Abstract
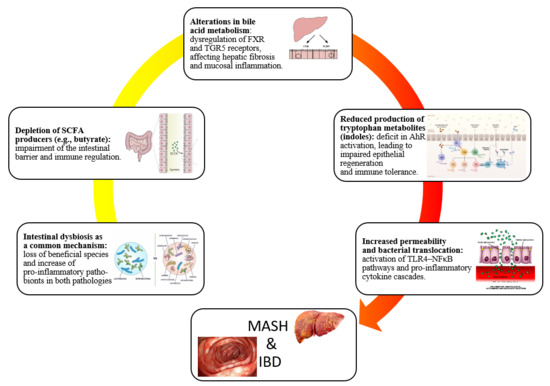
Gut microbiota changes have emerged as central players in the pathogenesis of both metabolic dysfunction-associated steatohepatitis (MASH) and inflammatory bowel disease (IBD). Although these diseases affect distinct primary organs, they share converging mechanisms driven by dysbiosis, including loss of beneficial short-chain fatty acid-producing taxa such as Faecalibacterium prausnitzii and Roseburia, enrichment of pro-inflammatory Enterobacteriaceae, and disruption of bile acid and tryptophan metabolism. These shifts compromise epithelial barrier integrity, promote the translocation of microbial products such as lipopolysaccharide, and trigger toll-like receptor 4-mediated activation of inflammatory cascades dominated by tumor necrosis factor-alpha, interleukin-6, and transforming growth factor-beta. In MASH, this dysbiotic environment fuels hepatic inflammation, insulin resistance, and fibrogenesis, while in IBD it sustains chronic mucosal immune activation. Shared features include impaired butyrate availability, altered bile acid pools affecting farnesoid X receptor and Takeda G protein-coupled Receptor 5 signaling, and defective aryl hydrocarbon receptor activation, all of which link microbial dysfunction to host metabolic and immune dysregulation. Understanding these overlapping pathways provides a deeper understanding of the role of the gut-liver and gut-immune axes as unifying frameworks in disease progression. This narrative review synthesizes current evidence on gut microbiota in MASH and IBD, underscoring the need for longitudinal, multi-omics studies and microbiome-targeted strategies to guide personalized therapeutic approaches.
1. Introduction
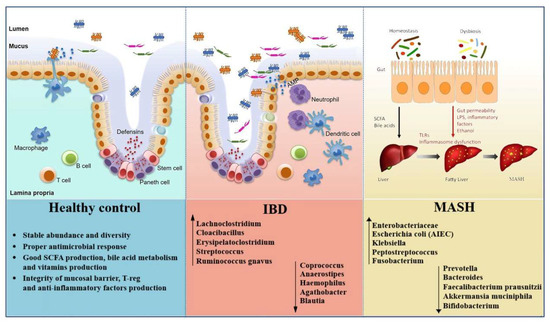
The human gut microbiota plays a critical role in maintaining intestinal homeostasis and regulating host metabolism and immune responses [1]. Disruptions in microbial composition and function, collectively referred to as gut dysbiosis, have been implicated in the development of various chronic inflammatory and metabolic disorders [2,3,4]. Among these, metabolic dysfunction-associated steatohepatitis (MASH) and inflammatory bowel disease (IBD) have gained increasing attention for their overlapping pathophysiological features linked to microbiota changes [5,6]. MASH, the progressive form of metabolic dysfunction-associated steatotic liver disease (MASLD), is characterized by hepatic steatosis, inflammation, and fibrosis, and is now one of the most common causes of chronic liver disease globally [7,8]. In this regard, MASLD is now recognized as the preferred term to describe hepatic steatosis linked to cardiometabolic abnormalities. It is diagnosed when evidence of liver fat accumulation, documented by imaging, histology, or validated biomarkers, coexists with at least one cardiometabolic risk factor such as overweight or obesity, type 2 diabetes mellitus (T2DM), hypertension, or dyslipidemia [9]. In contrast to the old non-alcoholic fatty liver disease definition, MASLD emphasizes the role of metabolic dysfunction rather than relying primarily on the exclusion of alcohol use [10]. Within the spectrum of fatty liver disease, MASH represents the progressive form, marked by steatosis along with hepatocyte ballooning and inflammation [11]. All individuals with MASH are included within the MASLD spectrum, yet only a subset of MASLD patients progress into MASH. The likelihood of progression is highest in those with cardiometabolic risk factors and genetic predispositions such as the Patatin-like phospholipase domain-containing protein 3 I148M variant [12]. Clinically, MASLD without inflammation is often stable, while MASH carries a much higher likelihood of advancing to fibrosis, cirrhosis, and hepatocellular carcinoma (HCC) [13]. IBD, which includes Crohn’s disease (CD) and ulcerative colitis (UC), is a group of chronic immune-mediated disorders affecting the gastrointestinal tract, driven by dysregulated immune responses to intestinal microbes in genetically susceptible individuals [14]. Despite differing in clinical manifestations, both MASH and IBD exhibit characteristic gut microbiota alterations. These include reduced microbial richness and alpha diversity, depletion of beneficial anti-inflammatory taxa such as Faecalibacterium prausnitzii and Akkermansia muciniphila, and an increased abundance of pro-inflammatory pathobionts such as Escherichia coli (E. coli) and members of the Enterobacteriaceae family [15,16,17]. Functional consequences of gut dysbiosis include impaired short-chain fatty acid (SCFA) production, disrupted bile acid metabolism, and increased intestinal permeability leading to bacterial translocation and systemic inflammation [18,19,20]. Crucially, these microbial shifts influence host physiology along the gut-liver axis, modulating hepatic inflammation and metabolic homeostasis, and along the gut-immune axis as well, promoting mucosal immune dysregulation [21,22]. Key mechanisms include toll-like receptor (TLR)-mediated signaling, compromised epithelial barrier integrity, and altered levels of microbial metabolites that impact immune tone and host metabolism [23,24]. A better understanding of these shared microbial and mechanistic pathways may shed light on how microbiota-driven inflammation and metabolic dysfunction interplay in MASH and IBD. As such, the present narrative review aims to synthesize current evidence on gut dysbiosis in both conditions, highlighting common pathogenic ways.
