Multi Omics Applications in Biological Systems
Abstract
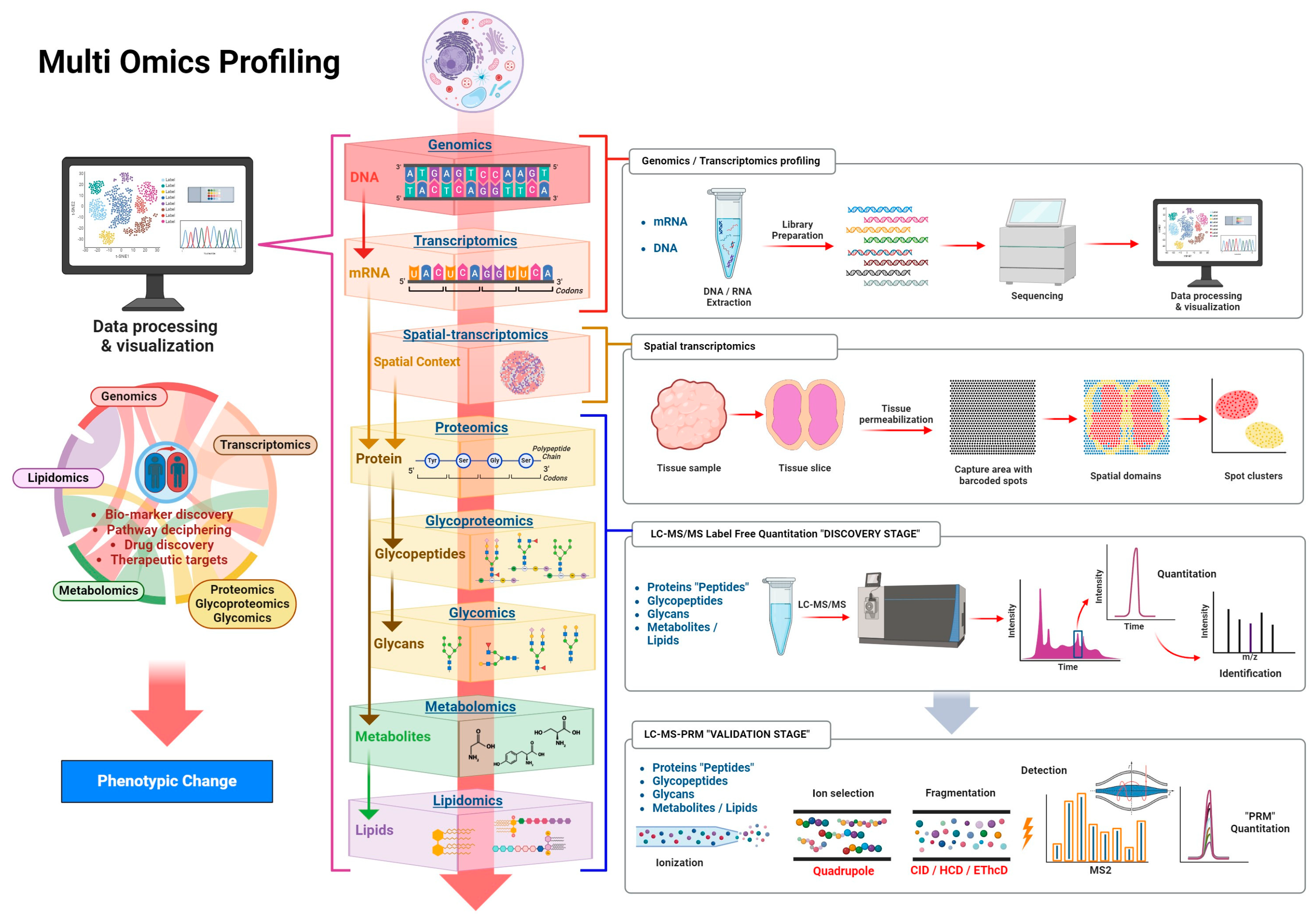
1. Introduction
2. Genomics and Transcriptomics
3. Proteomics, Glycoproteomics, and Glycomics
4. Metabolomics and Lipidomics
5. Multi Omics Integration
Foodomics
6. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Karahalil, B. Overview of Systems Biology and Omics Technologies. Curr. Med. Chem. 2016, 23, 4221–4230. [Google Scholar] [CrossRef] [PubMed]
- Westerhoff, H.V.; Palsson, B.O. The evolution of molecular biology into systems biology. Nat. Biotechnol. 2004, 22, 1249–1252. [Google Scholar] [CrossRef] [PubMed]
- Munung, N.S.; Ambele, M.A.; Moela, P. Advancing global equity in cancer genomics—Challenges and opportunities in Sub-Saharan Africa. Curr. Opin. Genet. Dev. 2021, 66, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Heidecker, B.; Hare, J.M. The use of transcriptomic biomarkers for personalized medicine. Heart Fail. Rev. 2007, 12, 1–11. [Google Scholar] [CrossRef]
- He, T. Implementation of Proteomics in Clinical Trials. Proteom. Clin. Appl. 2019, 13, e1800198. [Google Scholar] [CrossRef] [PubMed]
- Mechref, Y.; Peng, W.; Gautam, S.; Ahmadi, P.; Lin, Y.; Zhu, J.; Zhang, J.; Liu, S.; Singal, A.G.; Parikh, N.D.; et al. Mass spectrometry based biomarkers for early detection of HCC using a glycoproteomic approach. Adv. Cancer Res. 2023, 157, 23–56. [Google Scholar] [CrossRef]
- Peng, W.; Zhao, J.; Dong, X.; Banazadeh, A.; Huang, Y.; Hussien, A.; Mechref, Y. Clinical application of quantitative glycomics. Expert Rev. Proteom. 2018, 15, 1007–1031. [Google Scholar] [CrossRef]
- Dona, A.C.; Coffey, S.; Figtree, G. Translational and emerging clinical applications of metabolomics in cardiovascular disease diagnosis and treatment. Eur. J. Prev. Cardiol. 2016, 23, 1578–1589. [Google Scholar] [CrossRef]
- Astarita, G.; Stocchero, M.; Paglia, G. Unbiased Lipidomics and Metabolomics of Human Brain Samples. Methods Mol. Biol. 2018, 1750, 255–269. [Google Scholar] [CrossRef]
- Aizat, W.M.; Ismail, I.; Noor, N.M. Recent Development in Omics Studies. Adv. Exp. Med. Biol. 2018, 1102, 1–9. [Google Scholar] [CrossRef]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.R.; Beale, D.J.; Paten, A.M.; Kouremenos, K.; Swarup, S.; Schirra, H.J.; Wishart, D. Systems Biology and Multi-Omics Integration: Viewpoints from the Metabolomics Research Community. Metabolites 2019, 9, 76. [Google Scholar] [CrossRef]
- Dihazi, H.; Asif, A.R.; Beißbarth, T.; Bohrer, R.; Feussner, K.; Feussner, I.; Jahn, O.; Lenz, C.; Majcherczyk, A.; Schmidt, B.; et al. Integrative omics—From data to biology. Expert Rev. Proteom. 2018, 15, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Raufaste-Cazavieille, V.; Santiago, R.; Droit, A. Multi-omics analysis: Paving the path toward achieving precision medicine in cancer treatment and immuno-oncology. Front. Mol. Biosci. 2022, 9, 962743. [Google Scholar] [CrossRef] [PubMed]
