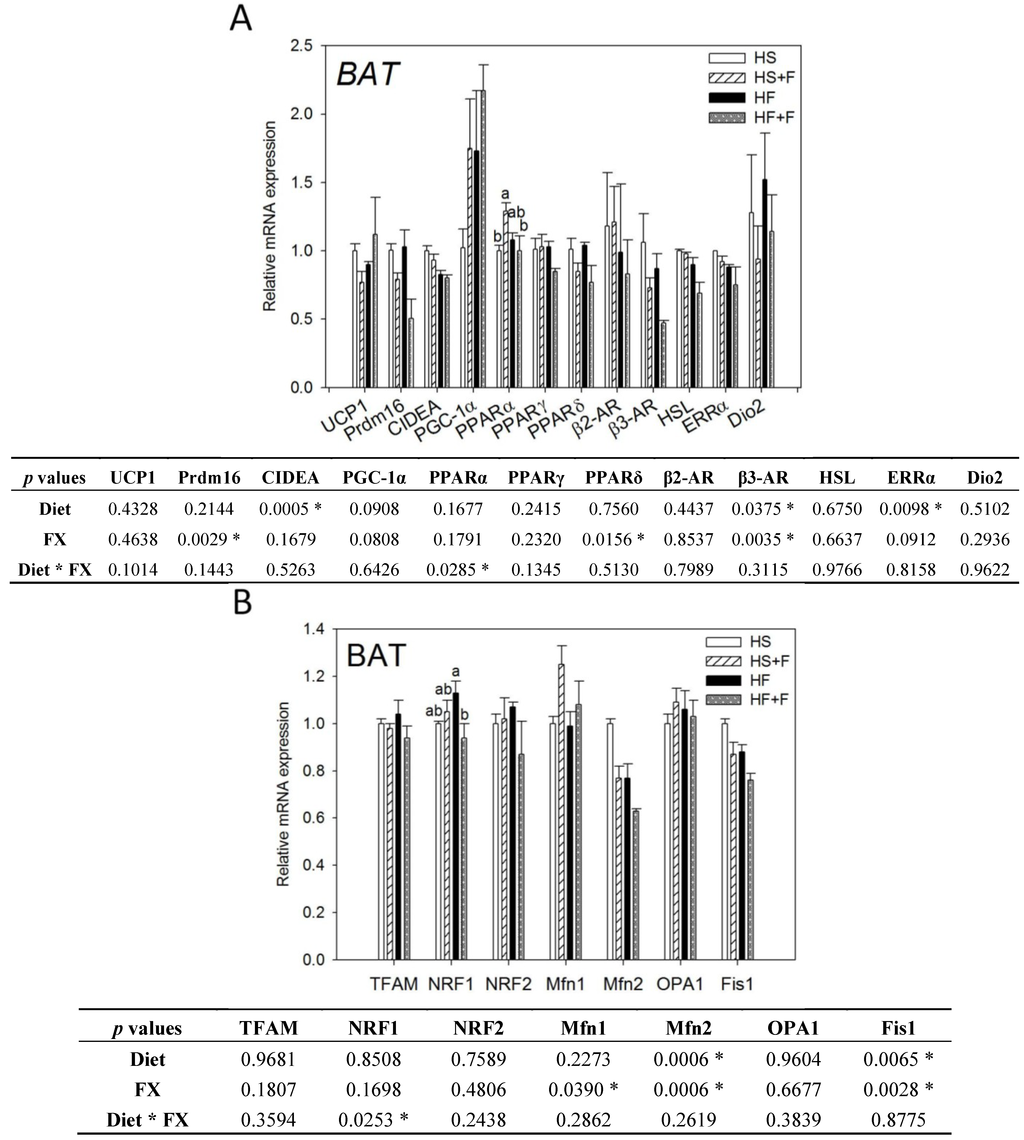
Figure 2.
Thermogenic (A) and mitochondrial homeostasis-related (B) mRNA levels in brown adipose tissue of mice fed test diets for five weeks. Values are means, and error bars are SEM (n = 4). * denotes significant effect by either dietary factor at p < 0.05 analyzed by two-way ANOVA. When an interaction (p < 0.05) between diet and FX existed, the significance of differences among the HS, HS + F, HF and HF + F groups was further analyzed by Duncan’s multiple range test. HS, HS + F, HF and HF + F: As indicated in Figure 1. Relative mRNA expression was measured by real-time qRT-PCR using β-actin as the internal control and normalized to group HS.
2.3. Effect of FX on Thermogenic and Mitochondrial Homeostasis-Related Gene Expressions in BAT and Serum Hormone Concentration
Both high fat diet and FX lowered β3-AR, Mfn2 and Fis1 mRNA levels in the BAT (p < 0.05). Mice fed high fat diets had significantly lower CIDEA and ERRα mRNA expression in the BAT (p < 0.05). FX also decreased Prdm16 and PPARδ but increased Mfn1 mRNA in the BAT. Neither diet nor FX changed PGC-1α UCP1, PPARα, PPARγ, β2-AR, HSL, Dio2, TFAM, NRF1, NRF2 and OPA1 mRNA expressions in BAT. Among them, diet and FX had an interaction effect on the PPARα and NRF1 mRNA expression (p < 0.05). FX increased PPARα mRNA expression in the high sucrose diet-fed mice, but not in the high fat diet-fed mice. FX reduced NRF1 mRNA expression in the high fat diet-fed mice, but not in the high sucrose diet-fed mice (Figure 2).
In this study, BAT mass was significantly increased by FX as in some of previous reports [23,24,25], although other studies did not observe such an effect [31,38]. This discrepancy might be associated with the different mouse strain, gender and diet formula used. Although BAT mass was increased by FX and positively correlated to carbon dioxide production (p = 0.02) during the dark period (Supplementary Figure S1), we did not observe any thermogenic genes upregulated in BAT. β3-AR, Prdm16 and PPARδ mRNA were even downregulated by FX. To validate these results, we even conducted the Housekeeping Genes PCR array (Qiagen, Germantown, MD, USA) and employed three popular software packages (GeNorm, NormFinder and BestKeeper) to confirm the use of β-actin as the most stable housekeeping gene.
On the other hand, our serum hormone analysis showed that the norepinephrine concentration was decreased by FX (Table 4). Norepinephrine, the sympathetic neurotransmitter, is the main in vivo stimulator of the adrenergic signaling mediating the BAT thermogenic machinery. It is not known whether this low serum norepinephrine in FX-fed mice is related to the very minor change in the BAT gene expression irrespective of the enlarged mass observed in this study. Moreover, no significant correlation between BAT mass and O2 consumption was observed (Supplementary Figure S1). Taken together, it is speculated that BAT contributes little, if any, to the FX enhanced O2 consumption.