2. Gut Microbiota and MASH
The disequilibrium of gut microbiota has emerged as playing a key role in the pathogenesis of MASH onset and development [6]. Indeed, MASH is associated with a distinct microbial signature, involving not only changes in taxonomic composition, but also in the functional activity of the microbiota. These changes influence hepatic inflammation, insulin resistance, and fibrogenesis through complex interactions along the gut-liver axis [25].
2.1. Taxonomic Shifts of Gut Microbiota in MASH
As reported in the following lines, several studies using 16S ribosomal RNA (16S rRNA) gene sequencing and metagenomic approaches have reported taxonomic alterations in MASH. Zhu et al. analyzed children with MASH, obesity, and healthy controls, finding distinct enterotypes by disease status. While healthy and obese groups differed markedly, the microbiota of obese and MASH patients overlapped. Importantly, MASH patients were enriched in Proteobacteria, Enterobacteriaceae, and E. coli, and uniquely showed elevated blood ethanol levels, suggesting endogenous ethanol production may contribute to liver injury [26]. Del Chierico et al. compared stool samples from obese, MASLD, MASH, and healthy children. Reduced diversity was observed in obesity and MASLD. In MASH, Lachnospiraceae, Ruminococcus, and Dorea were increased, while Oscillospira decreased in steatosis. Gas chromatography–mass spectrometry identified differences in volatile organic compounds (2-butanone, 4-methyl-2-pentanone) between MASH and controls. Microbiota-metabolite signatures distinguished healthy subjects from diseased ones, although MASLD and MASH could not be reliably separated [27,28]. Loomba et al. developed a metagenomic machine-learning model to predict MASH with fibrosis. An increase in Proteobacteria and Enterobacteriaceae, as well as a reduction in Ruminococcus obeum and Eubacterium rectale, were the strongest features. The model achieved an area under the curve (AUC) of 0.92, outperforming clinical indices [29]. In a study of biopsy-proven MASH stratified by body mass index (BMI), fibrosis ≥ F2 was linked to increased Lactobacillus. Compared with controls, MASH patients showed differences in Faecalibacterium, Ruminococcus, Lactobacillus, and Bifidobacterium. Lean patients had a threefold reduction in Faecalibacterium and Ruminococcus; obese patients were enriched in Lactobacillus; overweight patients had reduced Bifidobacterium. Interestingly, alpha diversity in lean MASH resembled healthy controls, pointing to distinct microbiota mechanisms in lean versus obese disease [30]. Caussy et al. combined metagenomic data from MASLD cohorts, identifying a fibrosis signature with reduced SCFA-producers (Oscillospiraceae, Lachnospiraceae, Ruminococcus) and enrichment of pro-inflammatory genera (Veillonella, Streptococcus, Klebsiella). These taxa correlated with histological severity across cohorts [31]. MASH can present in both obese and lean individuals, reflecting heterogeneity in its metabolic background. Obese MASH is typically associated with dyslipidemia, T2DM, and a stronger inflammatory and fibrotic profile, while lean MASH often occurs in individuals with normal BMI but shares metabolic risk factors such as dyslipidemia or visceral adiposity [32,33]. Relating to this, Sookoian et al. found bacterial DNA from Roseibacillus, Peptostreptococcus, Bifidobacterium, and Streptomyces enriched in obese MASLD, with the greatest increase in MASH. Proteobacteria abundance was associated with more severe disease [34]. Another study showed progressive loss of diversity from MASLD to MASH. At phylum level, both groups had more Bacteroidetes and Fusobacteria and fewer Firmicutes. At genus level, Prevotella decreased in MASLD, while Megamonas and Fusobacterium increased in MASH. Functional analyses revealed altered glucose metabolism and reduced flavonoid/flavonol biosynthesis in MASH [35]. Finally, in pediatric patients, MASH was linked to increased Alistipes and reduced Peptostreptococcaceae. Species such as Bacteroides uniformis, Lachnospiraceae bacterium 7_1_58FAA, and Eubacterium ventriosum were significantly higher in MASH than MASLD. The authors suggested these shifts may drive progression and could support microbiota-based diagnostic profiling [36] (Table 1).

Table 1.
Summary of the studies about the gut microbiota composition in MASH.
2.2. Functional Alterations in Microbial Metabolism in MASH
These compositional changes suggest a loss of beneficial anti-inflammatory taxa and an enrichment of pathobionts capable of driving endotoxemia and low-grade inflammation. Taxonomic shifts in MASH are accompanied by profound functional alterations in microbial metabolism, which can be understood as being subdivided into three main areas.
