Abstract
Genomic mining revealed one major nonribosomal peptide synthetase (NRPS) phylogenetic cluster in 12 marine sponge species, one ascidian, an actinobacterial isolate and seawater. Phylogenetic analysis predicts its taxonomic affiliation to the actinomycetes and hydroxy-phenyl-glycine as a likely substrate. Additionally, a phylogenetically distinct NRPS gene cluster was discovered in the microbial metagenome of the sponge Aplysina aerophoba, which shows highest similarities to NRPS genes that were previously assigned, by ways of single cell genomics, to a Chloroflexi sponge symbiont. Genomic mining studies such as the one presented here for NRPS genes, contribute to on-going efforts to characterize the genomic potential of sponge-associated microbiota for secondary metabolite biosynthesis.
1. Introduction
Sponges (phylum Porifera) are an extraordinarily rich source for bioactive metabolites [1]. Current hypothesis holds that because sponges lack physical defenses, they are exposed to an enormous predator and epibiont pressure, which, in turn, has provoked the evolution of structurally highly diverse, effective and sophisticated chemical defenses. Many sponges also contain massive amounts of microorganisms extracellularly within the mesohyl matrix, which may constitute up to one third of the animal’s biomass [2]. There is increasing evidence that important marine natural product classes, complex polyketides and nonribosomal peptides, are truly synthesized by symbiotic bacteria rather than by the sponge itself [3]. However, since the vast majority of sponge symbionts, much like most environmental bacteria, are still refractory to cultivation, new experimental approaches are needed to provide information about their genomic potential for secondary metabolite biosynthesis. Methods such as metagenomics, and more recently single-cell genomics were developed to access the DNA pool of complex environmental microbial consortia in a cultivation-independent manner [4].
Nonribosomal peptide synthetases (NRPS) are large, multimodular enzymes that are organized in modules containing specific domains that sequentially incorporate amino acid building blocks into a growing peptide chain [5,6]. A typical NRPS module contains an adenylation (A) domain, a peptidyl carrier protein domain and a condensation domain. A thioesterase domain frequently terminates chain elongation. NRPS gene clusters encode for a wide range of nonribosomal peptides, ranging from antibiotics (e.g., penicillin, vancomycin [7]), toxins (e.g., kahalalide F [8]), siderophores (enterobactin, vibriobactin [9]) to anti-inflammatorials and immunosuppressants (e.g., cyclosporin A [10]). These pharmacologically relevant bioactivities have motivated extensive searches for novel NRPS genes in microbial isolates and in environmental samples. For example, degenerate oligonucleotide primers that target the conserved region of the A domain have been used to unravel the diversity of NRPS genes in actinobacterial and fungal endophytes of plants [11,12], in actinobacterial [13,14,15] and fungal [16] isolates of marine sponges, and in free-living freshwater cyanobacteria [17], and marine actinobacteria [18,19]. Mixed PKS-NRPS gene clusters from the marine sponge Discodermia dissoluta were furthermore reported using a metagenomic approach [20] and a bimodular NRPS gene cluster was cloned from a Chloroflexi symbiont of the marine sponge Aplysina aerophoba by phi29-mediated whole genome amplification [21].
The aim of the current study was to explore the presence and diversity of NRPS genes in the microbial metagenomes of marine sponges. PCR screening of diverse marine sponge species and of metagenomic libraries, followed by cloning of a NRPS gene cluster, were employed towards this goal. This study represents a continuation of previous efforts where the diversity, distribution and genomic context of gene clusters relevant for secondary metabolism, such as polyketide synthases [22,23] and halogenases [24], were investigated in sponge associated microbial consortia.
2. Results and Discussion
2.1. NRPS Gene Diversity
We first aimed to investigate the NRPS gene diversity in twelve marine sponge species from the Bahamas and the Mediterranean. For this purpose NRPS A domain gene fragments were amplified as previously described [25] using degenerate primers A3 and A7R. Altogether 62 A domain DNA sequences (ca. 750 bp) were amplified from the metagenomes of all 12 sponge species, from the ascidian Ecteinascidia turbinata and from seawater. No PCR product was obtained from the sediment sample. Additionally, a NRPS A domain from the bacterium Streptomyces sp. Aer003, which had previously been isolated from Mediterranean Aplysina aerophoba (Acc# JN830622) was amplified and sequenced. Sequences from each sponge metagenome that exhibited ≥98% sequence identities were presumed to have been amplified from identical genes, thus taking into account the PCR-induced errors that may arise from using degenerate primers [20]. Using this criterion, a total of 24 sequences were considered different (Acc# JN815085–JN815111). A neighbor-joining tree was constructed including related sequences from BLAST analysis (Figure 1).
