Abstract
Lipid A, the hydrophobic anchor of lipopolysaccharide, is an essential component in the outer membrane of most Gram-negative bacteria. Food-borne pathogen Cronobacter sakazakii synthesizes two lipid A species, differing by the length of the secondary acyl chain. In this work, we identified three genes ESA02293, ESA02951 and ESA01386 encoding for the late acyltransferases of lipid A biosynthesis pathway in C. sakazakii. Based on the sequence alignment, proteins YP_001438378.1 encoded by ESA02293, YP_001439016.1 encoded by ESA02951, and YP_001437482.1 encoded by ESA01386 are homologous to E. coli LpxL, LpxP and LpxM, respectively. Functions of the three acyltransferases were confirmed by overexpressing the genes in E. coli, isolating lipid As and analyzing their structures using an ESI/MS. C. sakazakii LpxL and LpxM transfer a C14:0 secondary acyl chain to the 2′- and 3′-position of lipid A, respectively. C. sakazakii LpxP can transfer either a C16:1 or a C14:0 secondary acyl chains to the 2′-position of lipid A.
1. Introduction
Lipid Ais the hydrophobic anchor of lipopolysaccharide in the outer membrane of Gram-negative bacteria [1]. It is recognized by the TLR4/MD2 receptor of the innate immune system, which triggers an inflammatory response accompanied by massive over-production of cytokines and leads to Gram-negative septic shock [2,3,4]. Cronobacter sakazakii is a Gram-negative bacterial pathogen associated with contaminated, temperature-abused, powdered infant formula, which can cause meningitis and enterocolitis in neonates. Cronobacter sakazakii synthesizes two lipid A species, but the biosynthesis of lipid A in C. sakazakii has not been well characterized [5,6].
In E. coli, lipid A contains four 3-hydroxyacyl chains at the 2-, 3-, 2′-, and 3′-positions, and two acyl chains linked to the 2′-, and 3′-hydroxyacyl chains, forming acyloxyacyl moieties [7]. The four primary 3-hydroxyacyl chains are added by acyltransferases LpxA [8] and LpxD [9,10], and the two secondary acyl chains are added by late acyltransferases LpxL, LpxP and LpxM. E. coli LpxL transfers a C12:0 secondary acyl chain to the 2′-position of lipid A [11,12,13], LpxP has high similarity to LpxL, and can transfer a C16:0 secondary acyl chain to the 2′-position of lipid A at low temperature (12 °C) [14,15], LpxM transfers a C14:0 secondary acyl chain to the 3′-position of lipid A [16]. In contrast to E. coli, which synthesizes only one lipid A species, C. sakazakii, synthesizes two lipid A species, differing on the length of the secondary acyl chains at 2′- and 3′-positions. Late acyltransferases of lipid A in C. sakazakii have not been reported.
In this work, we identified three genes encoding the late acyltransferases of lipid A in C. sakazakii BAA894. Three enzymes encoded by genes ESA02293, ESA02952 and ESA01386 are homologous to E. coli LpxL, LpxP and LpxM, respectively. Like E. coli LpxM, C. sakazakii LpxM transfers a C14:0 secondary acyl chain to the 3′-position of lipid A, C. sakazakii LpxL and LpxP, however, functions differently from E. coli LpxL and LpxP. C. sakazakii LpxL transfers a C14:0 secondary acyl chain to the 2′-position of lipid A, while C. sakazakii LpxP can transfer either a C16:1 or a C14:0 secondary acyl chain to the 2′-position of lipid A even at ambient temperatures. These three acyltransferases might play important roles in biosynthesis of lipid A in C. sakazakii.
2. Results and Discussion
2.1. Three Genes Encoding Late Acyltransferases Were Found in C. sakazakii BAA894
Lipid A samples were extracted from E. coli W3110 and C. sakazakii BAA894, respectively, and analyzed using ESI/MS in the negative ion mode (Figure 1). Two majorions at m/z 1796.1 and 1716.2 were observed in the spectrum of E. coli Lipid A as expected (Figure 1A). The peak at m/z 1796.1 was created by the molecular ion [M − H]− of lipid A; the peak at m/z 1716.2 arisen by loss of a phosphate from the lipid A during the extraction procedure [5]. The spectrum of C. sakazakii lipid A showed four major peaks at m/z 1716.1, 1744.1, 1796.1 and 1824.1, respectively (Figure 1B). Two of the peaks (m/z 1716.1 and 1796.1) are the same to that of E. coli lipid A, while the other two peaks (m/z 1744.1 and 1824.1) differ from the first two by 28 Da, suggesting that C. sakazakii BAA894 synthesizes two different species of lipid A which differ at the length of secondary fatty acyl chains at 2′ and 3′-position [6].