Table 4.
Serum thyroid hormone, (nor)epinephrine and corticosterone concentrations of C57BL/6J male mice fed test diets for five weeks. Values are the means ± SD (n = 4). * denotes a significant effect by either dietary factor at p < 0.05 analyzed by two-way ANOVA. T4, thyroxine; T3, triiodothyronine; NE, norepinephrine; E, epinephrine; Cort, corticosterone.
| HS | HS + F | HF | HF + F | p-values | |||
|---|---|---|---|---|---|---|---|
| Diet | FX | Diet * FX | |||||
| T4, nM | 24.59 ± 3.69 | 20.73 ± 5.22 | 18.94 ± 2.88 | 17.55 ± 3.79 | 0.0467 * | 0.2117 | 0.5464 |
| T3, nM | 0.80 ± 0.21 | 0.87 ± 0.34 | 0.80 ± 0.14 | 0.82 ± 0.21 | 0.8297 | 0.6998 | 0.8432 |
| T4/T3 | 32.70 ± 11.11 | 27.42 ± 14.18 | 23.80 ± 1.92 | 21.62 ± 2.66 | 0.1346 | 0.4312 | 0.7415 |
| NE, nM | 92.26 ± 16.52 | 66.75 ± 11.69 | 105.72 ± 20.01 | 72.05 ± 10.17 | 0.2383 | 0.0021 * | 0.5989 |
| E, nM | 3.05 ± 0.75 | 3.52 ± 1.21 | 5.61 ± 3.09 | 4.98 ± 3.03 | 0.0949 | 0.9931 | 0.5768 |
| NE/E | 31.87 ± 10.69 | 20.62 ± 7.69 | 22.09 ± 7.96 | 17.53 ± 6.75 | 0.1512 | 0.0843 | 0.4413 |
| Cort, ng/mL | 71.11 ± 10.15 | 63.99 ± 20.76 | 55.55 ± 32.38 | 58.69 ± 23.35 | 0.3898 | 0.8384 | 0.6634 |
2.4. FX Induced Thermogenic-Related Gene Expressions in eWAT and iWAT
While the high fat diet decreased CIDEA mRNA expression, FX significantly increased mRNA expressions of CIDEA, PGC-1α, ERRα, PPARγ, β3-AR, Dio2, PPARα and HSL in eWAT (Figure 3A). The high fat diet decreased UCP1, CIDEA, Prdm16, ERRα, PPARγ, Dio2 and PPARα mRNA expression, but FX increased CIDEA, PGC-1α, ERRα, β3-AR, Dio2 and PPARα mRNA expression in iWAT (Figure 3B). FX did not significantly affect mRNA expressions of these genes in the abdominal rWAT (data not shown), except for upregulating PGC-1α.
Expressions of UCP1, Prdm16, CIDEA and PGC-1α in WAT are indicators for the “browning” of WAT, which could reduce the adverse effects of WAT and help to improve metabolic health [34,39]. In this study, we observed increases in the mRNA of CIDEA and PGC-1α, but not UCP1 and Prdm16 in eWAT and iWAT in mice fed the FX diets. The expression of UCP1 is transcriptionally regulated by the adrenergic signaling (β-AR, cAMP-dependent protein kinase A (PKA), etc.) coupled to PGC-1α and PPAR [40]. PGC-1α has been shown to be required for exercise-induced UCP1 expression in WAT [41]. Prdm16, by co-activating PGC-1α increased mitochondrial content, as well as enhanced uncoupled respiration. Prdm16 activates a robust brown fat phenotype, including the induction of PGC-1α, UCP1 and Dio2 [42,43,44]. In this study, although UCP1 mRNA in these WATs of FX-fed mice was slightly higher than that of the respective controls, the difference did not reach statistical significance, due to the low number of animals per group (n = 4), as well as the large individual variations. Like our study, Woo et al. did not observe increased UCP1 in eWAT of mice fed the 0.2% FX diet, although mice fed the 0.05% FX diet did show an increase of UCP1 in eWAT [31]. CIDEA has been shown to inhibit uncoupling activity of UCP1 when co-expressed in yeast and increases mitochondrial coupling by suppressing UCP1 expression [45]. Therefore, the unchanged UCP1 mRNA level in this study could also be related to the dose of FX and the elevated CIDEA mRNA expression in WAT. Induction of CIDEA has been used as a marker for the emergence of brown adipocyte-like cells in WAT [46]. As we found that only two of the four “beige” adipocyte-specific genes were upregulated, whether FX induces the whole program of WAT “browning” cannot be confirmed.
On the other hand, we observed that β3-AR, Dio2, PGC-1α, ERRα and PPARα mRNA in both eWAT and iWAT and HSL in eWAT were elevated by FX. The adrenergic signaling (β3-AR, PKA, etc.) coupled to PGC-1α and PPAR regulates UCP1 expression [40]. PGC-1α also controls mitochondria biogenesis and respiratory function through targeting multiple transcription factors, like NRF1, NRF2 and ERRα. β3-AR is expressed abundantly and predominantly in brown and white adipocytes. Treatment of mice with β3-AR agonists increases oxygen consumption [47]. Under β3-AR stimulation, PKA is activated and further turns on the transcription of Dio2 and PGC-1α and phosphorylation of HSL [48,49]. Dio2 catalyzes the conversion of T4 to T3 (the active form of thyroid hormone) in most peripheral tissues and is essential for the adrenergic receptor mediated thermogenesis in BAT [50]. T3 increases oxygen consumption, which is mediated through PGC-1α and NRF1 [51]. In this study, increased Dio2 mRNA in WATs is speculated to increase local T3 content and further stimulate PGC-1α mRNA expression and the metabolic rate of mice.
Fucoxanthinol and amarouciaxanthin A are two major metabolites of FX in mouse plasma and liver [52]. In mouse adipose tissue, amarouciaxanthin A is the major metabolite of FX [53]. In vitro, amarouciaxanthin A showed a higher activity in suppressing 3T3-L1 adipocyte differentiation than FX, fucoxanthinol and amarouciaxanthin B [54]. The suppression is associated with downregulations of PPARγ and CCAAT-enhancer-binding protein α (C/EBPα) mRNA levels. In the present in vivo study, however, PPARγ mRNA expression in eWAT was upregulated by FX, implying that inhibition of adipocyte differentiation might have a minor role on the low WAT mass observed in our in vivo study.