The majority of these sequences (22/24) formed one large distinct cluster with 97–99% in-cluster amino acid identity. The cluster also included the NRPS A domain sequences from the isolate Streptomyces sp. Aer003 and from seawater. The closest relatives were all Actinobacteria with the A domain sequences from Streptomyces roseosporus (ZP_04696845) and S. fungicidus (ABD65957) being the closest relatives (64, 65% sequence identity). The substrate for the NRPS adenylation domains appears to be hydoxy-phenyl-glycine (hpg) as predicted by NRPSpredictor2 [26]. Notably, Xestospongia muta clone 8 and Aplysina cauliformis clone 19 fell outside of this large cluster with the A domain sequences of Stenotrophomonas maltophila (YP_002028658) and Pseudomonas putida (YP_001750394) being their closest phylogenetic neighbors, respectively and with valine being the likely substrate. Overall, the NRPS gene diversity discovered in this study was very low. However, a bias cannot be ruled out as primers targeting specifically NRPS systems from actinomycetes were used [25]. If primer sets targeting different groups of microorganisms were employed, a higher diversity of NRPS genes would be expected. The fact that a closely related NRPS sequence clade was found in all sponge samples independent of their geographic location, in Caribbean seawater and in a Mediterranean streptomycete isolate suggests a wide geographic distribution of this NRPS-bearing bacterium.
2.2. NRPS-Containing Metagenomic Cosmid Clone
PCR screening using degenerate primers [25] targeting two independently constructed metagenomic libraries harboring altogether ca. 2.4 Gb Aplysina aerophoba microbial community DNA [22] resulted in the identification of 14 NRPS-positive library pools, that is, at least 14 NRPS-containing cosmid clones. Sequence analysis of the NRPS PCR products from AApAY1 [22] NRPS-positive, metagenomic cosmid clones revealed nearly identical DNA sequences (data not shown). Two overlapping cosmid clones were sequenced and assembled into a 57031 bp long DNA fragment (AANRPS; Acc# HQ456128) with an overall GC content of 67%. Remarkably, the entire DNA fragment did not exhibit significant similarity on the DNA level to any known sequence in the database, except for NRPS identified previously in the Chloroflexi symbiont of A. aerophoba [21]. Phylogenetic analysis of the translated NRPS A domains confirmed the NRPS gene of a Chloroflexi symbiont of A. aerophoba (“uncultured sponge symbiont cosmid clone ln22” (ACX49739)) as the nearest phylogenetic relative (Figure 2) [21]. Those two NRPS genes share high protein identity (82%) and high similarity (87%) on the DNA level. Beside the NRPS gene and the efflux protein (lubB), there are however no further similarities in the gene neighborhood between the two cosmid clones.