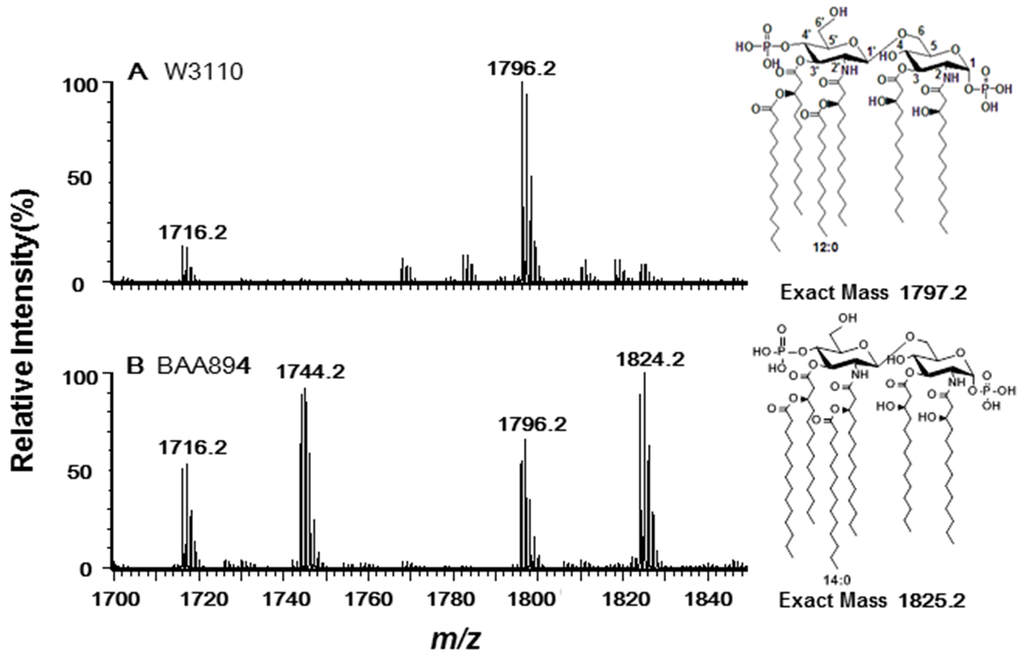
Figure 1.
Negative ion electrospray ionization (ESI)/MS analysis of lipid A purified from E. coli W3110 (A) and C. sakazakii BAA894 (B). The chemical structures of the major lipid A species are shown on the right.
To explore the nature of diversity of the secondary fatty acyl chains of lipid A in C. sakazakii, BLASTp search against the genome of C. sakazakii BAA894 was performed, using E. coli LpxL, LpxP and LpxM as queries. Three late acyltransferases YP_001438378.1, YP_001439016.1, and YP_001437482.1 were revealed and the sequence alignments of all these proteins are shown in Figure 2. YP_001438378.1 is encoded by ESA02293, contains 306 amino acids, and shares 79%, 51% and 31% identity to E. coli LpxL, LpxP and LpxM, respectively. YP_001439016.1 is encoded by ESA02951, contains 307 amino acids, and shares 55%, 71% and 28% identity to E. coli LpxL, LpxP and LpxM, respectively. YP_001437482.1 is encoded by ESA01386, contains 322 amino acids, and shares 30%, 28% and 75% identity to E. coli LpxL, LpxP and LpxM, respectively. Based on the sequence similarity, YP_001438378.1 is proposed as C. sakazakii LpxL, YP_001439016.1 as C. sakazakii LpxP and YP_001437482.1 as C. sakazakii LpxM.
To confirm the function of the three putative acyltransferases, genes ESA02293, ESA02951 and ESA01386 were cloned into pWSK29, and transformed into E. coli mutant MLK1067 and MKV15b. MLK1067 was constructed by inactivating lpxM in E. coli and can only synthesize pentaacylated lipid A [16]; MKV15b is a Lpxl, LpxM, LpxP triple E. coli mutant and could only synthesize tetraacylated lipid A [17].