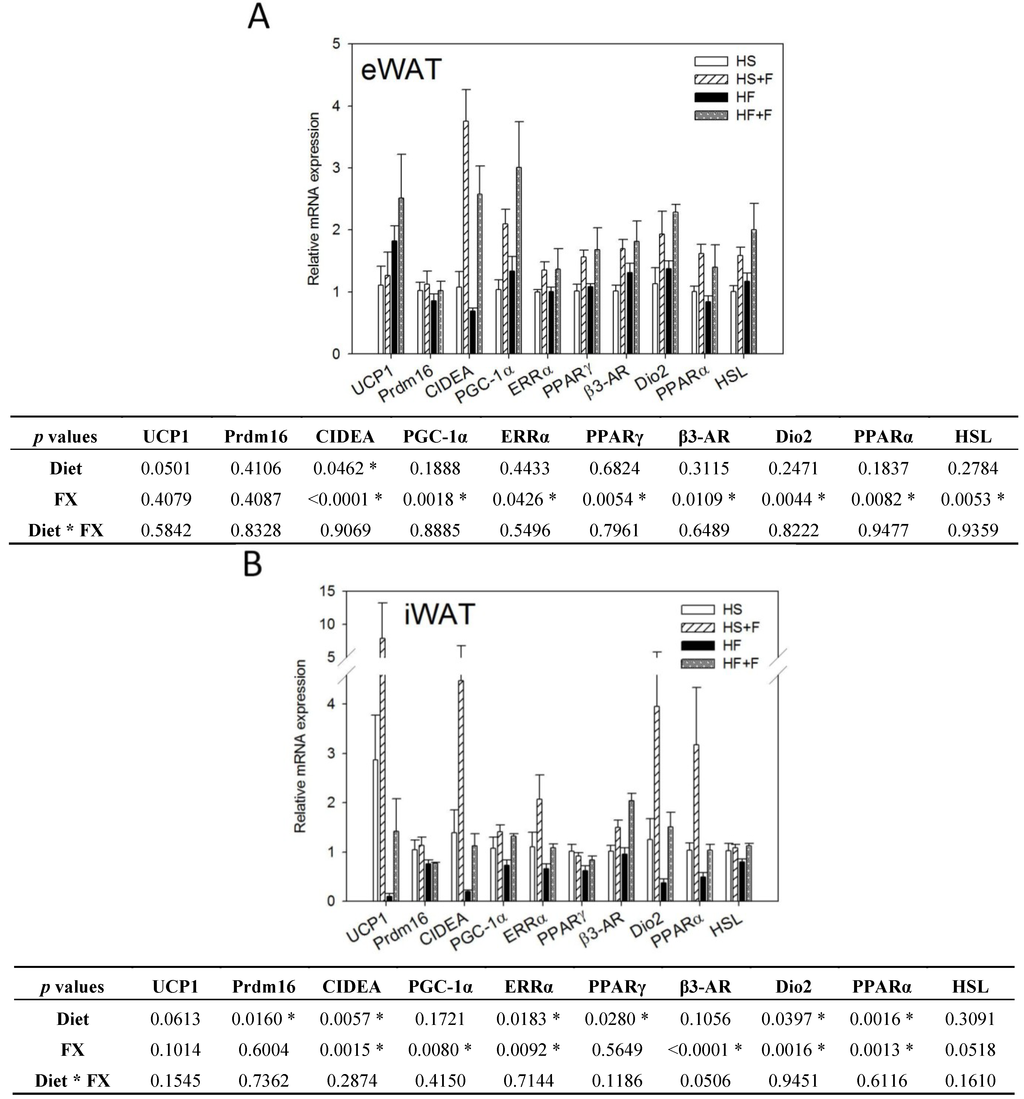
Figure 3.
Thermogenic gene expressions in epididymal (A) and inguinal (B) white adipose tissue. Values are means, and error bars are SEM (n = 4). * denotes significant effect by either dietary factor at p < 0.05 analyzed by two-way ANOVA. HS, HS + F, HF and HF + F: as indicated in Figure 1. The measurement of relative mRNA expression is as indicated in Figure 2.
2.5. FX Increased Mitochondrial Biogenesis and Fusion-Related Gene Expressions in eWAT and iWAT
FX increased NRF1 and NRF2 mRNA expressions in eWAT, but not iWAT. Diet did not affect TFAM, NRF1 and NRF2 mRNA expressions in eWAT and iWAT (Figure 4). The high fat diet decreased Mfn1, Mfn2, OPA1 and Fis1 mRNA levels in iWAT, but not eWAT. In contrast, FX increased mitochondrial fusion-related genes, including Mfn1, Mfn2 and OPA1, but not fission-related gene Fis1 and mRNA expressions in both eWAT and iWAT (Figure 5). Mitochondria provide ~90% of the cellular energy supply and are also the most important organelle of metabolism. PGC-1α is a strong promoter of mitochondrial biogenesis and oxidative metabolism through NRF1, NRF2, ERRα, PPARγ and PPARα [13,14,15]. PGC-1α targets NRF1 and NRF2 directly or indirectly through ERRα in stimulating nuclear genes required for mitochondrial biogenesis and respiration function [14]. PGC-1α interacts with PPARα in the transcription control of genes encoding mitochondrial fatty acid oxidation enzymes [55]. PGC-1α acts as a partner of PPARγ in the induction of adaptive thermogenesis in brown fat and adipocyte differentiation [14]. The increased metabolic rate of mice fed the FX diets in this study coincides with increased PGC-1α networks. ERRα further activates the transcriptional activity of the Mfn2 promoter, and the effect was synergic with those of PGC-1α [16]. In addition to Mfn2, other mitochondrial fusion-related genes (Mfn1 and OPA1) were also increased by FX in this study. In contrast, mitochondrial fission-related gene Fis1 in WATs was not affected by FX. Mitochondrial fusion is frequently found in metabolically active cells [56], and elevated mitochondrial fusion genes agreed with enhanced VO2 and VCO2 and the decreased WAT mass of mice in the present study. Among these genes, Mfn2 was shown to be stimulated under cold, adrenergic agonist or exercise treatment through the regulation of PGC-1α and ERRα [16,57]. Therefore, the elevated mitochondrial fusion genes by FX were in accordance with increased PGC-1α and ERRα mRNA in WATs.