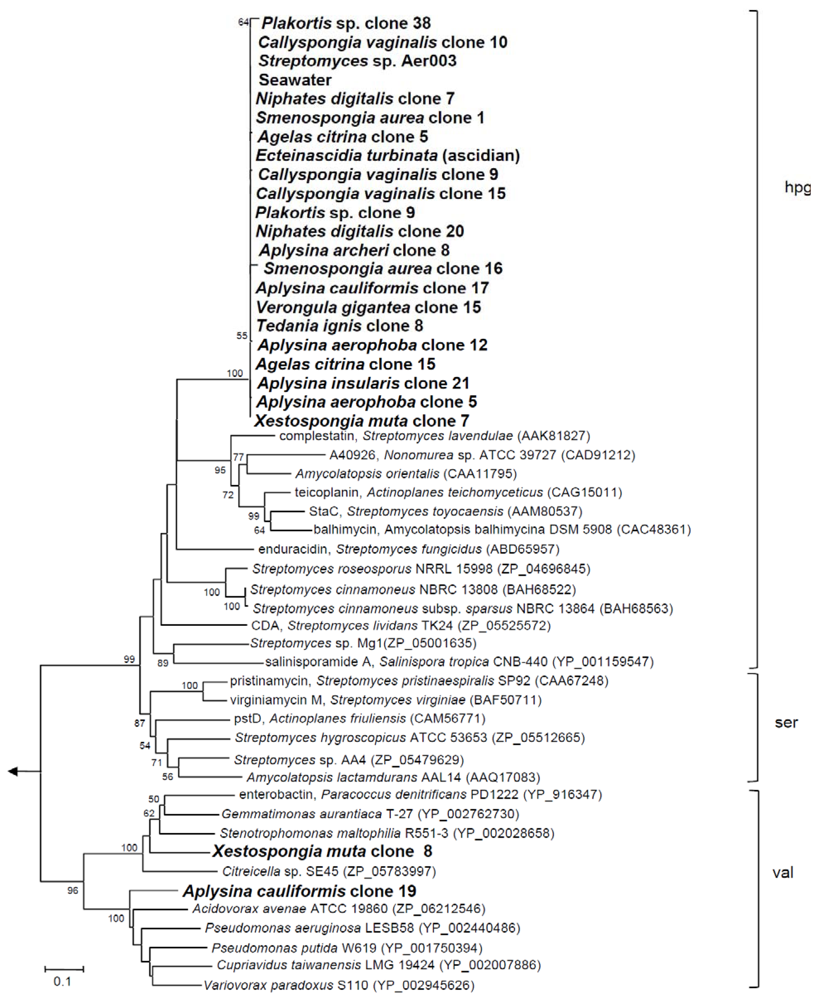
Figure 1.
Neighbor-joining tree of translated nonribosomal peptide synthetase (NRPS) A domains. Bootstrap values greater than 50% are at the nodes. The arrow points to the archaeal outgroup, Methanothermobacter thermoautotrophicus (NP_275799). The scale bar indicates 0.1 substitutions per nucleotide position. Substrate specificities correspond to 4-hydroxy-phenyl-glycine (hpg), serine (ser), and valine (val). Phylogenetic trees were constructed using MEGA version 4 with Poisson correction model for amino acids, complete deletion of gaps and bootstrap consisting of 1000 replications [27].
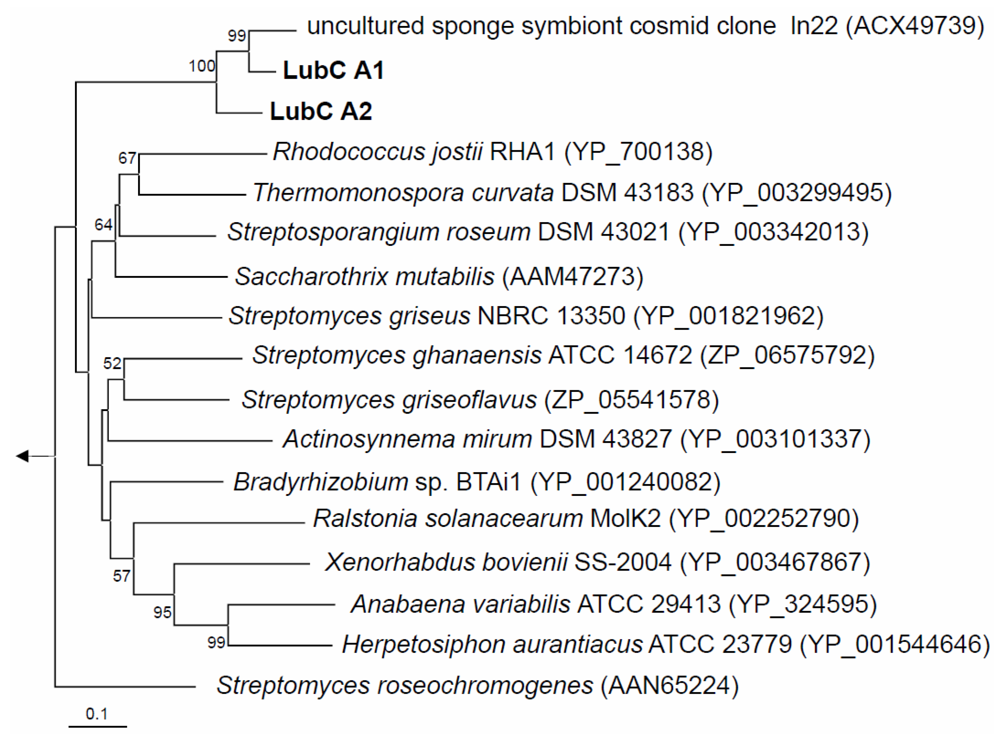
Figure 2.