- Sen, P.; Orešič, M. Integrating Omics Data in Genome-Scale Metabolic Modeling: A Methodological Perspective for Precision Medicine. Metabolites 2023, 13, 855. [Google Scholar] [CrossRef]
- Misra, B.B.; Langefeld, C.D.; Olivier, M.; Cox, L.A. Integrated Omics: Tools, Advances, and Future Approaches. J. Mol. Endocrinol. 2018, 62, R21–R45. [Google Scholar] [CrossRef]
- Katara, A.; Chand, S.; Chaudhary, H.; Chaudhry, V.; Chandra, H.; Dubey, R.C. Evolution and applications of Next Generation Sequencing and its intricate relations with chromatographic and spectrometric techniques in modern day sciences. J. Chromatogr. Open 2024, 5, 100121. [Google Scholar] [CrossRef]
- McGuire, A.L.; Gabriel, S.; Tishkoff, S.A.; Wonkam, A.; Chakravarti, A.; Furlong, E.E.M.; Treutlein, B.; Meissner, A.; Chang, H.Y.; López-Bigas, N.; et al. The road ahead in genetics and genomics. Nat. Rev. Genet. 2020, 21, 581–596. [Google Scholar] [CrossRef]
- Cano-Gamez, E.; Trynka, G. From GWAS to Function: Using Functional Genomics to Identify the Mechanisms Underlying Complex Diseases. Front. Genet. 2020, 11, 505357. [Google Scholar] [CrossRef]
- Jerkovic’, I.; Cavalli, G. Understanding 3D genome organization by multidisciplinary methods. Nat. Rev. Mol. Cell Biol. 2021, 22, 511–528. [Google Scholar] [CrossRef]
- Schneider, M.; Radoux, C.J.; Hercules, A.; Ochoa, D.; Dunham, I.; Zalmas, L.-P.; Hessler, G.; Ruf, S.; Shanmugasundaram, V.; Hann, M.M.; et al. The PROTACtable genome. Nat. Rev. Drug Discov. 2021, 20, 789–797. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Song, J.; Kong, L.; Yuan, T.; Li, W.; Zhang, W.; Hou, B.; Lu, Y.; Du, G. The strategies and techniques of drug discovery from natural products. Pharmacol. Ther. 2020, 216, 107686. [Google Scholar] [CrossRef] [PubMed]
- Kiriiri, G.K.; Njogu, P.M.; Mwangi, A.N. Exploring different approaches to improve the success of drug discovery and development projects: A review. Future J. Pharm. Sci. 2020, 6, 27. [Google Scholar] [CrossRef]
- Goel, M.; Sun, H.; Jiao, W.-B.; Schneeberger, K. SyRI: Finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome Biol. 2019, 20, 277. [Google Scholar] [CrossRef]
- Blair, C.; Ané, C. Phylogenetic Trees and Networks Can Serve as Powerful and Complementary Approaches for Analysis of Genomic Data. Syst. Biol. 2019, 69, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, C.L.M.; Booth, T.J.; van Wersch, B.; van Grieken, L.; Medema, M.H.; Chooi, Y.-H. Cblaster: A remote search tool for rapid identification and visualization of homologous gene clusters. Bioinform. Adv. 2021, 1, vbab016. [Google Scholar] [CrossRef]
- Vishwanath, P.P.; Bidaramali, V.; Lata, S.; Yadav, R.K. Transcriptomics: Illuminating the molecular landscape of vegetable crops: A review. J. Plant Biochem. Biotechnol. 2024. [Google Scholar] [CrossRef]