2.6. Overall Discussion
Using the 2 × 2 factorial design, we demonstrated that irrespective of the diet, FX increased both O2 consumption and CO2 production, indicating that FX enhanced metabolic rate. This increase in energy expenditure agreed with smaller adipose mass and increases of the PGC-1α network gene expressions in iWAT and eWAT. These included β3-AR, PGC-1α, CIDEA, Dio2, PPARα and genes regulating mitochondria biogenesis and fusion (ERRα, NRF1, NRF2, Mfn1, Mfn2 and OPA1). We speculate that dietary FX might increase the energy expenditure through changes in mitochondria biogenesis, homeostasis and/or activity mediated by the PGC-1α network in eWAT and iWAT.
PGC-1α is a master regulator of energy metabolism that orchestrates cellular responses to various types of metabolic stress, such as fasting, cold temperature and physical exertion. The expression of PGC-1α is induced by such factors as β3-AR and Dio2. It in turn activates the expressions of transcription factors, including PPARs, NRF1/2, ERRα and Mfn2. In this study, FX upregulated PGC-1α and both of its upstream and downstream genes in iWAT and eWAT, implying that the “PGC-1α network” is upregulated. As this network is known to regulate energy metabolism, it coincides with our observation that FX increased energy expenditure.
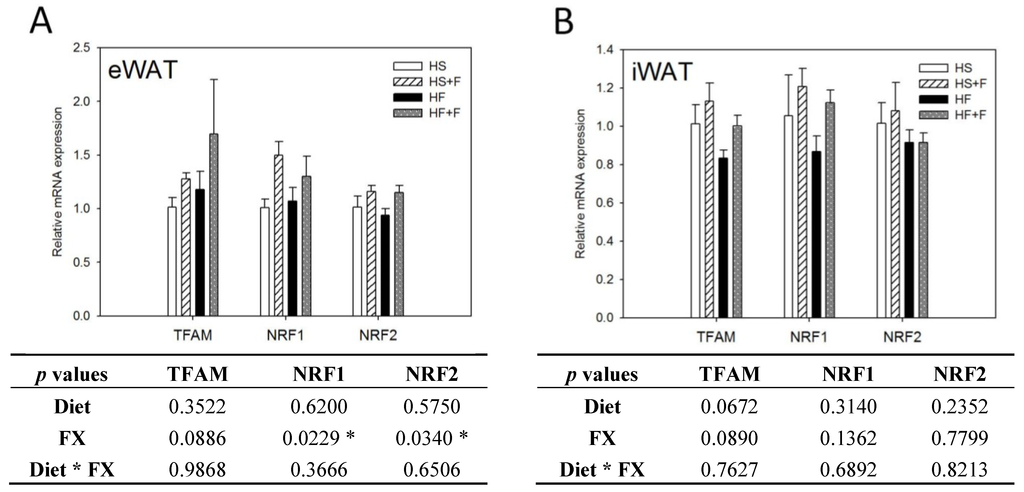
Figure 4.
Mitochondrial biogenic gene expressions in epididymal (A) and inguinal (B) white adipose tissue. Values are means, and error bars are SEM (n = 4). * denotes significant effect by either dietary factor at p < 0.05 analyzed by two-way ANOVA. HS, HS + F, HF and HF + F: As indicated in Figure 1. The measurement of relative mRNA expression is as indicated in Figure 2.
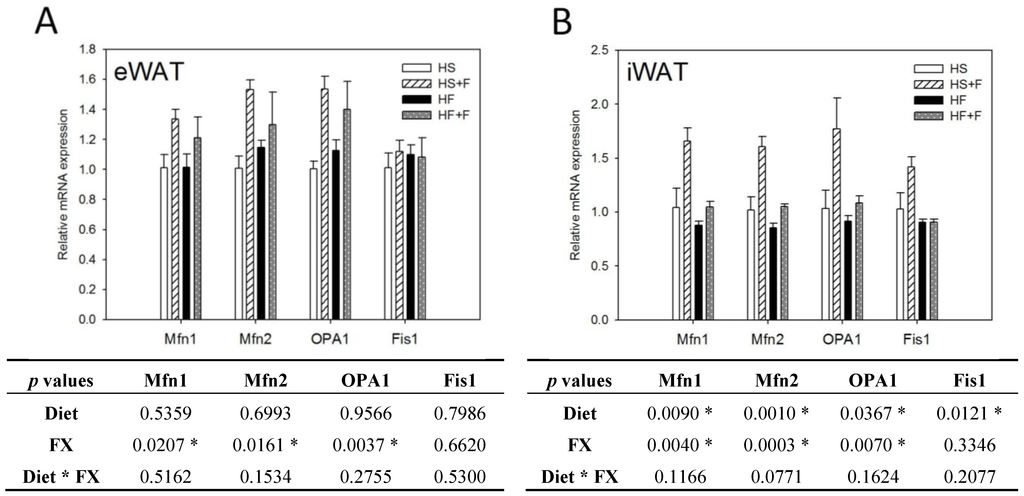
Figure 5.
Mitochondrial homeostatic gene expressions in epididymal (A) and inguinal (B) white adipose tissue. Values are means, and error bars are SEM (n = 4). * denotes significant effect by either dietary factor at p < 0.05 analyzed by two-way ANOVA. HS, HS + F, HF and HF + F: As indicated in Figure 1. The measurement of relative mRNA expression is as indicated in Figure 2.
As we only measured the whole body O2 consumption, it is difficult to specify the contribution of different tissues by FX. However, comparison of changes in the gene expression pattern in WATs and BAT prompt us to speculate that WAT might contribute more than BAT in promoting O2 consumption.
Moreover, our data together with data of previous reports did not support a major role of BAT in the FX enhanced oxygen consumption. These include: (1) FX downregulated β3-AR, Mfn2, Fis1, Prdm16 and PPARδ mRNA levels in the BAT and did not change other thermogenic genes in this study; (2) some studies [31,38] found an anti-obesity effect of FX without BAT enlargement; (3) oxygen consumption did not correlate to BAT mass in our data (Supplementary Information, Figure S1); (4) FX lowered both serum NE (norepinephrine) and BAT β3-AR expression, which are key regulators of BAT adaptive thermogenesis. Lowered serum NE might be related to a decreased sympathetic activity or an increased NE degradation. It is not known why and how FX lowered serum NE and downregulated BAT β3-AR expression. Although it might not be applicable to FX, there has been a study showing that beta-carotene suppresses exhaustive exercise-induced plasma levels of adrenocorticotropic hormone, norepinephrine and epinephrine by inhibiting the secretion of corticotropin-releasing hormone [58]. Although NE concentration is very important for the activation of PGC-1alpha networks in whole body, factors other than NE, such as PPARs and thyroid receptor activation, etc., can also lead to the activation of the PGC-1α network.