Neighbor-joining tree of translated NRPS A domains. Bootstrap values greater than 50% are indicated at the nodes. LubC A1 and A2 are the two adenylation domains from the NRPS structural gene lubC, which contains the two modules each bearing a single adenylation domain (see Figure 3). The arrow points to the archaeal outgroup, Methanothermobacter thermoautotrophicus (NP_275799). The scale bar indicates 0.1 substitutions per amino acid position.
The genomic organization of the NRPS-containing cosmid clone is shown in Figure 3. At least 26 putative ORFs were identified (Table 1). Of these, four ORFs are proposed to represent the gene cluster coding for NRPS related proteins, which were termed lubA, B, C, D. The proposed gene cluster consists of 15752 bp and has a G + C content of 68%. The gene lubA encodes for a putative transcriptional regulator of NRPS expression, and is located upstream of the putative efflux protein-encoding gene lubB. The NRPS structural gene lubC contains two complete NRPS modules and is therefore predicted to encode biosynthesis of the dipeptide [28]. Both A1 and A2 adenylation domains probably use aromatic amino acids, such as phenylalanine and tyrosine, as substrates (NRPSpredictor2; [26]). Furthermore, three TonB-dependent receptors and a phosphopantetheinyl transferase (lubD) are contained on the metagenomic cosmid fragment (Table 1). TonB-dependent transporters are frequently involved in iron uptake via siderophores, as well as other substrates including heme, vitamin B12, proteins or polysaccharides [29]. The phosphopantetheinyl transferase enzyme (PPT) activates a carrier protein by the transfer of a phosphopantetheinyl moiety to a serine residue.
The postulated chemical product of this novel NRPS gene cluster remains elusive, owing to a lack of related NRPS genes in the databases and the lack of robust prediction tools. However, it has become clear that NRPS gene clusters are widespread in actinomycete strain collections with recoveries of NRPS genes from more than half of the isolates screened [18,19] as well as being abundant in microbial genomes/metagenomes. With ever cheaper sequencing technologies and improved bioinformatic prediction tools, genomic mining approaches will undoubtedly be instrumental for the identification of sources suitable for natural product discovery.
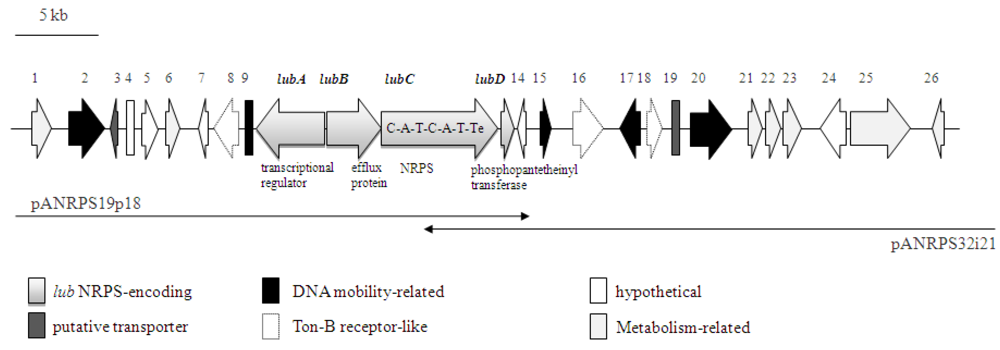
Figure 3.
Genetic organization of the NRPS containing metagenomic cosmid clone from Aplysina aerophoba (AANRPS; Acc# HQ456128). Overlapping cosmids pANRPS19p18 and pANRPS32i21 and ORF numbers are indicated. Putative functions of lub genes-encoded NRPS proteins, as well as domain organization of lubC are depicted.

Table 1.