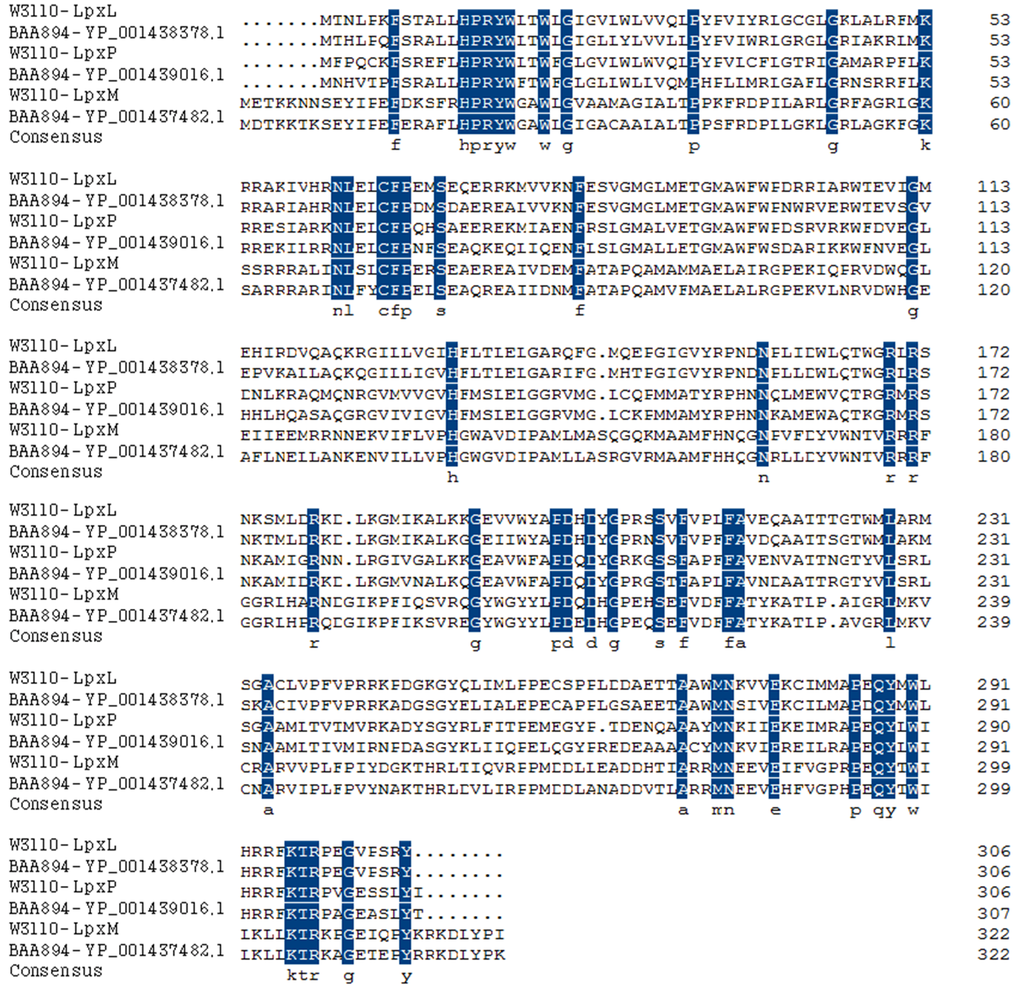
Figure 2.
Sequence alignment of three putative late acyltransferases in C. sakazakii BAA894 with E. coli LpxL, LpxM, and LpxP. The consensus residues are emphasized.
2.2. Acyltransferases Encoded by ESA02293 and ESA02952 Are Homologous to E. coli LpxL and LpxP, Respectively
Lipid A and its derivatives were isolated from MKV15b/PWSK29, MK15b/pWSK29-ESA02293, MK15b/pWSK29-ESA02951, and MK15b/pWSK29-ESA01386, respectively, and analyzed by ESI/MS (Figure 3).
Two major ions at m/z 1403.9 and 1323.9 were observed in the spectrum of lipid A from the control MKV15b/pWSK29 (Figure 3A). The peak at m/z 1403.9 is created by the molecular ion [M − H]− of lipid A containing two phosphate groups and four primary fatty acid chains; the peak at m/z 1323.9 arises by loss of a phosphate from the molecular ion during the extraction procedure. In the spectrum of lipid A sample from MKV15b/pWSK29-ESA02293, two major ions were also observed (Figure 3B). The molecular ion at m/z 1614.1 differs from that of the lipid A from MKV15b/pWSK29 by 210 amu, that is, a C14:0 fatty acid unit. The peak at m/z 1534.1 arises by loss of a phosphate from the molecular ion. The results indicate that the enzyme encoded by ESA02293 transfers a C14:0 secondary acyl chain to the 2′-position of lipid A.