- Gut microbiota and SCFAs: one of the most consistent findings is the reduced microbial capacity to produce SCFAs, particularly butyrate, which preserves epithelial barrier integrity and exerts anti-inflammatory effects via G-protein–coupled receptor 41/43 (GPR41/43) activation and histone deacetylase (HDAC) inhibition [37,38].
- Bile acid metabolism: MASH-associated dysbiosis is also characterized by disruption of bile acid metabolism. Microbes expressing bile salt hydrolase and 7α-dehydroxylase convert primary bile acids into secondary bile acids, such as deoxycholic acid (DCA). Elevated fecal DCA levels in MASH patients, together with a reduced abundance of Bacteroides, promote activation of hepatic farnesoid X receptor (FXR) and Takeda G protein–coupled receptor 5 (TGR5), enhancing pro-inflammatory and pro-fibrotic signaling [39].
- Microbial translocation and inflammatory mediators: dysbiosis further increases intestinal permeability, as shown by downregulation of tight junction proteins (occludin, claudin-1) in both human MASH and high-fat-diet-fed mouse models. This results in higher circulating levels of lipopolysaccharide (LPS) [40]. LPS activates TLR-4 on Kupffer and hepatic stellate cells, stimulating the release of tumor necrosis factor-alpha (TNF-α), interleukin-6 (IL-6), and pro-fibrotic mediators such as transforming growth factor-beta (TGF-β), thereby linking microbial products directly to liver inflammation and fibrogenesis [41].
Overall, MASH-associated dysbiosis is defined by a concurrent reduction in microbial diversity and metabolic functionality [6]. These alterations highlight the key role of the gut microbiota in the pathogenesis of MASH and provide a rationale for the development of microbiome-targeted therapeutic strategies.
3. Gut Microbiota and IBD
Multiple studies employing 16S rRNA gene sequencing, shotgun metagenomics, and multi-omics approaches have consistently demonstrated that patients with IBD exhibit significant alterations in gut microbial composition when compared to healthy individuals.
3.1. Taxonomic Shifts of Gut Microbiota in IBD
Alterations in IBD extend beyond microbial diversity, including phylum-level shifts, expansion of pathobionts, and loss of immunomodulatory taxa. Indeed, Darfeuille-Michaud et al. first identified adherent-invasive E. coli (AIEC) from ileal biopsies of CD patients, showing that these strains invade epithelial cells, persist in macrophages, and trigger TNF-α, thus sustaining chronic inflammation [42]. Sokol et al. later reported a selective reduction of F. prausnitzii in CD, especially in post-operative recurrence, and demonstrated its ability to induce IL-10, supporting an anti-inflammatory role [43]. In treatment-naïve pediatric CD, Gevers et al. showed that biopsy microbiota better detected disease than stool, with enrichment of Enterobacteriaceae, Fusobacterium, and Peptostreptococcus, and depletion of Bacteroides and Faecalibacterium [44]. Machiels et al. found UC patients had reduced butyrate producers (Roseburia hominis, F. prausnitzii) and increased mucin-degraders (Ruminococcus gnavus) [45]. In mice, germ-free conditions prevented ileitis, antibiotics reduced it, and microbiota transfer from diseased donors induced it. Gut dysbiosis was associated with loss of Paneth cell antimicrobial function, while E. coli LF82 alone was insufficient, highlighting the causal role of complex microbial communities [46]. Forbes et al. confirmed reduced Clostridia and Bacteroides with increased Enterobacteriaceae across immune-mediated disorders but also identified IBD-specific signatures [47]. Data from the IBD Multi-omics Database showed depletion of Firmicutes (F. prausnitzii, Roseburia, Eubacterium hallii) and expansion of Proteobacteria (E. coli, K. pneumoniae), correlating with disease activity and flares [48]. A meta-analysis of ~2000 metagenomes supported disease-specific functional profiles across gut–liver axis disorders. Patients showed reduced pathways for SCFA and bile acid metabolism and enrichment of genes related to oxidative stress and pro-inflammatory metabolites. While some alterations overlapped with other gut-liver disorders, IBD had a distinct loss of symbiotic taxa and expansion of pathobionts, reinforcing the role of the microflora in intestinal inflammation [49]. Network-based analyses further showed reduced Firmicutes, Bacteroidetes, and Actinobacteria in IBD, with depleted taxa highly connected in microbial networks, suggesting that their loss destabilizes community structure [50]. In remission, IBD patients still showed lower alpha diversity, distinct beta diversity, and enrichment of flavonoid-degraders, while healthy controls had more Akkermansia, Oscillibacter, and Coprococcus. Enterobacteriaceae remained central in microbial networks, supporting persistent low-grade inflammation [51]. A recent study confirmed reduced richness and depletion of Subdoligranulum, Ruminococcus, Anaerostipes, and Lachnospira. UC patients showed more Streptococcus but fewer Alistipes, while CD patients were enriched in Lachnoclostridium, Fusobacterium, Cloacibacillus, and Erysipelatoclostridium, with reduced Faecalibacterium, Roseburia, and Haemophilus. Active disease showed further depletion of anti-inflammatory taxa (Roseburia, Coprococcus, Ruminiclostridium) [52]. Moreover, extraintestinal manifestations were linked to decreased Agathobacter and Blautia and increased Eggerthella [53]. Finally, Italian data showed an increased Firmicutes/Bacteroidetes (F/B) ratio in IBD, suggesting its potential role as a biomarker [54,55] (Table 2).

Table 2.
Summary of the studies about the gut microbiota composition in IBD.
