Abstract
Background and Objectives: Alzheimer’s disease (AD) is the most common progressive neurodegenerative disorder, characterized by the accumulation of amyloid-beta (Aβ) in the brain. A recent study reported that the interferon-induced transmembrane protein 3 (IFITM3) protein plays a pivotal role in Aβ processing by the γ-secretase complex. Since several single nucleotide polymorphisms (SNPs) of the IFITM3 gene are related to the function and expression levels of the IFITM3 gene, the relationship between genetic polymorphisms in the IFITM3 gene and susceptibility to AD needs to be investigated. Materials and Methods: We investigated the genotype and allele frequencies of IFITM3 polymorphisms in 177 AD patients and 233 matched healthy controls by amplicon sequencing. In addition, we compared the genotype, allele and haplotype frequencies between AD patients and matched controls and performed an association analysis. Results: There were no significant differences in the genotype, allele or haplotype frequency distributions of the IFITM3 polymorphisms between AD patients and matched controls. Conclusions: To the best of our knowledge, this is the first case-control association study of the IFITM3 gene in AD.
1. Introduction
Alzheimer’s disease (AD) is the most common progressive neurodegenerative disease among the several types of dementia [1,2,3,4]. AD is characterized by the accumulation of amyloid beta-peptide (Aβ) derived from amyloid precursor protein (APP) in brain lesions. Aβ is produced by cleavage of the APP protein by the β-secretase encoded by the beta-site amyloid precursor protein cleaving enzyme 1 (BASE1) gene and γ-secretase, a complex that is comprised of a total of 4 high molecular weight subunits, including presenilin (PS), presenilin enhancer 2 (PEN-2), nicastrin and anterior pharynx defective 1 (APH-1) [5,6]. Several genetic variations of these genes affect the production of Aβ via the processing of APP protein. Thus, previous studies have reported that genetic variations in the APP, presenilin-1 (PSEN1) and presenilin-2 (PSEN2) genes are related to susceptibility to AD. In the APP gene, the genotype and allele frequencies of the -877T > C and -955A > G single nucleotide polymorphisms (SNPs) located in the promoter region were significantly associated with susceptibility to AD via transcription efficiency [7]. In addition, Q665D, K670M, N671L A692G, E693G, I716V and V717I germline mutations of the APP gene affect the onset of AD [8,9,10]. In the PSEN1 and PSEN2 genes, over 30 genetic variants are associated with elevated levels of Aβ40 and Aβ42 [8,9,10,11,12,13]. Based on this knowledge, several drugs targeting the APP and γ-secretase proteins have been introduced. However, there was no clear effect for any of the drugs tried [1,4]. Thus, it is necessary to discover specific targets to develop precise therapeutic agents for AD.
Interferon-induced transmembrane protein 3 (IFITM3) is a member of the innate immune system that is expressed primarily in the late endosome or lysosome and limits the entry of enveloped viruses [14,15,16,17,18,19,20]. In addition, a recent study have reported that upregulation of the IFITM3 protein affects γ-secretase, leading to increased Aβ production, and plays a pivotal role in the onset of AD [21]. In previous studies, the rs12252, rs34481144 and rs6598045 SNPs of the IFITM3 gene were found to be involved in the gene expression level and function of the IFITM3 protein. The rs12252 C allele is related to cleavage of the 21 amino acids at the N-terminus of the IFITM3 protein and produces an isoform of the IFITM3 protein. The truncated IFITM3 protein induced by the rs12252 C allele has been shown to decrease the inhibition capacity of viral replication [16,22]. The A allele of the rs34481144 SNP inhibits the expression of the IFITM3 gene by increasing the CCCTC-binding factor (CTCF) binding ability at the promoter site of the IFITM3 gene [17]. The recent study has reported that rs12252 and 34481144 SNPs showed no correlation with the susceptibility of AD and the expression level of the IFITM3 protein. However, this study has performed association analysis with a relatively small sample size (control, n = 9; AD, n = 18) [21], and has not investigated rs6598045 SNP that modulates promoter activity [18]. Since several SNPs of the IFITM3 gene are related to the function and expression level of the IFITM3 gene and the IFITM3 protein plays a pivotal role in the processing of Aβ, further investigation of the relationship between genetic polymorphisms in IFITM3 and susceptibility to AD is necessary.