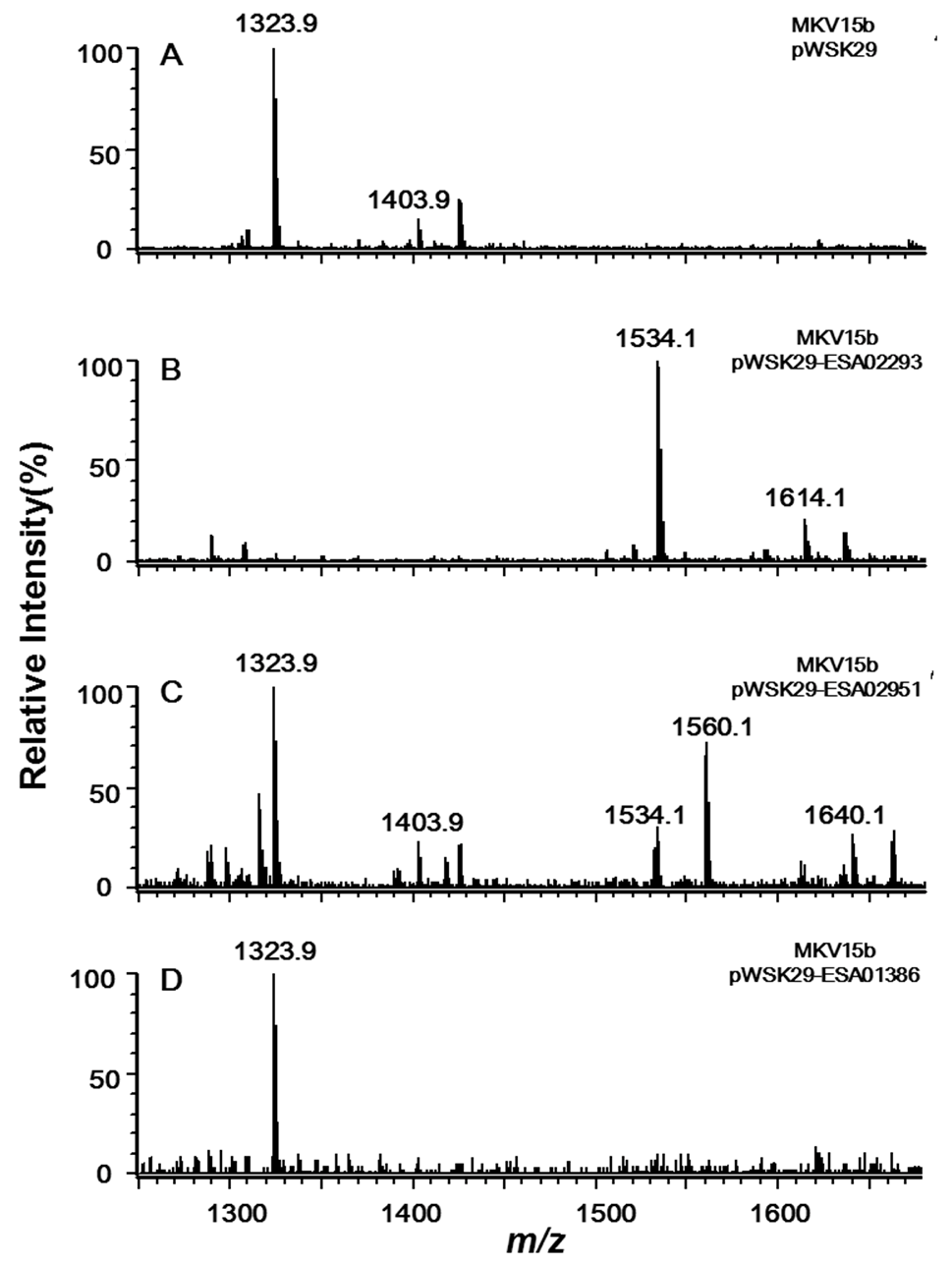
Figure 3.
Negative ion ESI/MS analysis of lipid A isolated from E. coli MKV15b/pWSK29 (A), MKV15b/pWSK29-ESA02293 (B), MKV15b/pWSK29-ESA02951 (C) and MKV15b/pWSK29-ESA01386 (D).
Six peaks were observed in the spectrum of lipid A sample from MKV15b/pWSK29-ESA02951 (Figure 3C). Peaks at m/z 1323.9 and 1403.9 are the same to that found in spectrum of lipid A from both MKV15b/pWSK29. Peaks at m/z 1534.1 and 1614.1 are the same to that found in spectrum of lipid A from MKV15b/pWSK29-ESA02293. This indicates that the enzyme encoded by ESA02951 can transfer a C14:0 secondary acyl chain to the 2′-position of lipid A, but its efficiency is not as high as the enzyme encoded by ESA02293. In addition, there were two new peaks at m/z 1560.1 and 1640.1 in the spectrum of lipid A from MKV15b/pWSK29-ESA02951; the former might arise by loss of a phosphate form the later. The molecular ion at m/z 1640.1 differs from that of the molecular ion of lipid A from MKV15b/pWSK29 by 236 amu moiety, that is, a C16:1 fatty acid unit. The results indicate that the enzyme encoded by ESA02951 could transfer a C14:0 or a C16:1 secondary acyl chain to the 2′-position of lipid A.
The spectrum of lipid A sample from MK15b/pWSK29-ESA01386 only showed a peak at m/z 1323.9 created by the molecular ion losing a phosphate, which is exactly the same as that of the control strain MKV15b/pWSK29 (Figure 3D), suggesting the enzyme encoded by the gene ESA01386 does not function on the substrate lipid A with only four primary fatty acyl chains. Based on the analysis of sequence and function, the acyltransferase encoded by ESA02293 should be the LpxL in C. sakazakii, and the acyltransferase encoded by ESA_02952 should be the LpxP in C. sakazakii.