- Lee, J.-Y. The Principles and Applications of High-Throughput Sequencing Technologies. Dev. Reprod. 2023, 27, 9–24. [Google Scholar] [CrossRef]
- Nguyen, H.L.; Duviau, M.-P.; Cocaign-Bousquet, M.; Nouaille, S.; Girbal, L. Multiplexing polysome profiling experiments to study translation in Escherichia coli. PLoS ONE 2019, 14, e0212297. [Google Scholar] [CrossRef]
- Pringle, E.S.; McCormick, C.; Cheng, Z. Polysome Profiling Analysis of mRNA and Associated Proteins Engaged in Translation. Curr. Protoc. Mol. Biol. 2019, 125, e79. [Google Scholar] [CrossRef]
- Brar, G.A.; Weissman, J.S. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat. Rev. Mol. Cell Biol. 2015, 16, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Bellato, H.M.; Lorent, J.; Lupinacci, F.C.S.; Oertlin, C.; van Hoef, V.; Andrade, V.P.; Roffé, M.; Masvidal, L.; Hajj, G.N.M.; et al. Polysome-profiling in small tissue samples. Nucleic Acids Res. 2018, 46, e3. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, G.; Voichek, Y.; Rabani, M.; Benjamin, S.; Gilad, S.; Amit, I.; Oren, M. Simultaneous measurement of genome-wide transcription elongation speeds and rates of RNA polymerase II transition into active elongation with 4sUDRB-seq. Nat. Protoc. 2015, 10, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, G.; Voichek, Y.; Benjamin, S.; Gilad, S.; Amit, I.; Oren, M. 4sUDRB-seq: Measuring genomewide transcriptional elongation rates and initiation frequencies within cells. Genome Biol. 2014, 15, R69. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-O.; Fu, Y.; Mou, H.; Xue, W.; Weng, Z. The temporal landscape of recursive splicing during Pol II transcription elongation in human cells. PLoS Genet. 2018, 14, e1007579. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, H.; Kohnen, M.V.; Prasad, K.V.S.K.; Gu, L.; Reddy, A.S.N. Analysis of Transcriptome and Epitranscriptome in Plants Using PacBio Iso-Seq and Nanopore-Based Direct RNA Sequencing. Front. Genet. 2019, 10, 430951. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Xu, X.; Wang, W.; Tian, T.; Zhu, Z.; Yang, C. Single cell transcriptomics: Moving towards multi-omics. Analyst 2019, 144, 3172–3189. [Google Scholar] [CrossRef] [PubMed]
- Adil, A.; Kumar, V.; Jan, A.T.; Asger, M. Single-Cell Transcriptomics: Current Methods and Challenges in Data Acquisition and Analysis. Front. Neurosci. 2021, 15, 591122. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Li, G.; Huang, T.; Geng, W.; Pei, H.; Yang, J.; Zhu, M.; Zhang, P.; Hou, R.; Tian, G.; et al. Single-cell technologies: From research to application. Innovation 2022, 3, 100342. [Google Scholar] [CrossRef]
- Salcher, S.; Heidegger, I.; Untergasser, G.; Fotakis, G.; Scheiber, A.; Martowicz, A.; Noureen, A.; Krogsdam, A.; Schatz, C.; Schäfer, G.; et al. Comparative analysis of 10X Chromium vs. BD Rhapsody whole transcriptome single-cell sequencing technologies in complex human tissues. Heliyon 2024, 10, e28358. [Google Scholar] [CrossRef]