In this study, FX did not significantly increase UCP1 mRNA expression in iWAT, eWAT and BAT (p > 0.05). In contrast, expressions of CIDEA and the PGC-1α network in eWAT and iWAT were upregulated, although to different extents. These changes were not noted in liver and skeletal muscle. Moreover, differential extents of induction were also observed in other study. For example, mRNA expression levels of adipogenic marker genes (PPARγ, aP2, adiponectin, C/EBPα, FATP1, LPL and UCP2) and thermogenic genes and mitochondrial biogenesis genes (UCP1, PGC-1α, NRF1, TFAM, Prdm16, CIDEA and Elovl3) induced by triiodothyronine (T3) were also to different extents [59].
3. Experimental Section
3.1. Preparation of FX
FX was isolated from the dried Hincksia mitchellae (Harvey) P. C. Silva brown algae. The brown algae, Hincksia mitchellae (Harvey) P. C. Silva, was originally collected from the Taiwan coast and characterized. The material used in this study was obtained by cultivation in enriched seawater medium (SWM-III) in the lab. The dried powder was extracted with acetone in a brown bottle for 24 h, filtered (No. 2, Whatman filter paper, Maidstone, Kent, UK) and the solvent removed, with a temperature lower than 35 °C. The crude extracts were separated by using a silica gel flash column chromatography (Geduran® Si 60, 0.040–0.063 mm, Merck, Darmstadt, Germany), eluted with ethyl acetate/n-hexane (4.5:5.5, v/v). The red-orange color fraction was collected and passed through a second silica gel column eluted with ethanol/acetone/n-hexane (1:19:80, v/v). After removing the solvent, the residue was re-dissolved in acetone/n-hexane (45:55, v/v). An equal volume of Millipore-Q water was added, and the mixture was standing at −20 °C for 4 h until a red precipitate was produced. The precipitate was filtered and dried in a freeze drier. The obtained solid was confirmed as FX by H-NMR and the purity checked by HPLC (PU-980 pump and UV-visible absorbance detector, Jasco, Japan and LiChrospher® 100 RP-18 column, 5 μm, Merck, Darmstadt, Germany). The mobile phase used was methanol/H2O (90:10, v/v) with a flow rate of 1 mL/min. The FX was detected at 450 nm and quantified from the peak area by using a standard curve with previous purified and identified FX (95%). The purity of FX prepared was >95% by this HPLC analysis.
3.2. Animals and Diets
The animal study was approved by the Institution Animal Care and Use Committee of National Taiwan University (No.NTU-98-EL-106). Three-week old male C57BL/6J mice were purchased from the National Laboratory Animal Center (Taipei, Taiwan) and housed individually in stainless steel wire cages in an animal room with a 12-hr light, 12-hr dark cycle (light period: 0800–2000) and constant temperature (22 ± 2 °C). Mice were fed a non-purified diet (LabDiet® 5001, PMI® Nutrition International Inc., Brentwood, MO, USA) for 1 week and switched to the high sucrose (HS, Table 1) diet for another week before the experiment. After the 2-week acclimation, mice were randomly assigned into four groups (4 mice/group) and respectively fed: HS (50% sucrose, 7% soybean oil), HF (high-fat diet, 23% butter plus 7% soybean oil), HS + F (HS diet supplemented with 0.2% FX) or HF + F (HF diet supplemented with 0.2% FX) test diets for 5 weeks. The composition of the 4 test diets was modified from AIN-93G [37] and our previous studies [60,61] (Table 1). The amounts of casein, cellulose, vitamin and mineral mixtures in the high-fat diets were adjusted to make the nutrient/energy ratios equivalent to those of the HS diet. Mice had free access to the diet and water. Body weight and food intake were recorded weekly.
3.3. Metabolic Rate Measurement
After feeding test diets for 2 weeks, mice were placed in the respiratory chamber individually (AccuScan Instruments, Inc., Columbus, OH, USA) with free access to their respective diet food and water. After a 24-h acclimation, mice were continuously monitored in the metabolic chambers for 6 days. Gas samples were collected every minute per mouse, and the data were averaged for each hour. Output parameters from the average of 6 days included the O2 consumption (VO2), CO2 production (VCO2) and respiratory quotients (RQ = VO2/VCO2).
3.4. Tissue and Serum Collection
After feeding test diets for 5 weeks, mice were deprived of diet for 12 h and killed by CO2 asphyxiation. Blood was withdrawn through retro-orbital. Serum was obtained after centrifugation at 12,000 rpm for 10 min at 4 °C and stored at −80 °C. Liver, spleen, kidney, heart, lung, gastrocnemius muscle, retroperitoneal WAT (rWAT), eWAT, inguinal (iWAT) and intra-scapular BAT were excised, weighed and immediately frozen in liquid nitrogen and stored at −80 °C for mRNA analysis.
3.5. Housekeeping Genes PCR Array
rWAT and BAT total RNA (0.5 μg) of 2 mice per group were reversed transcribed using the RT2 First Strand Kit and further analyzed by the Mouse Housekeeping Genes PCR array (Qiagen, Germantown, MD, USA), which analyzed 12 commonly used housekeeping genes in a 96-well plate. Briefly, 1 μL cDNA (1 ng/μL), 12.5 μL RT2 SYBR Green Mastermix and 11.25 μL water were loaded into primer-precoated wells, and PCR was performed using a StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Data were analyzed by three popular algorithms: GeNorm [62], NormFinder [63] and BestKeeper [64]. β-actin was shown to be the most stable and suitable reference gene in our study.
3.6. RNA Extraction and Quantitative Real-Time RT-PCR
Total RNA of eWAT, rWAT, iWAT and BAT were isolated by an RNeasy® Plus Universal Mini Kit (Qiagen, Stockach, Germany), according to the manufacturer’s instruction. Total RNA (2 μg) was reverse transcribed to cDNA using a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). PCR was performed in a final volume of 25 μL containing 10 μL cDNA (1 ng/μL), 12.5 μL TaqMan® Gene Expression Master Mix, 1.25 μL probe/primer assay mix and 1.25 μL water. PCR primers and probes were purchased from Applied Biosystems: UCP1, CIDEA (cell death-inducing DFFA-like effecter a), Prdm16 (PR domain containing 16), PGC-1α, NRF1, NRF2, TFAM, ERRα, PPARα, PPARγ, HSL (hormone-sensitive lipase), β3-AR (β3-adrenergic receptor), Dio2 (deiodinase 2), Mfn1, Mfn2, OPA1 (optic atrophy 1), Fis1 (fission 1) and β-actin. These genes are associated with thermogenesis, mitochondrial biogenesis, browning of WAT and mitochondrial fusion and fission. The mRNA expression levels were determined by quantitative real-time RT-PCR using the StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The expression levels were normalized to β-actin. Data were analyzed by StepOne Software (v2.2.2, Applied Biosystems, Foster City, CA, USA).