3.2. Functional Alterations in Microbial Metabolism in IBD
Beyond compositional changes, IBD-associated dysbiosis is also characterized by functional disruptions in microbial metabolism, which can be described as being divided into four main areas.
- Gut microbiota and SCFAs: metagenomic and metabolomic analyses consistently demonstrate reduced production of SCFAs, particularly butyrate and propionate [56,57]. Butyrate, mainly produced by F. prausnitzii, Roseburia, and Eubacterium rectale, serves as the primary energy source for colonocytes and exerts potent anti-inflammatory effects. Its depletion impairs epithelial barrier integrity and exacerbates mucosal inflammation in IBD [58].
- Redox imbalance: IBD microbiota displays increased oxidative stress and preferential use of nitrate and sulfate respiration, creating conditions that favor facultative anaerobes such as Enterobacteriaceae over obligate anaerobes. This shift in redox balance reinforces microbial instability and drives chronic immune activation [59].
- Bile acid metabolism: in healthy individuals, microbial deconjugation and 7α-dehydroxylation convert primary bile acids into secondary ones such as DCA and lithocholic acid. In IBD, these pathways are impaired, resulting in an accumulation of conjugated bile acids that promote barrier dysfunction and mucosal inflammation through FXR and TGR5 signaling [60].
- Tryptophan metabolism and immune regulation: microbial catabolism of tryptophan into indole derivatives is a major source of ligands for the aryl hydrocarbon receptor (AhR), which maintains intestinal immune tolerance and epithelial repair. Loss of this pathway in IBD reduces AhR activation, contributing to chronic inflammation [61]. In addition, multi-omics evaluations reveal that strain-level variation modulates functional outputs. For example, Ruminococcus gnavus, commonly enriched in IBD, may exert either pro- or anti-inflammatory effects depending on capsular polysaccharide expression [62].
These findings suggest that microbial function, and not only taxonomy, plays a pivotal role in modulating host inflammation and disease onset.
5. Conclusions and Future Perspectives
To the best of our knowledge, this narrative review is the first to specifically address the changes in gut microbiota in the context of both MASH and IBD. While previous reviews have examined these conditions separately, our work uniquely integrates them within a shared pathogenic framework, emphasizing overlapping microbial, metabolic, and immune pathways. As such, the present review could prove interesting to both researchers and clinicians. The complex interplay between gut dysbiosis, intestinal barrier dysfunction, and immune-metabolic signaling emerges as a shared pathogenic framework linking MASH and IBD. Despite differences in their primary target organs, both conditions converge on common microbial, metabolic, and inflammatory pathways that perpetuate chronic tissue injury. Advances in multi-omics technologies have deepened our understanding of these shared mechanisms, yet translational integration into clinical practice remains limited. Future research should prioritize large-scale, longitudinal studies in diverse populations to dissect the temporal sequence of dysbiosis, barrier disruption, and host response in MASH–IBD coexistence. In this regard, comparative studies evaluating microbiota-targeted therapies, ranging from dietary interventions, prebiotics to live biotherapeutics, bile acid modulators and fecal microbiota transplantation, are warranted to define personalized treatment strategies [75,76,77]. Furthermore, leveraging machine learning-driven microbiome profiling may enable early disease detection, risk stratification, and therapy optimization [78,79]. Closing these knowledge gaps will play an essential role in translating mechanistic insights into effective interventions, ultimately improving outcomes for patients affected by these interconnected gut–liver–immune disorders.
Author Contributions
Conceptualization: G.G.M.S. and L.A.; formal analysis: G.G.M.S. and D.M., data curation: A.I. and R.S.; writing—original draft: G.G.M.S. and L.A.; writing, reviewing, and editing: A.I., R.S., F.L. and D.L.D. supervision: L.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
We would like to thank Simone Scarlata for his critical review of the English language.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| 16S rRNA | 16S ribosomal RNA |
| AhR | Aryl hydrocarbon Receptor |
| AUC | Area Under the Curve |