- Qiu, Q.; Hu, P.; Qiu, X.; Govek, K.W.; Cámara, P.G.; Wu, H. Massively parallel and time-resolved RNA sequencing in single cells with scNT-seq. Nat. Methods 2020, 17, 991–1001. [Google Scholar] [CrossRef]
- Andreatta, M.; Corria-Osorio, J.; Müller, S.; Cubas, R.; Coukos, G.; Carmona, S.J. Interpretation of T cell states from single-cell transcriptomics data using reference atlases. Nat. Commun. 2021, 12, 2965. [Google Scholar] [CrossRef]
- Adema, K.; Schon, M.A.; Nodine, M.D.; Kohlen, W. Lost in space: What single-cell RNA sequencing cannot tell you. Trends Plant Sci. 2024. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, B.; Zhao, G.; Lee, Y.; Buzdin, A.; Mu, X.; Zhao, J.; Chen, H.; Li, X. Spatial transcriptomics: Technologies, applications and experimental considerations. Genomics 2023, 115, 110671. [Google Scholar] [CrossRef]
- Chen, T.-Y.; You, L.; Hardillo, J.A.U.; Chien, M.-P. Spatial Transcriptomic Technologies. Cells 2023, 12, 2042. [Google Scholar] [CrossRef]
- Piñeiro, A.J.; Houser, A.E.; Ji, A.L. Research Techniques Made Simple: Spatial Transcriptomics. J. Investig. Dermatol. 2022, 142, 993–1001.e1001. [Google Scholar] [CrossRef]
- Shi, W.; Zhang, J.; Huang, S.; Fan, Q.; Cao, J.; Zeng, J.; Wu, L.; Yang, C. Next-Generation Sequencing-Based Spatial Transcriptomics: A Perspective from Barcoding Chemistry. JACS Au 2024, 4, 1723–1743. [Google Scholar] [CrossRef]
- Chen, A.; Liao, S.; Cheng, M.; Ma, K.; Wu, L.; Lai, Y.; Qiu, X.; Yang, J.; Xu, J.; Hao, S.; et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 2022, 185, 1777–1792.e1721. [Google Scholar] [CrossRef]
- Al-Amrani, S.; Al-Jabri, Z.; Al-Zaabi, A.; Alshekaili, J.; Al-Khabori, M. Proteomics: Concepts and applications in human medicine. World J. Biol. Chem. 2021, 12, 57–69. [Google Scholar] [CrossRef]
- Gutierrez Reyes, C.D.; Jiang, P.; Donohoo, K.; Atashi, M.; Mechref, Y.S. Glycomics and glycoproteomics: Approaches to address isomeric separation of glycans and glycopeptides. J. Sep. Sci. 2021, 44, 403–425. [Google Scholar] [CrossRef]
- Peng, W.; Gutierrez Reyes, C.D.; Gautam, S.; Yu, A.; Cho, B.G.; Goli, M.; Donohoo, K.; Mondello, S.; Kobeissy, F.; Mechref, Y. MS-based glycomics and glycoproteomics methods enabling isomeric characterization. Mass. Spectrom. Rev. 2023, 42, 577–616. [Google Scholar] [CrossRef] [PubMed]
- Alejo-Jacuinde, G.; Nájera-González, H.-R.; Chávez Montes, R.A.; Gutierrez Reyes, C.D.; Barragán-Rosillo, A.C.; Perez Sanchez, B.; Mechref, Y.; López-Arredondo, D.; Yong-Villalobos, L.; Herrera-Estrella, L. Multi-omic analyses reveal the unique properties of chia (Salvia hispanica) seed metabolism. Commun. Biol. 2023, 6, 820. [Google Scholar] [CrossRef]