3.7. Serum Hormone Analysis
Commercially available ELISA kits were used to measure serum thyroxine (T4) and triiodothyronine (T3) (Calbiotech, Spring Valley, CA, USA), norepinephrine and epinephrine (Labor Diagnostika Nord, Nordhorn, Germany), as well as corticosterone (Cayman chemical company, Ann Arbor, MI, USA).
3.8. Statistical Analysis
Data are expressed as the means ± SD or SEM. Statistical analysis were performed using SAS 9.1 (SAS Institute Inc., Cary, NC, USA). For all statistical analyses, data were transformed logarithmically if the variances were non-homogeneous. In order to examine the effect of diet, FX and their interaction in this 2 × 2 factorial design, two-way ANOVAs were used. The effect is considered significant if p < 0.05. When an interaction (p < 0.05) existed between diet and FX, data were further analyzed by one-way ANOVA and Duncan’s multiple range test. In the metabolic rate study, differences of VO2, VCO2 and RQ at each time point between HS and HS + F and between HF and HF + F were respectively analyzed by the Student’s t-test. Areas under curve (AUCs) of VO2 and VCO2 were analyzed by two-way ANOVA. The AUC of RQ was analyzed by the Wilcoxon rank-sum test.
4. Conclusions
In conclusion, we demonstrated that dietary FX elevated the metabolic rate and reduced both subcutaneous and abdominal WAT mass irrespective of the diet used. These were associated with upregulated genes of the PGC-1α network, including those regulating mitochondrial biogenesis and fusion in iWAT and eWAT.
Supplementary Files
Acknowledgments
This study was supported by a grant from the National Science Council of Taiwan (NSC 99-2313-B-002-013-MY3).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Malik, V.S.; Willett, W.C.; Hu, F.B. Global obesity: Trends, risk factors and policy implications. Nat. Rev. Endocrinol. 2013, 9, 13–27. [Google Scholar] [CrossRef]
- Medina-Gomez, G. Mitochondria and endocrine function of adipose tissue. Best Pract. Res. Clin. Endocrinol. Metab. 2012, 26, 791–804. [Google Scholar] [CrossRef]
- Bach, D.; Pich, S.; Soriano, F.X.; Vega, N.; Baumgartner, B.; Oriola, J.; Daugaard, J.R.; Lloberas, J.; Camps, M.; Zierath, J.R.; et al. Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. A novel regulatory mechanism altered in obesity. J. Biol. Chem. 2003, 278, 17190–17197. [Google Scholar]
- Patti, M.E.; Corvera, S. The role of mitochondria in the pathogenesis of type 2 diabetes. Endocr. Rev. 2010, 31, 364–395. [Google Scholar] [CrossRef]
- Lowell, B.B.; Shulman, G.I. Mitochondrial dysfunction and type 2 diabetes. Science 2005, 307, 384–387. [Google Scholar] [CrossRef]
- Liu, J.; Shen, W.; Zhao, B.; Wang, Y.; Wertz, K.; Weber, P.; Zhang, P. Targeting mitochondrial biogenesis for preventing and treating insulin resistance in diabetes and obesity: Hope from natural mitochondrial nutrients. Adv. Drug Deliv. Rev. 2009, 61, 1343–1352. [Google Scholar] [CrossRef]
- Joseph, A.M.; Joanisse, D.R.; Baillot, R.G.; Hood, D.A. Mitochondrial dysregulation in the pathogenesis of diabetes: Potential for mitochondrial biogenesis-mediated interventions. Exp. Diabetes Res. 2012, 2012, 642038. [Google Scholar]
- Kusminski, C.M.; Scherer, P.E. Mitochondrial dysfunction in white adipose tissue. Trends Endocrinol. Metab. 2012, 23, 435–443. [Google Scholar] [CrossRef]
- Youle, R.J.; van der Bliek, A.M. Mitochondrial fission, fusion, and stress. Science 2012, 337, 1062–1065. [Google Scholar] [CrossRef]
- Liesa, M.; Shirihai, O.S. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013, 17, 491–506. [Google Scholar] [CrossRef]
- Bach, D.; Naon, D.; Pich, S.; Soriano, F.X.; Vega, N.; Rieusset, J.; Laville, M.; Guillet, C.; Boirie, Y.; Wallberg-Henriksson, H.; et al. Expression of Mfn2, the Charcot-Marie-Tooth neuropathy type 2A gene, in human skeletal muscle: Effects of type 2 diabetes, obesity, weight loss, and the regulatory role of tumor necrosis factor alpha and interleukin-6. Diabetes 2005, 54, 2685–2693. [Google Scholar] [CrossRef]
- Zorzano, A.; Liesa, M.; Palacin, M. Role of mitochondrial dynamics proteins in the pathophysiology of obesity and type 2 diabetes. Int. J. Biochem. Cell Biol. 2009, 41, 1846–1854. [Google Scholar] [CrossRef]
- Wu, Z.; Puigserver, P.; Andersson, U.; Zhang, C.; Adelmant, G.; Mootha, V.; Troy, A.; Cinti, S.; Lowell, B.; Scarpulla, R.C.; et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999, 98, 115–124. [Google Scholar] [CrossRef]
- Scarpulla, R.C. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochimi. Biophys. Acta 2001, 1813, 1269–1278. [Google Scholar] [CrossRef]
- Lin, J.; Handschin, C.; Spiegelman, B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005, 1, 361–370. [Google Scholar] [CrossRef]
- Soriano, F.X.; Liesa, M.; Bach, D.; Chan, D.C.; Palacin, M.; Zorzano, A. Evidence for a mitochondrial regulatory pathway defined by peroxisome proliferator-activated receptor-gamma coactivator-1 alpha, estrogen-related receptor-alpha, and mitofusin 2. Diabetes 2006, 55, 1783–1791. [Google Scholar]
- Clayton, D.A. Replication and transcription of vertebrate mitochondrial DNA. Annu. Rev. Cell Biol. 1991, 7, 453–478. [Google Scholar] [CrossRef]