| BMI | Body mass index |
| CD | Crohn’s disease |
| DCA | Deoxycholic Acid |
| E. coli | Escherichia coli |
| F/B | Firmicutes/Bacteroidetes ratio |
| FXR | Farnesoid X Receptor |
| GPR41/43 | G-protein–coupled Receptor 41/43 |
| HCC | Hepatocellular carcinoma |
| HDAC | Histone Deacetylase |
| IBD | Inflammatory Bowel Disease |
| IL | Interleukin |
| LPS | Lipopolysaccharide |
| MAMPs | Microbe-Associated Molecular Patterns |
| MASH | Metabolic dysfunction-Associated Steatohepatitis |
| MASLD | Metabolic dysfunction-Associated Steatotic Liver Disease |
| SCFA | Short-Chain Fatty Acid |
| T2DM | Type 2 diabetes mellitus |
| TGF-β | Transforming Growth Factor-beta |
| TGR5 | Takeda G protein-coupled Receptor 5 |
| TLR | Toll-Like Receptor |
| TNF-α | Tumor Necrosis Factor-alpha |
| UC | Ulcerative Colitis |
References
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Shen, Y.; Fan, N.; Ma, S.X.; Cheng, X.; Yang, X.; Wang, G. Gut Microbiota Dysbiosis: Pathogenesis, Diseases, Prevention, and Therapy. MedComm (2020) 2025, 6, e70168. [Google Scholar] [CrossRef] [PubMed]
- Acevedo-Román, A.; Pagán-Zayas, N.; Velázquez-Rivera, L.I.; Torres-Ventura, A.C.; Godoy-Vitorino, F. Insights into Gut Dysbiosis: Inflammatory Diseases, Obesity, and Restoration Approaches. Int. J. Mol. Sci. 2024, 25, 9715. [Google Scholar] [CrossRef] [PubMed]
- Bandopadhyay, P.; Ganguly, D. Gut dysbiosis and metabolic diseases. Prog. Mol. Biol. Transl. Sci. 2022, 191, 153–174. [Google Scholar] [PubMed]
- Scarlata, G.G.M.; Abenavoli, L. Gut microbiota: The pathogenetic bridge between inflammatory bowel disease and metabolic-associated steatotic liver disease. Expert. Rev. Gastroenterol. Hepatol. 2025, 19, 85–88. [Google Scholar]
- Ohtani, N.; Kamiya, T.; Kawada, N. Recent updates on the role of the gut-liver axis in the pathogenesis of NAFLD/NASH, HCC, and beyond. Hepatol. Commun. 2023, 7, e0241. [Google Scholar]
- Rinella, M.E.; Sookoian, S. From NAFLD to MASLD: Updated naming and diagnosis criteria for fatty liver disease. J. Lipid Res. 2024, 65, 100485. [Google Scholar]
- Guo, Z.; Wu, D.; Mao, R.; Yao, Z.; Wu, Q.; Lv, W. Global burden of MAFLD, MAFLD related cirrhosis and MASH related liver cancer from 1990 to 2021. Sci. Rep. 2025, 15, 7083. [Google Scholar] [CrossRef]
- Rinella, M.E.; Lazarus, J.V.; Ratziu, V.; Francque, S.M.; Sanyal, A.J.; Kanwal, F.; Romero, D.; Abdelmalek, M.F.; Anstee, Q.M.; Arab, J.P.; et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. Hepatology 2023, 78, 1966–1986. [Google Scholar] [CrossRef]
- Gastaldelli, A.; Newsome, P.N. NAFLD vs MASLD (Metabolic Dysfunction-Associated Steatotic Liver Disease)-Why the Need for a Change of Nomenclature? J. Clin. Endocrinol. Metab. 2025, 110, e2407–e2410. [Google Scholar]
- Sergi, C.M. NAFLD (MASLD)/NASH (MASH): Does It Bother to Label at All? A Comprehensive Narrative Review. Int. J. Mol. Sci. 2024, 25, 8462. [Google Scholar] [CrossRef] [PubMed]
- Jegodzinski, L.; Rudolph, L.; Castven, D.; Sayk, F.; Rout, A.K.; Föh, B.; Hölzen, L.; Meyhöfer, S.; Schenk, A.; Weber, S.N.; et al. PNPLA3 I148M variant links to adverse metabolic traits in MASLD during fasting and feeding. JHEP Rep. 2025, 7, 101450. [Google Scholar] [CrossRef] [PubMed]
- Armandi, A.; Rosso, C.; Caviglia, G.P.; Bugianesi, E. An updated overview on hepatocellular carcinoma in patients with Metabolic dysfunction-Associated Steatotic Liver Disease: Trends, pathophysiology and risk-based surveillance. Metabolism 2025, 162, 156080. [Google Scholar] [CrossRef] [PubMed]
- Calvez, V.; Puca, P.; Di Vincenzo, F.; Del Gaudio, A.; Bartocci, B.; Murgiano, M.; Iaccarino, J.; Parand, E.; Napolitano, D.; Pugliese, D.; et al. Novel Insights into the Pathogenesis of Inflammatory Bowel Diseases. Biomedicines 2025, 13, 305. [Google Scholar] [CrossRef]
- Cao, Y.; Shen, J.; Ran, Z.H. Association between Faecalibacterium prausnitzii Reduction and Inflammatory Bowel Disease: A Meta-Analysis and Systematic Review of the Literature. Gastroenterol. Res. Pract. 2014, 2014, 872725. [Google Scholar]
- Zheng, M.; Han, R.; Yuan, Y.; Xing, Y.; Zhang, W.; Sun, Z.; Liu, Y.; Li, J.; Mao, T. The role of Akkermansia muciniphila in inflammatory bowel disease: Current knowledge and perspectives. Front. Immunol. 2023, 13, 1089600. [Google Scholar] [CrossRef]
- Mirsepasi-Lauridsen, H.C.; Vallance, B.A.; Krogfelt, K.A.; Petersen, A.M. Escherichia coli Pathobionts Associated with Inflammatory Bowel Disease. Clin. Microbiol. Rev. 2019, 32, e00060-18. [Google Scholar] [CrossRef]