In the present study, we investigated genotype, allele and haplotype frequencies of the IFITM3 polymorphisms of 177 AD patients and 233 matched controls by amplicon sequencing. To find an association between IFITM3 polymorphisms and susceptibility to AD, we compared genotype, allele, and haplotype frequencies of IFITM3 polymorphisms between the two groups and performed association analysis.
2. Materials and Methods
2.1. Subjects
A total of blood samples from 177 AD patients were provided by the Pusan National University Hospital Biobank, a member of the Korea Biobank Network. A total of blood samples from 233 healthy Korean subjects were obtained from the Korea Biobank Network at the Centers for Disease Control and Prevention. The sample size used in this study may be enough to identify rare polymorphisms, including below 1% genotype frequency [22]. In addition, the sample size of healthy Korean subjects used in this study can also represent the total population of Korea with a 95% confidence level and a confidence interval of 6.42. The sample size of AD patients used in this study can also represent the total Korean AD patients with a 95% confidence level and a confidence interval of 7.36. All experimental procedures were approved according to the guidelines of the institutional review board of Jeonbuk National University and the 1964 Helsinki declaration and its later amendments or comparable ethical standards (approval number: JBNU 2019-03-009). Information for all samples was anonymized prior to investigation.
2.2. Genomic DNA Extraction
Genomic DNA was extracted from 200 µL of blood using the BeadTM Genomic DNA Prep Kit (Biofact, Daejeon, Korea) according to the manufacturer’s instructions.
2.3. Polymerase Chain Reaction (PCR)
The human IFITM3 gene was amplified using PCR with the gene-specific forward primer: 5′-CAGGGGAAGTCTCCAGGACC-3′ and reverse primer: 5′-CCAAGCCACACACACACACA-3′. The PCR mixture consisted of 1 µL of genomic DNA, 10 pmol of each primer, 2.5 µL of 10× Taq DNA polymerase buffer, 0.5 µL of a 0.2 µM dNTP mixture, 5 µL of 5× Band Helper, and 0.25 µL Taq DNA polymerase. The PCR conditions followed the manufacturer’s recommendations. The annealing temperature of the IFITM3 gene primers was 68 °C.
2.4. Amplicon Sequencing and Genotyping
All PCR products were purified using a FavorPrep GEL/PCR purification Mini Kit (FAVORGEN, Pingtung City, Taiwan) and sequenced on an ABI 3730 sequencer (ABI, Foster City, CA, USA). Sequencing results were visualized using Finch TV software (Geospiza Inc., Seattle, WA, USA), and genotyping was performed.
2.5. Statistical Analysis
The genotype and allele frequencies of the IFITM3 gene were compared between the AD patients and healthy controls by the chi-square (χ2) test using SAS 9.4 software. Analysis of the Hardy-Weinberg equilibrium (HWE) test and haplotypes were performed using Haploview Version 4.2 (Broad Institute, Cambridge, MA, USA).
3. Results
3.1. Subject Description
A total of 177 Korean AD patients and 233 healthy Koreans without dementia were included in the association analysis. Detailed information on the study population is given in Table 1. The 177 AD patients included 128 females and 49 males. The 233 healthy individuals included 103 females and 130 males. The mean age of AD patients at diagnosis was 73.34 ± 7 years, and the mean age of healthy individuals at sample collection was 63.75 ± 9.74 years.

Table 1.
Detailed information on the study population.