- Reyes, C.D.G.; Hakim, M.A.; Atashi, M.; Goli, M.; Gautam, S.; Wang, J.; Bennett, A.I.; Zhu, J.; Lubman, D.M.; Mechref, Y. LC-MS/MS Isomeric Profiling of N-Glycans Derived from Low-Abundant Serum Glycoproteins in Mild Cognitive Impairment Patients. Biomolecules 2022, 12, 1657. [Google Scholar] [CrossRef] [PubMed]
- Cho, B.G.; Gutierrez Reyes, C.D.; Mechref, Y. N-Glycomics of Cerebrospinal Fluid: Method Comparison. Molecules 2021, 26, 1712. [Google Scholar] [CrossRef]
- Gonçalves, E.; Poulos, R.C.; Cai, Z.; Barthorpe, S.; Manda, S.S.; Lucas, N.; Beck, A.; Bucio-Noble, D.; Dausmann, M.; Hall, C.; et al. Pan-cancer proteomic map of 949 human cell lines. Cancer Cell 2022, 40, 835–849.e838. [Google Scholar] [CrossRef]
- Rontogianni, S.; Synadaki, E.; Li, B.; Liefaard, M.C.; Lips, E.H.; Wesseling, J.; Wu, W.; Altelaar, M. Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping. Commun. Biol. 2019, 2, 325. [Google Scholar] [CrossRef]
- Dupree, E.J.; Jayathirtha, M.; Yorkey, H.; Mihasan, M.; Petre, B.A.; Darie, C.C. A Critical Review of Bottom-Up Proteomics: The Good, the Bad, and the Future of this Field. Proteomes 2020, 8, 14. [Google Scholar] [CrossRef]
- Toby, T.K.; Fornelli, L.; Srzentić, K.; DeHart, C.J.; Levitsky, J.; Friedewald, J.; Kelleher, N.L. A comprehensive pipeline for translational top-down proteomics from a single blood draw. Nat. Protoc. 2019, 14, 119–152. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Kashina, A. Post-translational Modifications of the Protein Termini. Front. Cell Dev. Biol. 2021, 9, 719590. [Google Scholar] [CrossRef]
- Lee, J.M.; Hammarén, H.M.; Savitski, M.M.; Baek, S.H. Control of protein stability by post-translational modifications. Nat. Commun. 2023, 14, 201. [Google Scholar] [CrossRef]
- Johnson, E.C.B.; Bian, S.; Haque, R.U.; Carter, E.K.; Watson, C.M.; Gordon, B.A.; Ping, L.; Duong, D.M.; Epstein, M.P.; McDade, E.; et al. Cerebrospinal fluid proteomics define the natural history of autosomal dominant Alzheimer’s disease. Nat. Med. 2023, 29, 1979–1988. [Google Scholar] [CrossRef] [PubMed]
- Palstrøm, N.B.; Matthiesen, R.; Rasmussen, L.M.; Beck, H.C. Recent Developments in Clinical Plasma Proteomics-Applied to Cardiovascular Research. Biomedicines 2022, 10, 162. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Huo, S.; Zhang, M.; Qian, S.; Zhu, X.; Pu, J.; Rasam, S.; Xue, C.; Shen, S.; An, B.; et al. In-depth mapping of protein localizations in whole tissue by micro-scaffold assisted spatial proteomics (MASP). Nat. Commun. 2022, 13, 7736. [Google Scholar] [CrossRef] [PubMed]
- Kangas, P.; Nyman, T.A.; Metsähonkala, L.; Burns, C.; Tempest, R.; Williams, T.; Karttunen, J.; Jokinen, T.S. Towards optimised extracellular vesicle proteomics from cerebrospinal fluid. Sci. Rep. 2023, 13, 9564. [Google Scholar] [CrossRef] [PubMed]