- Weiss, G.A.; Hennet, T. Mechanisms and consequences of intestinal dysbiosis. Cell Mol. Life Sci. 2017, 74, 2959–2977. [Google Scholar] [CrossRef]
- Peterson, D.; Weidenmaier, C.; Timberlake, S.; Gura Sadovsky, R. Depletion of key gut bacteria predicts disrupted bile acid metabolism in inflammatory bowel disease. Microbiol. Spectr. 2025, 13, e0199924. [Google Scholar] [CrossRef]
- Camilleri, M. What is the leaky gut? Clinical considerations in humans. Curr. Opin. Clin. Nutr. Metab. Care 2021, 24, 473–482. [Google Scholar] [CrossRef]
- Hsu, C.L.; Schnabl, B. The gut-liver axis and gut microbiota in health and liver disease. Nat. Rev. Microbiol. 2023, 21, 719–733. [Google Scholar] [CrossRef]
- Dawson, S.L.; Todd, E.; Ward, A.C. The Interplay of Nutrition, the Gut Microbiota and Immunity and Its Contribution to Human Disease. Biomedicines 2025, 13, 329. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Whitley, C.S.; Haribabu, B.; Jala, V.R. Regulation of Intestinal Barrier Function by Microbial Metabolites. Cell Mol. Gastroenterol. Hepatol. 2021, 11, 1463–1482. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, L.; Hua, H.; Liu, L.; Mao, Y.; Wang, R. Interactions between toll-like receptors signaling pathway and gut microbiota in host homeostasis. Immun. Inflamm. Dis. 2024, 12, e1356. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, S.G.; van der Merwe, S.; Krag, A.; Wiest, R. Gut-liver axis: Pathophysiological concepts and medical perspective in chronic liver diseases. Semin. Immunol. 2024, 71, 101859. [Google Scholar] [CrossRef]
- Zhu, L.; Baker, S.S.; Gill, C.; Liu, W.; Alkhouri, R.; Baker, R.D.; Gill, S.R. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 2013, 57, 601–609. [Google Scholar]
- Del Chierico, F.; Nobili, V.; Vernocchi, P.; Russo, A.; De Stefanis, C.; Gnani, D.; Furlanello, C.; Zandonà, A.; Paci, P.; Capuani, G.; et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology 2017, 65, 451–464. [Google Scholar] [CrossRef]
- Haslam, D.B. Nonalcoholic steatohepatitis and the intestinal microbiota. Hepatology 2017, 65, 401–403. [Google Scholar] [CrossRef]
- Loomba, R.; Seguritan, V.; Li, W.; Long, T.; Klitgord, N.; Bhatt, A.; Dulai, P.S.; Caussy, C.; Bettencourt, R.; Highlander, S.K.; et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2017, 25, 1054–1062.e5. [Google Scholar] [CrossRef]
- Duarte, S.M.B.; Stefano, J.T.; Miele, L.; Ponziani, F.R.; Souza-Basqueira, M.; Okada, L.S.R.R.; de Barros Costa, F.G.; Toda, K.; Mazo, D.F.C.; Sabino, E.C.; et al. Gut microbiome composition in lean patients with NASH is associated with liver damage independent of caloric intake: A prospective pilot study. Nutr. Metab. Cardiovasc. Dis. 2018, 28, 369–384. [Google Scholar] [CrossRef]
- Caussy, C.; Tripathi, A.; Humphrey, G.; Bassirian, S.; Singh, S.; Faulkner, C.; Bettencourt, R.; Rizo, E.; Richards, L.; Xu, Z.Z.; et al. A gut microbiome signature for cirrhosis due to nonalcoholic fatty liver disease. Nat. Commun. 2019, 10, 1406. [Google Scholar] [CrossRef]
- van Kleef, L.A.; Pustjens, J.; Savas, M.; Ayada, I.; Li, P.; Pan, Q.; van Rossum, E.F.C.; Janssen, H.L.A.; Brouwer, W.P. MASLD, At-Risk MASH and Increased Liver Stiffness Are Associated with Young Adulthood Obesity Without Residual Risk After Losing Obesity. Liver Int. 2025, 45, e16169. [Google Scholar] [CrossRef]
- Bhattacharya, I.; Maity, D.K.; Kumar, A.; Sarkar, S.; Bhattacharya, T.; Sahu, A.; Sreedhar, R.; Arumugam, S. Beyond obesity: Lean metabolic dysfunction-associated steatohepatitis from unveiling molecular pathogenesis to therapeutic advancement. Naunyn Schmiedebergs Arch. Pharmacol. 2025, 398, 13647–13665. [Google Scholar] [CrossRef]
- Sookoian, S.; Salatino, A.; Castaño, G.O.; Landa, M.S.; Fijalkowky, C.; Garaycoechea, M.; Pirola, C.J. Intrahepatic bacterial metataxonomic signature in non-alcoholic fatty liver disease. Gut 2020, 69, 1483–1491. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Lyu, B.; Xie, F.; Li, F.; Xing, Y.; Han, Z.; Lai, J.; Ma, J.; Zou, Y.; Zeng, H.; et al. From gut to liver: Unveiling the differences of intestinal microbiota in NAFL and NASH patients. Front. Microbiol. 2024, 15, 1366744. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shi, M.; Zhao, C.; Liang, G.; Li, C.; Ge, X.; Pei, C.; Kong, Y.; Li, D.; Yang, W.; et al. Role of intestinal flora in the development of nonalcoholic fatty liver disease in children. Microbiol. Spectr. 2024, 12, e0100623. [Google Scholar] [CrossRef] [PubMed]