3.2. Evaluation of the Association of the Susceptibility of AD with the Genotype, Allele and Haplotype Frequencies of the IFITM3 Gene in the Korean Population
To investigate genotype and allele frequencies of the IFITM3 gene, we performed direct sequencing in 233 healthy individuals and 177 AD patients and carried out genotyping and HWE analyses. Detailed information on the genotyping results and HWE values in the Korean population is described in Table 2.

Table 2.
Comparison of genotype and allele frequencies between healthy controls and Alzheimer’s disease (AD) patients in a Korean population.
The genotype frequencies of 10 polymorphisms of the IFITM3 gene were in HWE in the healthy controls. To estimate the association between susceptibility to AD and polymorphisms of the IFITM3 gene, we performed case-control association analysis. There were no significant differences (p > 0.05) in the genotype or allele distributions of the IFITM3 polymorphisms between healthy individuals and AD patients (Figure 1, Table 2).
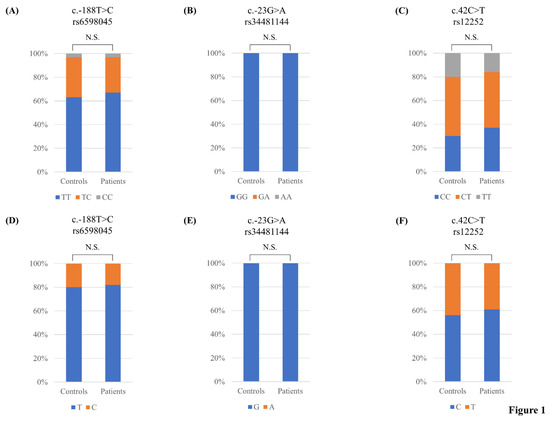
Figure 1.
Comparisons of the genotype and allele frequencies of rs6598045, rs34481144 and rs12252 single nucleotide polymorphisms (SNPs) of the interferon-induced transmembrane protein 3 (IFITM3) gene in AD patients and matched healthy controls: (A) Comparison of the genotype frequency of the rs6598045 SNP of the IFITM3 gene between these two groups; (B) Comparison of the genotype frequency of the rs34481144 SNP of the IFITM3 gene between these two groups; (C) Comparison of the genotype frequency of the rs12252 SNP of the IFITM3 gene in these two groups; (D) Comparison of the allele frequency of the rs6598045 SNP of the IFITM3 gene between these two groups; (E) Comparison of the allele frequency of the rs34481144 SNP of the IFITM3 gene between these two groups; (F) Comparison of the allele frequency of the IFITM3 rs12252 SNP between these two groups. N.S.: not significant.
Next, we performed haplotype analysis of 10 polymorphisms within the IFITM3 gene, namely, c.-223C > G, c.-204G > T, c.-188T > C, c.-181T > C, c.-178A > C, c.-175T > C, c.-128C > T, c.-13_-4DelTTCGCTGGAC, c.42C > T and c.165C > T, in AD patients and healthy controls. All haplotypes showed similar distributions (p value > 0.5) in AD patients and healthy controls (Table 3).

Table 3.
Haplotype frequencies of IFITM3 polymorphisms in the healthy controls and Alzheimer’s disease (AD) patients.
4. Discussion
AD is characterized by the M1 type of disease-associated microglia (DAM) and the elevation of several neuroinflammation-related genes in the brain [23,24]. In a previous study, upregulation of the IFITM3 protein was also observed with neuroinflammation-related proteins by large-scale proteomic profiling in Aβ-treated microglia cell lines [25]. In addition, a recent study has reported that IFITM3 protein plays a major role in Aβ production via activation of γ-secretase [21]. Thus, the IFITM3 gene was proposed as a novel therapeutic target for AD. A previous study did not find the association between AD and IFITM3 SNPs. However, this study was performed with a relatively small sample size and, has not investigated the association of the rs6598045 SNP affecting the expression level of the IFITM3 gene via TFII-I binding capacity [18,21]. Thus, we performed an association analysis of AD with IFITM3 polymorphisms in larger populations. However, in the present study, we did not find significant differences in genotype, allele or haplotype frequencies of the IFITM3 gene between AD patients and matched healthy controls (Table 2 and Table 3). In addition, when our data set were stratified by sex and APOE genotype, there were no differences in the genotype, allele or haplotype frequencies of the IFITM3 gene between AD patients and matched controls (data not shown).