Because there is a second molecular ion at m/z 1796.2 in the C. sakazakii lipid A mass spectrum (Figure 1B), there might be another acyltransferase in C. sakazakii which transfers a C12:0 secondary acyl chain to lipid A. This acyltransferase may have no sequence similarity to LpxL. This situation was observed for LpxH, a key enzyme in the lipid A biosynthesis of E. coli. A new enzyme LpxI which has no sequence similarity to LpxH but generates the same products by a different route was recently found in Caulobacter crescentus [18]. Unlike E. coli LpxP which transfers a C16:1 acyl chain only at low temperature (12 °C), C. sakazakii LpxP can function at ambient temperature and can transfer either a C14:0 or a C16:1 secondary acyl chains. It would be interesting to study the structural difference of these two LpxP proteins to figure out the machnism for their different substrate specificity and temperature regulation.
2.3. Acyltransferase Encoded by ESA01386 Is the Homologue of E. coli LpxM
Lipid A and its derivatives were isolated from MLK1067/PWSK29, MLK1067/pWSK29-ESA02293, MLK1067/pWSK29-ESA02951, and MLK1067/pWSK29-ESA01386, respectively, and analyzed by ESI/MS (Figure 4).
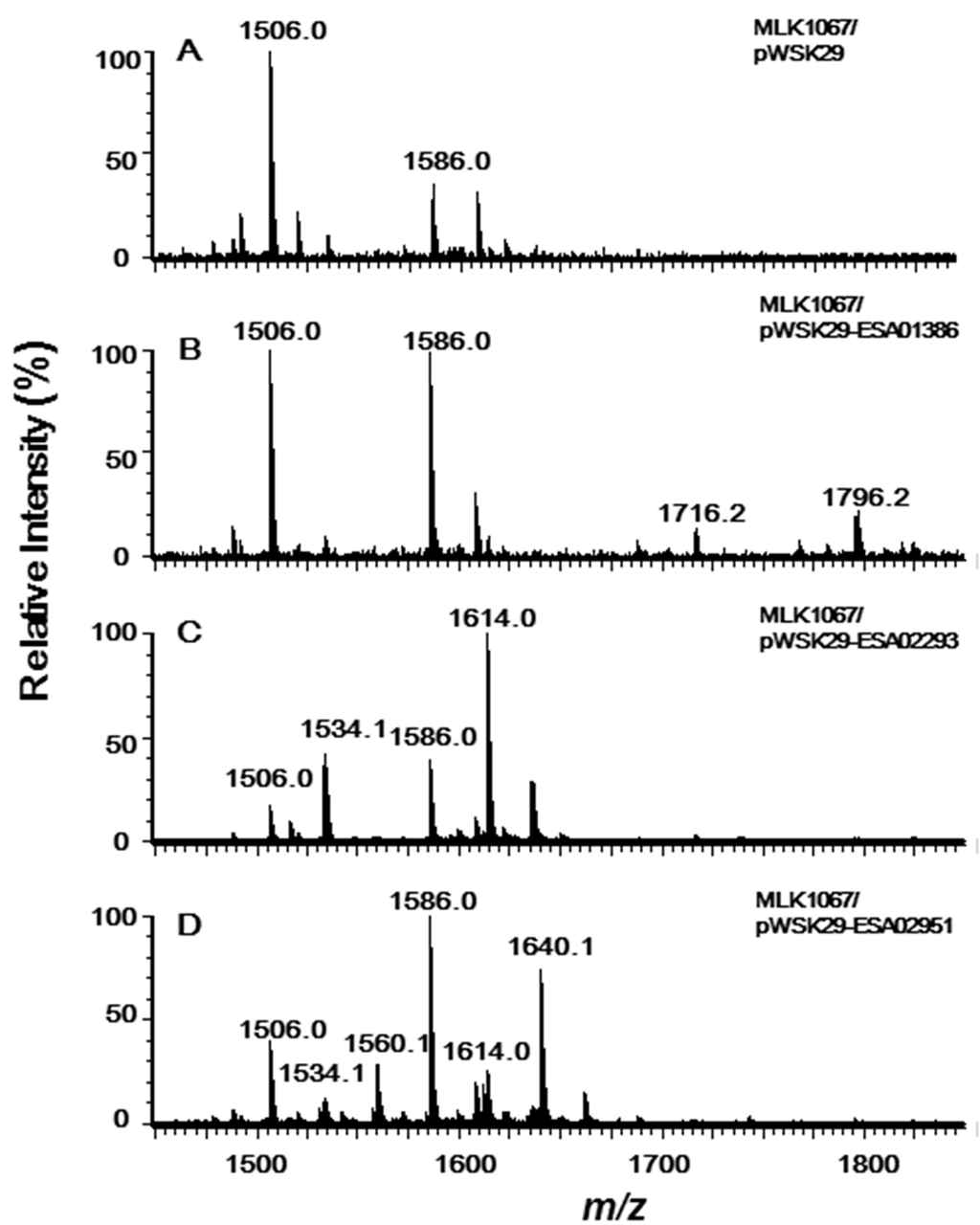
Figure 4.
Negative ion ESI/MS analysis of lipid A isolated from E. coli MLK1067/pWSK29 (A), MLK1067/pWSK29-ESA01386 (B), MLK1067/pWSK29-ESA02293 (C) and MLK1067/pWSK29-ESA02951 (D).