- Ríos-Covián, D.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M.; de Los Reyes-Gavilán, C.G.; Salazar, N. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front. Microbiol. 2016, 7, 185. [Google Scholar] [CrossRef]
- Du, Y.; He, C.; An, Y.; Huang, Y.; Zhang, H.; Fu, W.; Wang, M.; Shan, Z.; Xie, J.; Yang, Y.; et al. The Role of Short Chain Fatty Acids in Inflammation and Body Health. Int. J. Mol. Sci. 2024, 25, 7379. [Google Scholar] [CrossRef]
- Wahlström, A.; Sayin, S.I.; Marschall, H.U.; Bäckhed, F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef]
- Luther, J.; Garber, J.J.; Khalili, H.; Dave, M.; Bale, S.S.; Jindal, R.; Motola, D.L.; Luther, S.; Bohr, S.; Jeoung, S.W.; et al. Hepatic Injury in Nonalcoholic Steatohepatitis Contributes to Altered Intestinal Permeability. Cell Mol. Gastroenterol. Hepatol. 2015, 1, 222–232. [Google Scholar] [CrossRef]
- Gandhi, C.R. Pro- and Anti-fibrogenic Functions of Gram-Negative Bacterial Lipopolysaccharide in the Liver. Front. Med. 2020, 7, 130. [Google Scholar] [CrossRef] [PubMed]
- Darfeuille-Michaud, A.; Boudeau, J.; Bulois, P.; Neut, C.; Glasser, A.L.; Barnich, N.; Bringer, M.A.; Swidsinski, A.; Beaugerie, L.; Colombel, J.F. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 2004, 127, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [PubMed]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef]
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014, 63, 1275–1283. [Google Scholar] [CrossRef]
- Schaubeck, M.; Clavel, T.; Calasan, J.; Lagkouvardos, I.; Haange, S.B.; Jehmlich, N.; Basic, M.; Dupont, A.; Hornef, M.; von Bergen, M.; et al. Dysbiotic gut microbiota causes transmissible Crohn’s disease-like ileitis independent of failure in antimicrobial defence. Gut 2016, 65, 225–237. [Google Scholar] [CrossRef]
- Forbes, J.D.; Chen, C.Y.; Knox, N.C.; Marrie, R.A.; El-Gabalawy, H.; de Kievit, T.; Alfa, M.; Bernstein, C.N.; Van Domselaar, G. A comparative study of the gut microbiota in immune-mediated inflammatory diseases-does a common dysbiosis exist? Microbiome 2018, 6, 221. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef]
- Armour, C.R.; Nayfach, S.; Pollard, K.S.; Sharpton, T.J. A Metagenomic Meta-analysis Reveals Functional Signatures of Health and Disease in the Human Gut Microbiome. mSystems 2019, 4, e00332-18. [Google Scholar] [CrossRef]
- Alam, M.T.; Amos, G.C.A.; Murphy, A.R.J.; Murch, S.; Wellington, E.M.H.; Arasaradnam, R.P. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog. 2020, 12, 1. [Google Scholar] [CrossRef]
- Pisani, A.; Rausch, P.; Bang, C.; Ellul, S.; Tabone, T.; Marantidis Cordina, C.; Zahra, G.; Franke, A.; Ellul, P. Dysbiosis in the Gut Microbiota in Patients with Inflammatory Bowel Disease during Remission. Microbiol. Spectr. 2022, 10, e0061622. [Google Scholar] [CrossRef]
- Ma, J.; Wang, K.; Wang, J.; Zeng, Q.; Liu, K.; Zheng, S.; Chen, Y.; Yao, J. Microbial Disruptions in Inflammatory Bowel Disease: A Comparative Analysis. Int. J. Gen. Med. 2024, 17, 1355–1367. [Google Scholar] [CrossRef] [PubMed]
- Hertz, S.; Anderson, J.M.; Nielsen, H.L.; Schachtschneider, C.; McCauley, K.E.; Özçam, M.; Larsen, L.; Lynch, S.V.; Nielsen, H. Fecal microbiota is associated with extraintestinal manifestations in inflammatory bowel disease. Ann. Med. 2024, 56, 2338244. [Google Scholar] [CrossRef] [PubMed]
- De Caro, C.; Spagnuolo, R.; Quirino, A.; Mazza, E.; Carrabetta, F.; Maurotti, S.; Cosco, C.; Bennardo, F.; Roberti, R.; Russo, E.; et al. Gut Microbiota Profile Changes in Patients with Inflammatory Bowel Disease and Non-Alcoholic Fatty Liver Disease: A Metagenomic Study. Int. J. Mol. Sci. 2024, 25, 5453. [Google Scholar] [CrossRef] [PubMed]
- Scarlata, G.G.M.; Abenavoli, L. Should Routine Diagnostics Implement Gut Microbiota Analysis? Int. J. Gastroenterol. Hepatol. Dis. 2024, 3, e26662906338438. [Google Scholar] [CrossRef]
- Yan, D.; Ye, S.; He, Y.; Wang, S.; Xiao, Y.; Xiang, X.; Deng, M.; Luo, W.; Chen, X.; Wang, X. Fatty acids and lipid mediators in inflammatory bowel disease: From mechanism to treatment. Front. Immunol. 2023, 14, 1286667. [Google Scholar] [CrossRef]
- Lavelle, A.; Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 223–237. [Google Scholar] [CrossRef]
- Hodgkinson, K.; El Abbar, F.; Dobranowski, P.; Manoogian, J.; Butcher, J.; Figeys, D.; Mack, D.; Stintzi, A. Butyrate’s role in human health and the current progress towards its clinical application to treat gastrointestinal disease. Clin. Nutr. 2023, 42, 61–75. [Google Scholar] [CrossRef]