Putative genes identified on the genomic fragment AANRPS (pANRPS19p18 and pANRPS32i21) from Aplysina aerophoba.
| CDs | Position (nd) | Putative Function | Most Similar Homolog, (Acc#), origin | Identity/Similarity (%) | No. of Amino Acids | ||||
|---|---|---|---|---|---|---|---|---|---|
| ORF1 | 3626-4807 | Serine-threonine phosphatase | PrpA (NP_487771), Nostoc sp. PCC 7120 | 53/69 | 399 | ||||
| ORF2 | 5909-8170 | Helicase RecD/TraA | BAL199_00820, (ZP_02192076), Alpha proteobacterium BAL199 | 72/82 | 753 | ||||
| ORF3 | 8441-8953 | ABC transporter | Cagg_3718, (YP_002464990), Chloroflexus aggregans DSM 9485 | 42/51 | 170 | ||||
| ORF4 | 9894-10364 | Hypothetical protein | MC7420_1635, (ZP_05030609), Microcoleus chthonoplastes PCC 7420 | 40/51 | 156 | ||||
| ORF5 | 10853-11857 | Hypothetical protein | AM1_4437, (YP_001518731), Acaryochloris marina MBIC11017 | 34/52 | 334 | ||||
| ORF6 | 11903-12802 | Hydrolase | VEx25_1601, (ZP_04922735), Vibrio sp. Ex25 | 29/41 | 299 | ||||
| ORF7 | 13907-14512 | Hypothetical protein | BACCOPRO_03255, (ZP_03644864), Bacteroides coprophilus DSM 18228 | 26/40 | 201 | ||||
| ORF8 | 14867-16432 | TonB-dependent receptor | MXAN_6044, (YP_634179), Myxococcus xanthus DK 1622 | 35/54 | 521 | ||||
| ORF9 | 16943-17452 | Exonuclease | RLO149_22990, (ZP_02142576), Roseobacter litoralis Och 149 | 39/51 | 169 | ||||
| lubA | 17745-22022 | LuxR transcriptional regulator | HNE_2502, (YP_761196), Hyphomonas neptunium ATCC 15444 | 26/44 | 1429 | ||||
| lubB | 21850-25236 | Resistance protein | Sputw3181_3288, (YP_964656), Shewanella sp. W3-18-1 | 43/63 | 1128 | ||||
| lubC | 25260-32465 | NRPS (C-A-T-C-A-T-Te) | Siderophore, (ACX49739), uncultured marine bacterium 1n22 | 81/87 | 2401 | ||||
| lubD | 32655-33497 | Phosphopantetheinyl transferase | Mnod_1716, (YP_002497009), Methylobacterium nodulans ORS 2060 | 36/46 | 280 | ||||
| ORF14 | 33681-34217 | Hypothetical protein | MldDRAFT_3697, (ZP_01290808), Delta proteobacterium MLMS-1 | 63/77 | 178 | ||||
| ORF15 | 35067-35822 | Transposase | EbA6749, (YP_160886), Aromatoleum aromaticum EbN1 | 58/74 | 251 | ||||
| ORF16 | 37148-38974 | TonB-dependent receptor | Sama_2896, (YP_928768), Shewanella amazonensis SB2B | 65/82 | 608 | ||||
| ORF17 | 40041-41315 | Transposase | BAL199_06759, (ZP_02191799), Alpha proteobacterium BAL199 | 45/56 | 424 | ||||
| ORF18 | 41701-42630 | TonB-dependent receptor | GPB2148_3348, (ZP_05093557), marine Gamma proteobacterium HTCC2148 | 44/59 | 309 | ||||
| ORF19 | 43924-44364 | Membrane transport protein | Ykris0001_15620, (ZP_04625017), Yersinia kristensenii ATCC 33638 | 43/56 | 146 | ||||
| ORF20 | 44366-46975 | DNA invertase | NB231_12409, (ZP_01126794), Nitrococcus mobilis Nb-231 | 66/78 | 869 | ||||
| ORF21 | 47982-48890 | Methyltransferase | MaviaA2_010100001311, (ZP_05214826), Mycobacterium avium ATCC 25291 | 40/49 | 302 | ||||
| ORF22 | 49061-50023 | NADH-quinone oxidoreductase | Psta_3148, (YP_003371672), Pirellula staleyi DSM 6068 | 25/40 | 320 | ||||
| ORF23 | 50133-51254 | 2,3-Dihydroxybenzoic acid decarboxylase | PJE062_2683, (ZP_05084178), Pseudovibrio sp. JE062 | 65/75 | 373 | ||||
| ORF24 | 52396-54060 | Hypothetical protein | ZP_05710821, (ZP_05710821), Desulfurivibrio alkaliphilus AHT2 | 49/68 | 554 | ||||
| ORF25 | 54272-58006 | Cyclopropane-fatty-acyl-phospholipid synthase | ADG881_908, (ZP_05041385), Alcanivorax sp. DG881 | 51/66 | 1244 | ||||
| ORF26 | 59373-60101 | Nucleoside 2-deoxyribosyltransferase | P9211_14861, (YP_001551371), Prochlorococcus marinus str. MIT 9211 | 63/76 | 242 | ||||
3. Experimental Section
3.1. Sponge Collection