- Virbasius, J.V.; Scarpulla, R.C. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: A potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc. Natl. Acad. Sci. USA 1994, 91, 1309–1313. [Google Scholar] [CrossRef]
- Semple, R.K.; Crowley, V.C.; Sewter, C.P.; Laudes, M.; Christodoulides, C.; Considine, R.V.; Vidal-Puig, A.; O’Rahilly, S. Expression of the thermogenic nuclear hormone receptor coactivator PGC-1alpha is reduced in the adipose tissue of morbidly obese subjects. Int. J. Obes. Relat. Metab. Disord. 2004, 28, 176–179. [Google Scholar]
- Tiraby, C.; Tavernier, G.; Lefort, C.; Larrouy, D.; Bouillaud, F.; Ricquier, D.; Langin, D. Acquirement of brown fat cell features by human white adipocytes. J. Biol. Chem. 2003, 278, 33370–33376. [Google Scholar]
- Haugan, J.A.; Aakermann, T.; Liaaen-Jensen, S. Isolation of fucoxanthin and peridinin. Methods Enzymol. 1992, 213, 231–245. [Google Scholar] [CrossRef]
- Okada, T.; Mizuno, Y.; Sibayama, S.; Hosokawa, M.; Miyashita, K. Antiobesity Effects of Undaria Lipid Capsules Prepared with Scallop Phospholipids. J. Food Sci. 2011, 76, H2–H6. [Google Scholar] [CrossRef]
- Maeda, H.; Hosokawa, M.; Sashima, T.; Funayama, K.; Miyashita, K. Effect of medium-chain triacylglycerols on anti-obesity effect of fucoxanthin. J. Oleo Sci. 2007, 56, 615–621. [Google Scholar] [CrossRef]
- Maeda, H.; Hosokawa, M.; Sashima, T.; Miyashita, K. Dietary combination of fucoxanthin and fish oil attenuates the weight gain of white adipose tissue and decreases blood glucose in obese/diabetic KK-Ay mice. J. Agric. Food Chem. 2007, 55, 7701–7706. [Google Scholar] [CrossRef]
- Maeda, H.; Hosokawa, M.; Sashima, T.; Funayama, K.; Miyashita, K. Fucoxanthin from edible seaweed, Undaria pinnatifida, shows antiobesity effect through UCP1 expression in white adipose tissues. Biochem. Biophys. Res. Commun. 2005, 332, 392–397. [Google Scholar] [CrossRef]
- Maeda, H.; Tsukui, T.; Sashima, T.; Hosokawa, M.; Miyashita, K. Seaweed carotenoid, fucoxanthin, as a multi-functional nutrient. Asia Pac. J. Clin. Nutr. 2008, 17, 196–199. [Google Scholar]
- Maeda, H.; Hosokawa, M.; Sashima, T.; Murakami-Funayama, K.; Miyashita, K. Anti-obesity and anti-diabetic effects of fucoxanthin on diet-induced obesity conditions in a murine model. Mol. Med. Rep. 2009, 2, 897–902. [Google Scholar]
- Jeon, S.M.; Kim, H.J.; Woo, M.N.; Lee, M.K.; Shin, Y.C.; Park, Y.B.; Choi, M.S. Fucoxanthin-rich seaweed extract suppresses body weight gain and improves lipid metabolism in high-fat-fed C57BL/6J mice. Biotechnol. J. 2010, 5, 961–969. [Google Scholar] [CrossRef]
- Park, H.J.; Lee, M.K.; Park, Y.B.; Shin, Y.C.; Choi, M.S. Beneficial effects of Undaria pinnatifida ethanol extract on diet-induced-insulin resistance in C57BL/6J mice. Food Chem. Toxicol. 2011, 49, 727–733. [Google Scholar] [CrossRef]
- Woo, M.N.; Jeon, S.M.; Kim, H.J.; Lee, M.K.; Shin, S.K.; Shin, Y.C.; Park, Y.B.; Choi, M.S. Fucoxanthin supplementation improves plasma and hepatic lipid metabolism and blood glucose concentration in high-fat fed C57BL/6N mice. Chemico-Biol. Interact. 2010, 186, 316–322. [Google Scholar] [CrossRef]
- Woo, M.N.; Jeon, S.M.; Shin, Y.C.; Lee, M.K.; Kang, M.A.; Choi, M.S. Anti-obese property of fucoxanthin is partly mediated by altering lipid-regulating enzymes and uncoupling proteins of visceral adipose tissue in mice. Mol. Nutr. Food Res. 2009, 53, 1–9. [Google Scholar]
- Nishikawa, S.; Hosokawa, M.; Miyashita, K. Fucoxanthin promotes translocation and induction of glucose transporter 4 in skeletal muscles of diabetic/obese KK-A(y) mice. Phytomedicine 2012, 19, 389–394. [Google Scholar] [CrossRef]
- Wu, J.; Cohen, P.; Spiegelman, B.M. Adaptive thermogenesis in adipocytes: Is beige the new brown? Genes Dev. 2013, 27, 234–250. [Google Scholar] [CrossRef]
- Bartelt, A.; Heeren, J. Adipose tissue browning and metabolic health. Nature Rev. Endocrinol. 2014, 10, 24–36. [Google Scholar] [CrossRef]
- Yoneshiro, T.; Aita, S.; Matsushita, M.; Kayahara, T.; Kameya, T.; Kawai, Y.; Iwanaga, T.; Saito, M. Recruited brown adipose tissue as an antiobesity agent in humans. J. Clin. Investig. 2013, 123, 3404–3408. [Google Scholar] [CrossRef]
- Chan, D.C. Fusion and fission: interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 2012, 46, 265–287. [Google Scholar] [CrossRef]
- Reeves, P.G.; Nielsen, F.H.; Fahey, G.C., Jr. AIN-93 purified diets for laboratory rodents: Final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr. 1993, 123, 1939–1951. [Google Scholar]
- Hu, X.; Li, Y.; Li, C.; Fu, Y.; Cai, F.; Chen, Q.; Li, D. Combination of fucoxanthin and conjugated linoleic acid attenuates body weight gain and improves lipid metabolism in high-fat diet-induced obese rats. Arch. Biochemi. Biophys. 2012, 519, 59–65. [Google Scholar] [CrossRef]
- Shan, T.; Liang, X.; Bi, P.; Zhang, P.; Liu, W.; Kuang, S. Distinct populations of adipogenic and myogenic Myf5-lineage progenitors in white adipose tissues. J. Lipid Res. 2013, 54, 2214–2224. [Google Scholar] [CrossRef]
- Collins, S.; Yehuda-Shnaidman, E.; Wang, H. Positive and negative control of Ucp1 gene transcription and the role of beta-adrenergic signaling networks. Int. J. Obes. 2010, 34, S28–S33. [Google Scholar] [CrossRef]