Previous studies have reported that the rs12252 C allele of the IFITM3 gene was associated with the severity of infection for the influenza A H1N1 2009 pandemic virus [26,27,28,29]. In addition, it was suggested that the severity of influenza A 2009 virus infection varies among ethnic groups based on differences in the distribution of the IFITM3 rs12252 SNP. The genotype and allele frequencies of rs12252, rs34481144, and rs6598045 SNPs are significantly different between Asian and Caucasian populations. In particular, although the risk of A allele of the rs34481144 SNP exists in less than 1% of East Asian individuals, the A allele of this SNP is present in a high proportion of individuals in European populations and is involved in the severity of pandemic influenza A 2009 virus infection [15,17,18]. Thus, in this study, although the SNPs of the IFITM3 gene did not show an association with susceptibility to AD, the IFITM3 SNPs is likely to affect AD phenotypes, including cognitive ability and Aβ accumulation. Thus, further investigation of the effect of the IFITM3 SNPs on the phenotype of AD is needed. In addition, since this study investigated only a Korean population, further study is needed to validate the association between IFITM3 SNPs and susceptibility to AD in other ethnic groups. Furthermore, since the genetic mechanism of the IFITM3 gene was not related to susceptibility to AD, further investigation of the epigenetic mechanism of the IFITM3 gene with AD is needed in the future.
5. Conclusions
In this study, we investigated genotype, allele, and haplotype frequencies of the IFITM3 polymorphisms in AD patients and matched healthy controls. Notably, the genotype, allele, and haplotype distributions of the IFITM3 polymorphisms did not show significant differences between these two groups. To the best of our knowledge, this is the first report regarding the association analysis of the IFITM3 gene in AD patients.
Author Contributions
S.-Y.W., Y.-C.K. and B.-H.J. conceived and designed the experiment. S.-Y.W. and Y.-C.K. performed the experiments. S.-Y.W., Y.-C.K. and B.-H.J. analyzed the data. S.-Y.W., Y.-C.K. and B.-H.J. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
S.-Y.W. and Y.-C.K. was supported by the BK21 Plus Program in the Department of Bioactive Material Sciences. This research was supported by the Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education (2017R1A6A1A03015876). This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (2021R1A2C1013213). This work was supported by an NRF (National Research Foundation of Korea) Grant funded by the Korean Government (NRF-2019-Fostering Core Leaders of the Future Basic Science Program/Global Ph.D. Fellowship Program).
Institutional Review Board Statement
A total of blood samples from 177 AD patients were provided by the Pusan National University Hospital Biobank, a member of the Korea Biobank Network. A total of blood samples from 233 healthy Korean subjects were obtained from the Korea Biobank Network at the Centers for Disease Control and Prevention. All experimental procedures were approved according to the guidelines of the institutional review board of Jeonbuk National University and the 1964 Helsinki declaration and its later amendments or comparable ethical standards (approval number: JBNU 2019-03-009).
Informed Consent Statement
Informed consent for all samples was obtained prior to investigation.
Data Availability Statement
All data are available from the corresponding authors upon reasonable request.
Conflicts of Interest
The authors declared no conflict of interest.