Marine sponges were collected by SCUBA diving at a depth of 5 to 15 m: Aplysina aerophoba offshore from Banyuls sur mer, France (GPS: 42°29′N, 03°08′E); Agelas citrina, Aplysina archeri, Aplysina cauliformis, Aplysina insularis, Callyspongia vaginalis, Niphates digitalis, Plakortis sp., Smenospongia aurea, Tedania ignis, Verongula gigantea, Xestospongia muta offshore from Patch Reef, Bahamas (GPS: 24°14′N, 74°32′W). Additionally, the ascidian Ecteinascidia turbinata, sediment and seawater samples were collected from the sampling site at Patch Reef, Bahamas. Individual specimens were placed separately in plastic bags and brought to the surface. The sponge and ascidian tissues were cut into pieces and stored at −80 °C until use.
3.2. Cultivation and Identification of Sponge-Associated Bacteria
Strain Streptomyces sp. Aer003 was cultivated from the sponge Aplysina aerophoba using M1 [30] culture medium and identified by 16S rRNA gene sequencing as described previously by Hentschel et al. [31].
3.3. DNA Extraction, PCR Amplification and Sequencing of A Domains of NRPS Genes
Genomic DNA was isolated from freshly collected sponges, ascidian and seawater following the method as described previously by Fieseler et al. [22] using the FAST DNA Spin kit for Soil (Q-Biogene). Amplification of the A domains of NRPS gene fragments was performed as described previously [25] using degenerate primers A7R (5′-SASGTCVCCSGTSCGGTAS-3′) and A3 (5′-GCSTACSYSATSTACACSTCSGG-3′). PCR amplification products of ca. 750 bp in size were cloned into a pGEM-Teasy vector (Promega) and transformed into electrocompetent Escherichia coli XL1-Blue cells. Plasmid minipreps by alkaline lysis procedures and sequencing of the inserts were performed as described previously [22]. The same protocol was followed for the strain Streptomyces sp. Aer003.
3.4. Metagenomic Library Construction and Screening for NRPS-Encoding Clones
Two metagenomic libraries constructed from microbial cells of the marine sponge Aplysina aerophoba [22] using an E. coli-Streptomyces shuttle cosmid vector, pAY1, [32] were used for NRPS screening. The libraries represented a total of ca. 2.4 Gb of sponge-associated microbial DNA. Library pools were screened by PCR following the method of Piel et al. [33] using the degenerate primers targeting the A domains of NRPS genes (A7R and A3; see sequences above). Two PCR-positive overlapping cosmid clones were sequenced (pANRPS19p18 and pANRPS32i21).
3.5. Sequence Analysis
Sequencing analysis was performed by Agowa/LGC Genomics, Berlin, Germany. Sequence data were assembled and annotated using the Vector NTI software (Invitrogen) and analyzed using EMBOSS-Transeq and BLAST algorithms [34].
4. Conclusions
Genomic mining revealed the wide distribution of a single NRPS module that is phylogenetically related to actinomycetes and for which hydroxy-phenyl-glycine is the predicted substrate. This NRPS module has been identified in 12 marine sponge species from disparate geographic locations, in one ascidian, an actinobacterial isolate and in seawater. Two additional NRPS gene sequences with valine as predicted substrate were also identified. Metagenomic approaches furthermore revealed a phylogenetically different NRPS gene cluster that has previously been appointed to a Chloroflexi sponge symbiont. The chemical nature and putative bioactivity of the postulated NRPS as well as its possible role in the symbiosis context remain to be explored in future studies. The implementation of metagenomic approaches, such as presented here, are beginning to shed glimpses of light on the secondary metabolite biosynthesis gene repertoire of sponge symbionts, which are still inaccessible by conventional cultivation techniques.