- Ringholm, S.; Grunnet Knudsen, J.; Leick, L.; Lundgaard, A.; Munk Nielsen, M.; Pilegaard, H. PGC-1alpha is required for exercise- and exercise training-induced UCP1 upregulation in mouse white adipose tissue. PloS One 2013, 8, e64123. [Google Scholar]
- Kajimura, S.; Seale, P.; Tomaru, T.; Erdjument-Bromage, H.; Cooper, M.P.; Ruas, J.L.; Chin, S.; Tempst, P.; Lazar, M.A.; Spiegelman, B.M. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008, 22, 1397–1409. [Google Scholar]
- Seale, P.; Kajimura, S.; Yang, W.; Chin, S.; Rohas, L.M.; Uldry, M.; Tavernier, G.; Langin, D.; Spiegelman, B.M. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007, 6, 38–54. [Google Scholar]
- Seale, P.; Conroe, H.M.; Estall, J.; Kajimura, S.; Frontini, A.; Ishibashi, J.; Cohen, P.; Cinti, S.; Spiegelman, B.M. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J. Clin. Investig. 2011, 121, 96–105. [Google Scholar]
- Zhou, Z.; Yon Toh, S.; Chen, Z.; Guo, K.; Ng, C.P.; Ponniah, S.; Lin, S.C.; Hong, W.; Li, P. Cidea-deficient mice have lean phenotype and are resistant to obesity. Nature Genet. 2003, 35, 49–56. [Google Scholar]
- Meyer, C.W.; Willershauser, M.; Jastroch, M.; Rourke, B.C.; Fromme, T.; Oelkrug, R.; Heldmaier, G.; Klingenspor, M. Adaptive thermogenesis and thermal conductance in wild-type and UCP1-KO mice. Am. J. Physiol. 2010, 299, R1396–R1406. [Google Scholar]
- Susulic, V.S.; Frederich, R.C.; Lawitts, J.; Tozzo, E.; Kahn, B.B.; Harper, M.E.; Himms-Hagen, J.; Flier, J.S.; Lowell, B.B. Targeted disruption of the beta 3-adrenergic receptor gene. J. Biol. Chem. 1995, 270, 29483–29492. [Google Scholar]
- Lowell, B.B.; Spiegelman, B.M. Towards a molecular understanding of adaptive thermogenesis. Nature 2000, 404, 652–660. [Google Scholar]
- Mottillo, E.P.; Bloch, A.E.; Leff, T.; Granneman, J.G. Lipolytic products activate peroxisome proliferator-activated receptor (PPAR) alpha and delta in brown adipocytes to match fatty acid oxidation with supply. J. Biol. Chem. 2012, 287, 25038–25048. [Google Scholar]
- de Jesus, L.A.; Carvalho, S.D.; Ribeiro, M.O.; Schneider, M.; Kim, S.W.; Harney, J.W.; Larsen, P.R.; Bianco, A.C. The type 2 iodothyronine deiodinase is essential for adaptive thermogenesis in brown adipose tissue. J. Clin. Investig. 2001, 108, 1379–1385. [Google Scholar] [CrossRef]
- Weitzel, J.M.; Iwen, K.A.; Seitz, H.J. Regulation of mitochondrial biogenesis by thyroid hormone. Exp. Physiol. 2003, 88, 121–128. [Google Scholar]
- Asai, A.; Sugawara, T.; Ono, H.; Nagao, A. Biotransformation of fucoxanthinol into amarouciaxanthin A in mice and HepG2 cells: Formation and cytotoxicity of fucoxanthin metabolites. Drug Metab. Dispos. 2004, 32, 205–211. [Google Scholar] [CrossRef]
- Yonekura, L.; Kobayashi, M.; Terasaki, M.; Nagao, A. Keto-carotenoids are the major metabolites of dietary lutein and fucoxanthin in mouse tissues. J. Nutr. 2010, 140, 1824–1831. [Google Scholar] [CrossRef]
- Yim, M.J.; Hosokawa, M.; Mizushina, Y.; Yoshida, H.; Saito, Y.; Miyashita, K. Suppressive Effects of Amarouciaxanthin A on 3T3-L1 Adipocyte Differentiation through Down-regulation of PPARgamma and C/EBPalpha mRNA Expression. J. Agric. Food Chem. 2011, 59, 1646–1652. [Google Scholar]
- Vega, R.B.; Huss, J.M.; Kelly, D.P. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell. Biol. 2000, 20, 1868–1876. [Google Scholar] [CrossRef]
- Westermann, B. Molecular machinery of mitochondrial fusion and fission. J. Biol. Chem. 2008, 283, 13501–13505. [Google Scholar]
- Cartoni, R.; Leger, B.; Hock, M.B.; Praz, M.; Crettenand, A.; Pich, S.; Ziltener, J.L.; Luthi, F.; Deriaz, O.; Zorzano, A.; et al. Mitofusins 1/2 and ERRalpha expression are increased in human skeletal muscle after physical exercise. J. Phys. 2005, 567, 349–358. [Google Scholar]
- Hasegawa, T. Anti-stress effect of beta-carotene. Ann. N. Y. Acad. Sci. 1993, 691, 281–283. [Google Scholar] [CrossRef]
- Lee, J.Y.; Takahashi, N.; Yasubuchi, M.; Kim, Y.I.; Hashizaki, H.; Kim, M.J.; Sakamoto, T.; Goto, T.; Kawada, T. Triiodothyronine induces UCP-1 expression and mitochondrial biogenesis in human adipocytes. Am. J. Physiol. Cell Physiol. 2012, 302, C463–C472. [Google Scholar] [CrossRef]
- Hsu, S.C.; Huang, C.J. Reduced fat mass in rats fed a high oleic acid-rich safflower oil diet is associated with changes in expression of hepatic PPARalpha and adipose SREBP-1c-regulated genes. J. Nutr. 2006, 136, 1779–1785. [Google Scholar]
- Lu, K.N.; Hsu, C.; Chang, M.L.; Huang, C.J. Wild bitter gourd increased metabolic rate and upregulated genes related to mitochondria biogenesis and UCP-1 in mice. J. Funct. Foods 2013, 5, 668–678. [Google Scholar]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar]
Abbreviations
| DFFA | DNA fragmentation factor, alpha subunit |
| HF | High fat diet |
| HF + F | High fat diet supplemented with fucoxanthin |
| HS | High sucrose diet |
| HS + F | High sucrose diet supplemented with fucoxanthin |
| qRT | Quantitative real-time |
| RQ | Respiratory quotient |
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).