References
- Breijyeh, Z.; Karaman, R. Comprehensive Review on Alzheimer’s Disease: Causes and Treatment. Molecules 2020, 25, 5789. [Google Scholar] [CrossRef]
- Kim, Y.C.; Jeong, B.H. In Silico Evaluation of Acetylation Mimics in the 27 Lysine Residues of Human Tau Protein. Curr. Alzheimer Res. 2019, 16, 379–387. [Google Scholar] [CrossRef]
- Kim, Y.C.; Jeong, B.H. Identification of Somatic Mutations in Dementia-related Genes in Cancer Patients. Curr. Alzheimer Res. 2020, 17, 835–844. [Google Scholar] [CrossRef]
- Long, J.M.; Holtzman, D.M. Alzheimer Disease: An Update on Pathobiology and Treatment Strategies. Cell 2019, 179, 312–339. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gu, B.J.; Masters, C.L.; Wang, Y.J. A systemic view of Alzheimer disease-insights from amyloid-beta metabolism beyond the brain. Nat. Rev. Neurol. 2017, 13, 612–623. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, K.; Liu, F.; Gong, C.X.; Grundke-Iqbal, I. Tau in Alzheimer disease and related tauopathies. Curr. Alzheimer Res. 2010, 7, 656–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, H.; Jia, L.; Jia, J. Promoter polymorphisms which modulate APP expression may increase susceptibility to Alzheimer’s disease. Neurobiol. Aging 2008, 29, 194–202. [Google Scholar] [CrossRef]
- Bird, T.D. Early-Onset Familial Alzheimer Disease. In GeneReviews((R)); Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Stephens, K., Amemiya, A., Eds.; NCBI Bookshelf Site: Seattle, WA, USA, 1993. [Google Scholar]
- Beck, J.A.; Poulter, M.; Campbell, T.A.; Uphill, J.B.; Adamson, G.; Geddes, J.F.; Revesz, T.; Davis, M.B.; Wood, N.W.; Collinge, J.; et al. Somatic and germline mosaicism in sporadic early-onset Alzheimer’s disease. Hum. Mol. Genet. 2004, 13, 1219–1224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanoiselee, H.M.; Nicolas, G.; Wallon, D.; Rovelet-Lecrux, A.; Lacour, M.; Rousseau, S.; Richard, A.C.; Pasquier, F.; Rollin-Sillaire, A.; Martinaud, O.; et al. APP, PSEN1, and PSEN2 mutations in early-onset Alzheimer disease: A genetic screening study of familial and sporadic cases. PLoS Med. 2017, 14, e1002270. [Google Scholar] [CrossRef] [Green Version]
- Chavez-Gutierrez, L.; Bammens, L.; Benilova, I.; Vandersteen, A.; Benurwar, M.; Borgers, M.; Lismont, S.; Zhou, L.; Van Cleynenbreugel, S.; Esselmann, H.; et al. The mechanism of gamma-Secretase dysfunction in familial Alzheimer disease. EMBO J. 2012, 31, 2261–2274. [Google Scholar] [CrossRef]
- Houlden, H.; Crook, R.; Dolan, R.J.; McLaughlin, J.; Revesz, T.; Hardy, J. A novel presenilin mutation (M233V) causing very early onset Alzheimer’s disease with Lewy bodies. Neurosci. Lett. 2001, 313, 93–95. [Google Scholar] [CrossRef] [Green Version]
- Kelleher, R.J., 3rd; Shen, J. Presenilin-1 mutations and Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2017, 114, 629–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zani, A.; Yount, J.S. Antiviral Protection by IFITM3 In Vivo. Curr. Clin. Microbiol. Rep. 2018, 5, 229–237. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.C.; Jeong, B.H. Ethnic variation in risk genotypes based on single nucleotide polymorphisms (SNPs) of the interferon-inducible transmembrane 3 (IFITM3) gene, a susceptibility factor for pandemic 2009 H1N1 influenza A virus. Immunogenetics 2020, 72, 447–453. [Google Scholar] [CrossRef]