Acknowledgements
We thank J. R. Pawlik (University of North Carolina, Wilmington, NC, USA) for excellent cruise organization and S. Zea (Universidad Nacional de Colombia, INVEMAR, Bogotá D.C, Colombia) for sponge identification. This research was supported by the Deutsche Forschungsgemeinschaft-SFB630 (grant TP A5) to U.H.
References
- Blunt, J.W.; Copp, B.R.; Munro, M.H.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2010, 27, 165–237. [Google Scholar] [CrossRef]
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef]
- Piel, J. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 2009, 26, 338–362. [Google Scholar] [CrossRef]
- Piel, J. Approaches to capturing and designing biologically active small molecules produced by uncultured microbes. Ann. Rev. Microbiol. 2011, 65, 431–453. [Google Scholar] [CrossRef]
- Mootz, H.D.; Schwarzer, D.; Marahiel, M.A. Ways of assembling complex natural products on modular nonribosomal peptide synthetases. ChemBioChem 2001, 3, 490–504. [Google Scholar]
- Schwarzer, D.; Finking, R.; Marahiel, M.A. Nonribosomal peptides: From genes to products. Nat. Prod. Rep. 2003, 20, 275–287. [Google Scholar] [CrossRef]
- Cane, D.E.; Walsh, C.T.; Khosla, C. Harnessing the biosynthetic code: Combinations, permutations, and mutations. Science 1998, 282, 63–68. [Google Scholar] [CrossRef]
- Mansson, M.; Gram, L.; Larsen, T.O. Production of bioactive secondary metabolites by marine vibrionaceae. Mar. Drugs 2011, 9, 1440–1468. [Google Scholar] [CrossRef]
- Crosa, J.H.; Walsh, C.T. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 2002, 66, 223–249. [Google Scholar] [CrossRef]
- Cane, D.E.; Walsh, C.T. The parallel and convergent universes of polyketide synthases and nonribosomal peptide synthetases. Chem. Biol. 1999, 6, 319–325. [Google Scholar] [CrossRef]
- Janso, J.E.; Carter, G.T. Biosynthetic potential of phylogenetically unique endophytic actinomycetes from tropical plants. Appl. Environ. Microbiol. 2010, 76, 4377–4386. [Google Scholar] [CrossRef]
- Johnson, R.; Voisey, C.; Johnson, L.; Pratt, J.; Fleetwood, D.; Khan, A.; Bryan, G. Distribution of NRPS gene families within the Neotyphodium/Epichloe complex. Fungal Genet. Biol. 2007, 44, 1180–1190. [Google Scholar] [CrossRef]
- Schneemann, I.; Nagel, K.; Kajahn, I.; Labes, A.; Wiese, J.; Imhoff, J.F. Comprehensive investigation of marine Actinobacteria associated with the sponge Halichondria panicea. Appl. Environ. Microbiol. 2010, 76, 3702–3714. [Google Scholar] [CrossRef]
- Jiang, S.; Sun, W.; Chen, M.; Dai, S.; Zhang, L.; Liu, Y.; Lee, K.J.; Li, X. Diversity of culturable actinobacteria isolated from marine sponge Haliclona sp. Antonie Van Leeuwenhoek 2007, 92, 405–416. [Google Scholar] [CrossRef]
- Zhang, W.; Li, Z.; Miao, X.; Zhang, F. The screening of antimicrobial bacteria with diverse novel nonribosomal peptide synthetase (NRPS) genes from South China Sea sponges. Mar. Biotechnol. 2009, 11, 346–355. [Google Scholar] [CrossRef]
- Zhou, K.; Zhang, X.; Zhang, F.; Li, Z. Phylogenetically diverse cultivable fungal community and polyketide synthase (PKS), non-ribosomal peptide synthase (NRPS) genes associated with the South China Sea sponges. Microb. Ecol. 2011, 62, 644–654. [Google Scholar] [CrossRef]
- Ehrenreich, I.M.; Waterbury, J.B.; Webb, E.A. Distribution and diversity of natural product genes in marine and freshwater cyanobacterial cultures and genomes. Appl. Environ. Microbiol. 2005, 71, 7401–7413. [Google Scholar] [CrossRef]