Lipid A isolated from MLK1067/pWSK29 showed two major peaks in the spectrum; one is the molecular ion [M − H]− at m/z 1586.0 [6], and the other at m/z 1506.0 is derived by loss of a phosphate form the molecular ion (Figure 4A). Two additional peaks at m/z 1716.2 and 1796.2 were observed in the spectrum of lipid A samples from MLK1067/pWSK29-ESA01386 (Figure 4B). The peak at m/z 1796.2 should be from the molecular ion [M − H]−, and the peak at m/z 1716.2 is derived from the molecular ion by loss of a phosphate. The m/z value difference between the two molecular ions of lipid A from MLK1067/pWSK29 and MLK1067/pWSK29-ESA01386 is 210 amu, that is, a C14:0 acyl chain. This suggests that the enzyme encoded by ESA01386 could transfer a C14:0 fatty acids to the 3′-position of lipid A, but its efficiency is not as high as E. coli LpxM because the majority of lipid A in the cell are in the pentaacylated form. Considering C. sakazakii LpxL transfers a C14:0 secondary acyl chain to the 2′-position of lipid A (Figure 3B), the pentaacylated lipid A with a C12:0 secondary fatty acyl chain in E. coli MLK1067 might not the ideal substrate for this enzyme. Based on the analysis of sequence and function, the acyltransferase encoded by ESA01386 should be the LpxM in C. sakazakii.
The peaks at m/z 1796.2 and 1716.2 were not observed in the spectra of lipid A samples from MLK1067/pWSK29-ESA02293 and MLK1067/pWSK29-ESA02951, confirming that the enzymes encoded by ESA02293 and ESA02951 cannot add any acyl chains to the 3′-position of lipid A. In addition to the two peaks at m/z 1506.0 and 1586.0 observed in the control, two addition peaks at m/z 1534.1 and 1614.0 were observed in the spectrum of lipid A from MLK1067/pWSK29-ESA02293 (Figure 4C), confirming again that C. sakazakii LpxL prefers to transfer a C14:0 acyl chain to the 2′-position of lipid A.
Six peaks were observed in the spectrum of lipid A sample from MLK1067/pWSK29-ESA02951 (Figure 4D). Peaks at m/z 1506.0 and 1586.0 are the same to that found in spectrum of lipid A from MLK1067/pWSK29. Peaks at m/z 1534.1 and 1614.1 are the same to that found in spectrum of lipid A from MKV15b/pWSK29-ESA02293 or MLK1067/pWSK29-ESA02293. Peaks at m/z 1560.1 and 1640.1 are the same to that found in the spectrum of lipid A from MKV15b/pWSK29-ESA02951 (Figure 3C). These data further confirm that C. sakazakii LpxP can transfer either a C16:1 or a C14:0 secondary acyl chain to the 2′-position of lipid A.
3. Experimental Section
3.1. DNA Preparation and PCR Techniques
Plasmid DNA was prepared by using the EZ-10 spin column plasmid mini-preps kit from Bio Basic Inc. (Markham, Canada). Fifty microliters of PCR reaction mixture was used in this study, which contains 5 μL 10× Extaq buffer, 4 μL dNTP (2.5 mM each), 1 μL plasmid template, 1 μL forward primer (20 μM), 1 μL reverse primer (20 μM), and 0.5 μL Extaq polymerase. The PCR reaction was started at 94 °C for 5 min, followed by 35 cycles of denaturation (30 s at 94 °C), annealing, and extension. PCR products were purified using the TIAN gel purification kit from Tiangen (Beijing, China). Primers synthesis and DNA sequencing were performed by Sangon (Shanghai, China).
3.2. Construction of Plasmids Containing Genes ESA02293, ESA02951 or ESA01386
ESA02293, ESA02951, and ESA01386 genes were obtained by PCR, using the genome of C. sakazakii BAA-894 as a template. All the primers used for the clones were listed in Table 1. Briefly, all the forward primers contained an XbaI site, and all the reverse primers contained a BamHI site. The PCR products were purified, digested with XbaI and BamHI, and ligated into the vector pWSK29 which was similarly digested. The resulting plasmids were designated as pWSK29-ESA02293, pWSK29-ESA02951 and pWSK29-ESA01386, respectively. These plasmids were transformed into E. coli mutants MLK1067 [5] and MKV15b [17], resulting strains MLK1067/pWSK29-ESA02293, MLK1067/pWSK29-ESA02951, MLK1067/pWSK29-ESA01386, MLV15b/pWSK29-ESA02293, MLV15b/pWSK29-ESA02951, and MLV15b/pWSK29-ESA01386. All strains used in this study are listed in Table 1. All C. sakazakii and E. coli stains were grown in LB media at 37 °C. 100 μg/mL ampicillin, 50 μg/mL kanamycin, or 20 μg/mL chloramphenicol were used when necessary.

Table 1.