- Winter, S.E.; Winter, M.G.; Xavier, M.N.; Thiennimitr, P.; Poon, V.; Keestra, A.M.; Laughlin, R.C.; Gomez, G.; Wu, J.; Lawhon, S.D.; et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 2013, 339, 708–711. [Google Scholar] [CrossRef]
- Sinha, S.R.; Haileselassie, Y.; Nguyen, L.P.; Tropini, C.; Wang, M.; Becker, L.S.; Sim, D.; Jarr, K.; Spear, E.T.; Singh, G.; et al. Dysbiosis-Induced Secondary Bile Acid Deficiency Promotes Intestinal Inflammation. Cell Host Microbe 2020, 27, 659–670.e5. [Google Scholar] [CrossRef]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.B.; Yassour, M.; Sauk, J.; Garner, A.; Jiang, X.; Arthur, T.; Lagoudas, G.K.; Vatanen, T.; Fornelos, N.; Wilson, R.; et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017, 9, 103. [Google Scholar] [CrossRef] [PubMed]
- Neurath, M.F.; Artis, D.; Becker, C. The intestinal barrier: A pivotal role in health, inflammation, and cancer. Lancet Gastroenterol. Hepatol. 2025, 10, 573–592. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhang, Y.; Zhao, X.; Shang, C.; Xiang, M.; Li, L.; Cui, X. Microbiota-derived short-chain fatty acids: Implications for cardiovascular and metabolic disease. Front. Cardiovasc. Med. 2022, 9, 900381. [Google Scholar] [CrossRef]
- Yan, W.; Zhang, K.; Guo, J.; Xu, L. Bile acid-mediated gut-liver axis crosstalk: The role of nuclear receptor signaling in dynamic regulation of inflammatory networks. Front. Immunol. 2025, 16, 1595486. [Google Scholar] [CrossRef]
- He, Y.; Shaoyong, W.; Chen, Y.; Li, M.; Gan, Y.; Sun, L.; Liu, Y.; Wang, Y.; Jin, M. The functions of gut microbiota-mediated bile acid metabolism in intestinal immunity. J. Adv. Res. 2025; in press. [Google Scholar]
- Cebi, M.; Yilmaz, Y. Epithelial barrier hypothesis in the context of nutrition, microbial dysbiosis, and immune dysregulation in metabolic dysfunction-associated steatotic liver. Front. Immunol. 2025, 16, 1575770. [Google Scholar] [CrossRef]
- Wang, B.; Zhou, Z.; Li, L. Gut Microbiota Regulation of AHR Signaling in Liver Disease. Biomolecules 2022, 12, 1244. [Google Scholar] [CrossRef]
- Hou, J.J.; Ma, A.H.; Qin, Y.H. Activation of the aryl hydrocarbon receptor in inflammatory bowel disease: Insights from gut microbiota. Front. Cell Infect. Microbiol. 2023, 13, 1279172. [Google Scholar] [CrossRef]
- Abenavoli, L.; Giubilei, L.; Procopio, A.C.; Spagnuolo, R.; Luzza, F.; Boccuto, L.; Scarpellini, E. Gut Microbiota in Non-Alcoholic Fatty Liver Disease Patients with Inflammatory Bowel Diseases: A Complex Interplay. Nutrients 2022, 14, 5323. [Google Scholar] [CrossRef]
- Schnabl, B.; Damman, C.J.; Carr, R.M. Metabolic dysfunction-associated steatotic liver disease and the gut microbiome: Pathogenic insights and therapeutic innovations. J. Clin. Investig. 2025, 135, e186423. [Google Scholar] [CrossRef]
- Cui, C.; Gao, S.; Shi, J.; Wang, K. Gut-Liver Axis: The Role of Intestinal Microbiota and Their Metabolites in the Progression of Metabolic Dysfunction-Associated Steatotic Liver Disease. Gut Liver 2025, 19, 479–507. [Google Scholar]
- Yu, J.X.; Wu, J.; Chen, X.; Zang, S.G.; Li, X.B.; Wu, L.P.; Xuan, S.H. Gut microbiota in liver diseases: Initiation, development and therapy. Front. Med. 2025, 12, 1615839. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Yang, Y.; Lin, S.; Geller, D.A.; Yan, Y. The microenvironment in the development of MASLD-MASH-HCC and associated therapeutic in MASH-HCC. Front. Immunol. 2025, 16, 1569915. [Google Scholar] [CrossRef] [PubMed]
- Strati, F.; Lattanzi, G.; Amoroso, C.; Facciotti, F. Microbiota-targeted therapies in inflammation resolution. Semin. Immunol. 2022, 59, 101599. [Google Scholar] [CrossRef] [PubMed]
- Zikou, E.; Koliaki, C.; Makrilakis, K. The Role of Fecal Microbiota Transplantation (FMT) in the Management of Metabolic Diseases in Humans: A Narrative Review. Biomedicines 2024, 12, 1871. [Google Scholar] [CrossRef]
- Abenavoli, L.; Spagnuolo, R.; Scarlata, G.G.M.; Gambardella, M.L.; Boccuto, L.; Méndez-Sánchez, N.; Luzza, F. Metabolic Dysfunction-Associated Steatotic Liver Disease in Patients with Inflammatory Bowel Diseases: A Pilot Study. Life 2024, 14, 1226. [Google Scholar] [CrossRef]
- Chen, A.T.; Wu, X.; Ye, G.; Li, W. Editorial: Machine learning and deep learning applications in pathogenic microbiome research. Front. Cell Infect. Microbiol. 2024, 14, 1429197. [Google Scholar]
- Popa, S.L.; Ismaiel, A.; Cristina, P.; Cristina, M.; Chiarioni, G.; David, L.; Dumitrascu, D.L. Non-Alcoholic Fatty Liver Disease: Implementing Complete Automated Diagnosis and Staging. A Systematic Review. Diagnostics 2021, 11, 1078. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).