- Gontang, E.A.; Gaudêncio, S.P.; Fenical, W.; Jensen, P.R. Sequence-based analysis of secondary-metabolite biosynthesis in marine actinobacteria. Appl. Environ. Microbiol. 2010, 76, 2487–2499. [Google Scholar] [CrossRef]
- Hodges, T.W.; Slattery, M.; Olson, J.B. Unique actinomycetes from marine caves and coral reef sediments provide novel PKS and NRPS biosynthetic gene clusters. Mar. Biotechnol. 2012, 14, 270–280. [Google Scholar] [CrossRef]
- Schirmer, A.; Gadkari, R.; Reeves, C.D.; Ibrahim, F.; DeLong, E.F.; Hutchinson, C.R. Metagenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissoluta. Appl. Environ. Microbiol. 2005, 71, 4840–4849. [Google Scholar]
- Siegl, A.M.; Hentschel, U. PKS and NRPS gene clusters from microbial symbiont cells of marine sponges by whole genome amplification. Environ. Microbiol. Rep. 2010, 2, 507–513. [Google Scholar]
- Fieseler, L.; Hentschel, U.; Grozdanov, L.; Schirmer, A.; Wen, G.; Platzer, M.; Hrvatin, S.; Butzke, D.; Zimmermann, K.; Piel, J. Widespread occurrence and genomic context of unusually small polyketide synthase genes in microbial consortia associated with marine sponges. Appl. Environ. Microbiol. 2007, 73, 2144–2155. [Google Scholar]
- Hochmuth, T.; Niederkrüger, H.; Gernert, C.; Siegl, A.; Taudien, S.; Platzer, M.; Crews, P.; Hentschel, U.; Piel, J. Linking chemical and microbial diversity in marine sponges: Possible role for Poribacteria as producers of methyl-branched fatty acids. ChemBioChem. 2010, 11, 2572–2578. [Google Scholar] [CrossRef]
- Bayer, K.; Scheuermayer, M.; Fieseler, L.; Hentschel, U. Genomic mining for novel FADH2-dependent halogenases in marine sponge-associated microbial consortia. Mar. Biotechnol. 2012. [Google Scholar]
- Ayuso-Sacido, A.; Genilloud, O. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: Detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb. Ecol. 2005, 49, 10–24. [Google Scholar] [CrossRef]
- Ansari, M.Z.; Yadav, G.; Gokhale, R.S.; Mohanty, D. NRPS-PKS: A knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 2004, 1, 405–413. [Google Scholar]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Röttig, M.; Medema, M.H.; Blin, K.; Weber, T.; Rausch, C.; Kohlbacher, O. NRPSpredictor2-a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011, 39, 362–367. [Google Scholar]
- Hopkinson, B.M.; Barbeau, K.A. Iron transporters in marine prokaryotic genomes and metagenomes. Environ. Microbiol. 2012, 14, 114–128. [Google Scholar] [CrossRef]
- Mincer, T.J.; Jensen, P.R.; Kauffman, C.A.; Fenical, W. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl. Environ. Microbiol. 2002, 68, 5005–5011. [Google Scholar]
- Hentschel, U.; Schmid, M.; Wagner, M.; Fieseler, L.; Gernert, C.; Hacker, J. Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the Mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. FEMS Microbiol. Ecol. 2001, 35, 305–312. [Google Scholar] [CrossRef]
- Li, A.; Piel, J. A gene cluster from a marine Streptomyces encoding the biosynthesis of the aromatic spiroketal polyketide griseorhodin A. Chem. Biol. 2002, 9, 1017–1026. [Google Scholar] [CrossRef]
- Piel, J.; Hui, D.; Wen, G.; Butzke, D.; Platzer, M.; Fusetani, N.; Matsunaga, S. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc. Natl. Acad. Sci. USA 2004, 101, 16222–16227. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids. Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Samples Availability: Available from the authors.
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).