Stains and primers used in this study.
| Strains or oligonucleotides | Relevant characteristic or sequence | Source or purpose |
|---|---|---|
| Strains | ||
| ATCC BAA894 | Wild-type C. sakazakii | ATCC |
| W3110 | Wide-type E. coli | ATCC |
| MKV15b | W3110 lpxL::Tn10 lpxM::Ωcam lpxP::kan | [17] |
| MKV15b/pWSK29 | MKV15b harboring pWSK29 | This work |
| MKV15b/pWSK29-ESA02293 | MKV15b harboring pWSK29-ESA02293 | This work |
| MKV15b/pWSK29-ESA02951 | MKV15b harboring pWSK29-ESA02951 | This work |
| MKV15b/pWSK29-ESA01386 | MKV15b harboring pWSK29-ESA01386 | This work |
| MLK1067 | W3110 lpxM::Ωcam | [16] |
| MLK1067/pWSK29 | MLK1067 harboring pWSK29 | This work |
| MLK1067/pWSK29-ESA01386 | MLK1067 harboring pWSK29-ESA01386 | This work |
| MLK1067/pWSK29-ESA02293 | MLK1067 harboring pWSK29-ESA02293 | This work |
| MLK1067/pWSK29-ESA02951 | MLK1067 harboring pWSK29-ESA02951 | This work |
| Oligonucleotides | ||
| F-ESA02293 | GCTCTAGAATGACGCATTTACCGCAATT | Forward primer for cloning ESA-02293 |
| R-ESA02293 | CGGGATCCTTAGTAGCGGGACGGCACGC | Reverse primer for cloning ESA-02293 |
| F-ESA02951 | GCTCTAGAATGAATCACGTCACGCCTTTTTC | Forward primer for cloning ESA-02951 |
| R-ESA02951 | CGGGATCCTCAGGTATAGAGCGACGCTTC | Reverse primer for cloning ESA-02951 |
| F-ESA01386 | GCTCTAGAATGGACACAAAAAAAACAAAAAGTG | Forward primer for cloning ESA-01386 |
| R-ESA01386 | CGGGATCCTTATTTCGGATAAAGATCTTTGC | Reverse primer for cloning ESA-01386 |
3.3. Lipid A Isolation from Different E. coli Strains
Lipid A was isolated using the Bligh-Dyer method [19]. Briefly, 200 mL cultures were grown in LB broth at 37 °C to OD600 of 1.0. Cells were harvested by centrifugation at 8000 rpm and washed twice with ddH2O. The cell pellet was resuspended in 76 mL of a single-phase Bligh-Dyer mixture containing chloroform/methanol/H2O (1:2:0.8, v/v/v), incubated with stirring at room temperature for 1 h. The insoluble debris was collected by centrifugation at 2000 rpm for 20 min and washed three times with single-phase Bligh-Dyer mixture. The debris was resuspended in 27 mL of 12.5 mM sodium acetate (pH 4.5), heated at 100 °C for 30 min, and cooled to room temperature. The suspension was mixed with 30 mL chloroform and 30 mL methanol, and the two phases were separated by centrifugation. The lower phase contains lipid A, and was dried under a stream of nitrogen.
3.4. Mass Spectrometry Analysis
All the mass spectra of lipid A samples were acquired on a Waters SYNAPT quadrupole time-of-flight mass spectrometer equipped with an electrospray ionization (ESI) source. Lipid A samples were dissolved in chloroform/methanol (2:1, v/v) and subjected to ESI/MS in the negative ion mode. Data acquisition and analysis were performed using Mass Lynx V4.1 software [20].
4. Conclusions
Unlike E. coli which usually synthesizes only one lipid A species, C. sakazakii BAA894 synthesizes two different species of lipid A, differing on the length of the secondary acyl chain at 2′-position. Here, we identified three late acyltransferases in C. sakazakii BAA894. The enzyme encoded by the gene ESA02293 is homologous to E. coli LpxL, and transfers a C14:0 acyl chain to the 2′-position of lipid A. The enzyme encoded by the gene ESA_02953 is homologous to E. coli LpxP, and transfers either a C16:1 or a C14:0 acyl chains to the 2′-position of lipid A. The enzyme encoded by ESA01386 is homologous to E. coli LpxM, and transfer a C14:0 acyl chain to the lipid A at 3′-position. Since the number and length of secondary acyl chains of lipid A are important for the recognition of TLR4/MD2 to LPS, these late acyltransferases found in this study might be useful to construct various lipid A moieties for studying the immune responses.
Acknowledgments
Funding was provided by grants from National key Basic Research Program of China (973 Program 2012CB725202), the National Natural Science Foundation of China (NSFC31170069), the Specialized Research Fund for the Doctoral Program of Higher Education (SRFDP20100093110005).
- Samples Availability: Available from the authors.