- Everitt, A.R.; Clare, S.; Pertel, T.; John, S.P.; Wash, R.S.; Smith, S.E.; Chin, C.R.; Feeley, E.M.; Sims, J.S.; Adams, D.J.; et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature 2012, 484, 519–523. [Google Scholar] [CrossRef] [Green Version]
- Allen, E.K.; Randolph, A.G.; Bhangale, T.; Dogra, P.; Ohlson, M.; Oshansky, C.M.; Zamora, A.E.; Shannon, J.P.; Finkelstein, D.; Dressen, A.; et al. SNP-mediated disruption of CTCF binding at the IFITM3 promoter is associated with risk of severe influenza in humans. Nat. Med. 2017, 23, 975–983. [Google Scholar] [CrossRef]
- Kim, Y.C.; Jeong, M.J.; Jeong, B.H. Strong association of regulatory single nucleotide polymorphisms (SNPs) of the IFITM3 gene with influenza H1N1 2009 pandemic virus infection. Cell. Mol. Immunol. 2020, 17, 662–664. [Google Scholar] [CrossRef]
- Kim, Y.C.; Jeong, B.H. No Correlation of the Disease Severity of Influenza A Virus Infection with the rs12252 Polymorphism of the Interferon-Induced Transmembrane Protein 3 Gene. Intervirology 2017, 60, 69–74. [Google Scholar] [CrossRef]
- Kim, Y.C.; Jeong, M.J.; Jeong, B.H. Genetic association between the rs12252 SNP of the Interferon-Induced transmembrane protein gene and Influenza A Virus Infection in the Korean population. Mol. Cell. Toxicol. 2021, 17, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Hur, J.Y.; Frost, G.R.; Wu, X.; Crump, C.; Pan, S.J.; Wong, E.; Barros, M.; Li, T.; Nie, P.; Zhai, Y.; et al. The innate immunity protein IFITM3 modulates gamma-secretase in Alzheimer’s disease. Nature 2020, 586, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Kruglyak, L.; Nickerson, D.A. Variation is the spice of life. Nat. Genet. 2001, 27, 234–236. [Google Scholar] [CrossRef]
- Tang, Y.; Le, W. Differential Roles of M1 and M2 Microglia in Neurodegenerative Diseases. Mol. Neurobiol. 2016, 53, 1181–1194. [Google Scholar] [CrossRef]
- Correani, V.; Di Francesco, L.; Mignogna, G.; Fabrizi, C.; Leone, S.; Giorgi, A.; Passeri, A.; Casata, R.; Fumagalli, L.; Maras, B.; et al. Plasma Membrane Protein Profiling in Beta-Amyloid-Treated Microglia Cell Line. Proteomics 2017, 17, 1600439. [Google Scholar] [CrossRef]
- Xu, Y.; Jin, M.Z.; Yang, Z.Y.; Jin, W.L. Microglia in neurodegenerative diseases. Neural Regen. Res. 2021, 16, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.H.; Zhao, Y.; Li, N.; Peng, Y.C.; Giannoulatou, E.; Jin, R.H.; Yan, H.P.; Wu, H.; Liu, J.H.; Liu, N.; et al. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat. Commun. 2013, 4, 1418. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Xiao, M.; Yang, J.; Chen, Y.K.; Bai, T.; Tang, X.J.; Shu, Y.L. Association between rs12252 and influenza susceptibility and severity: An updated meta-analysis. Epidemiol. Infect. 2019, 147, E39. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.; Cao, B.; Ke, C.; Lu, H.; Hu, Y.; Tam, C.H.T.; Ma, R.C.W.; Guan, D.; Zhu, Z.; Li, H.; et al. IFITM3, TLR3, and CD55 Gene SNPs and Cumulative Genetic Risks for Severe Outcomes in Chinese Patients with H7N9/H1N1pdm09 Influenza. J. Infect. Dis. 2017, 216, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Mills, T.C.; Rautanen, A.; Elliott, K.S.; Parks, T.; Naranbhai, V.; Ieven, M.M.; Butler, C.C.; Little, P.; Verheij, T.; Garrard, C.S.; et al. IFITM3 and susceptibility to respiratory viral infections in the community. J. Infect. Dis. 2014, 209, 1028–1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).