References
- Wang, X.; Quinn, P.J. Lipopolysaccharide: Biosynthetic pathway and structure modification. Prog. Lipid Res. 2010, 49, 97–107. [Google Scholar] [CrossRef]
- Raetz, C.R.; Garrett, T.A.; Reynolds, C.M.; Shaw, W.A.; Moore, J.D.; Smith, D.C.; Ribeiro, A.A.; Murphy, R.C.; Ulevitch, R.J.; Fearns, C.; et al. Kdo2-Lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J. Lipid Res. 2006, 47, 1097–1111. [Google Scholar] [CrossRef]
- Miller, S.I.; Ernst, R.K.; Bader, M.W. LPS, TLR4 and infectious disease diversity. Nat. Rev. Microbiol. 2005, 3, 36–46. [Google Scholar] [CrossRef]
- Triantafilou, M.; Triantafilou, K. The dynamics of LPS recognition: Complex orchestration of multiple receptors. J. Endotoxin Res. 2005, 11, 5–11. [Google Scholar]
- Chen, J.; Tao, G.; Wang, X. Construction of an Escherichia coli mutant producing monophosphoryl lipid A. Biotechnol. Lett. 2011, 33, 1013–1019. [Google Scholar] [CrossRef]
- Zhang, C.; Li, Y.; Tao, G.; Li, Y.; Wang, X. Characterization of lipid A Cronobacter sakazakii. Eur. J. Mass Spectrom. 2010, 16, 531–538. [Google Scholar] [CrossRef]
- Raetz, C.R.; Guan, Z.; Ingram, B.O.; Six, D.A.; Song, F.; Wang, X.; Zhao, J. Discovery of new biosynthetic pathways: The lipid A story. J. Lipid Res. 2009, 50, S103–S108. [Google Scholar]
- Williams, A.H.; Raetz, C.R. Structural basis for the acyl chain selectivity and mechanism of UDP-N-acetylglucosamine acyltransferase. Proc. Natl. Acad. Sci. USA 2007, 104, 13543–13550. [Google Scholar]
- Buetow, L.; Smith, T.K.; Dawson, A.; Fyffe, S.; Hunter, W.N. Structure and reactivity of LpxD, the N-acyltransferase of lipid A biosynthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 4321–4326. [Google Scholar]
- Li, Y.; Powell, D.A.; Shaffer, S.A.; Rasko, D.A.; Pelletier, M.R.; Leszyk, J.D.; Scott, A.J.; Masoudi, A.; Goodlett, D.R.; Wang, X.; et al. LPS remodeling is an evolved survival strategy for bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 8716–8721. [Google Scholar]
- McLendon, M.K.; Schilling, B.; Hunt, J.R.; Apicella, M.A.; Gibson, B.W. Identification of LpxL, a late acyltransferase of Francisella tularensis. Infect. Immun. 2007, 75, 5518–5531. [Google Scholar]
- Clementz, T.; Bednarski, J.J.; Raetz, C.R. Function of the htrB high temperature requirement gene of Escherchia coli in the acylation of lipid A: HtrB catalyzed incorporation of laurate. J. Biol. Chem. 1996, 271, 12095–12102. [Google Scholar]
- Geurtsen, J.; Angevaare, E.; Janssen, M.; Hamstra, H.J.; ten Hove, J.; de Haan, A.; Kuipers, B.; Tommassen, J.; van der Ley, P. A novel secondary acyl chain in the lipopolysaccharide of Bordetella pertussis required for efficient infection of human macrophages. J. Biol. Chem. 2007, 282, 37875–37884. [Google Scholar]
- Carty, S.M.; Sreekumar, K.R.; Raetz, C.R. Effect of cold shock on lipid A biosynthesis in Escherichia coli. Induction At 12 degrees C of an acyltransferase specific for palmitoleoyl-acyl carrier protein. J. Biol. Chem. 1999, 274, 9677–9685. [Google Scholar]
- Vorachek-Warren, M.K.; Carty, S.M.; Lin, S.; Cotter, R.J.; Raetz, C.R. An Escherichia coli mutant lacking the cold shock-induced palmitoleoyltransferase of lipid A biosynthesis: Absence of unsaturated acyl chains and antibiotic hypersensitivity at 12 degrees C. J. Biol. Chem. 2002, 277, 14186–14193. [Google Scholar]
- Brozek, K.A.; Raetz, C.R. Biosynthesis of lipid A in Escherichia coli. Acyl carrier protein-dependent incorporation of laurate and myristate. J. Biol. Chem. 1990, 265, 15410–15417. [Google Scholar]
- Vorachek-Warren, M.K.; Ramirez, S.; Cotter, R.J.; Raetz, C.R. A triple mutant of Escherichia coli lacking secondary acyl chains on lipid A. J. Biol. Chem. 2002, 277, 14194–14205. [Google Scholar]
- Metzger, L.E.; Raetz, C.R. An alternative route for UDP-diacylglucosamine hydrolysis in bacterial lipid A biosynthesis. Biochemistry 2010, 49, 6715–6726. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- MassLynx Software, version 4.1, Waters Corporation: Milford, MA, USA, 2005.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).