- Van den Ackerveken, P.; Lobbens, A.; Turatsinze, J.-V.; Solis-Mezarino, V.; Völker-Albert, M.; Imhof, A.; Herzog, M. A novel proteomics approach to epigenetic profiling of circulating nucleosomes. Sci. Rep. 2021, 11, 7256. [Google Scholar] [CrossRef] [PubMed]
- Petrosius, V.; Schoof, E.M. Recent advances in the field of single-cell proteomics. Transl. Oncol. 2023, 27, 101556. [Google Scholar] [CrossRef]
- Liu, R.; Wu, J.; Guo, H.; Yao, W.; Li, S.; Lu, Y.; Jia, Y.; Liang, X.; Tang, J.; Zhang, H. Post-translational modifications of histones: Mechanisms, biological functions, and therapeutic targets. MedComm 2023, 4, e292. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Sabatier, P.; Hoeven, L.V.D.; Phlairaharn, T.; Hartlmayr, D.; Izaguirre, F.; Seth, A.; Joshi, H.J.; Bekker-Jensen, D.B.; Bache, N.; et al. High-throughput and scalable single cell proteomics identifies over 5000 proteins per cell. bioRxiv 2023. [Google Scholar] [CrossRef]
- Chandler, K.B.; Leon, D.R.; Kuang, J.; Meyer, R.D.; Rahimi, N.; Costello, C.E. N-Glycosylation regulates ligand-dependent activation and signaling of vascular endothelial growth factor receptor 2 (VEGFR2). J. Biol. Chem. 2019, 294, 13117–13130. [Google Scholar] [CrossRef]
- Perkey, E.; Maurice De Sousa, D.; Carrington, L.; Chung, J.; Dils, A.; Granadier, D.; Koch, U.; Radtke, F.; Ludewig, B.; Blazar, B.R.; et al. GCNT1-Mediated O-Glycosylation of the Sialomucin CD43 Is a Sensitive Indicator of Notch Signaling in Activated T Cells. J. Immunol. 2020, 204, 1674–1688. [Google Scholar] [CrossRef]
- Van Coillie, J.; Schulz, M.A.; Bentlage, A.E.H.; de Haan, N.; Ye, Z.; Geerdes, D.M.; van Esch, W.J.E.; Hafkenscheid, L.; Miller, R.L.; Narimatsu, Y.; et al. Role of N-Glycosylation in FcγRIIIa interaction with IgG. Front. Immunol. 2022, 13, 987151. [Google Scholar] [CrossRef]
- Ozdilek, A.; Paschall, A.V.; Dookwah, M.; Tiemeyer, M.; Avci, F.Y. Host protein glycosylation in nucleic acid vaccines as a potential hurdle in vaccine design for nonviral pathogens. Proc. Natl. Acad. Sci. USA 2020, 117, 1280–1282. [Google Scholar] [CrossRef]
- Onigbinde, S.; Reyes, C.D.G.; Fowowe, M.; Daramola, O.; Atashi, M.; Bennett, A.I.; Mechref, Y. Variations in O-Glycosylation Patterns Influence Viral Pathogenicity, Infectivity, and Transmissibility in SARS-CoV-2 Variants. Biomolecules 2023, 13, 1467. [Google Scholar] [CrossRef]
- Munkley, J.; Mills, I.G.; Elliott, D.J. The role of glycans in the development and progression of prostate cancer. Nat. Rev. Urol. 2016, 13, 324–333. [Google Scholar] [CrossRef]
- Zhou, S.; Veillon, L.; Dong, X.; Huang, Y.; Mechref, Y. Direct comparison of derivatization strategies for LC-MS/MS analysis of N-glycans. Analyst 2017, 142, 4446–4455. [Google Scholar] [CrossRef]
- Jiang, P.; Huang, Y.; Gutierrez Reyes, C.D.; Zhong, J.; Mechref, Y. Isomeric Separation of α2,3/α2,6-Linked 2-Aminobenzamide (2AB)-Labeled Sialoglycopeptides by C18-LC-MS/MS. Anal. Chem. 2023, 95, 18388–18397. [Google Scholar] [CrossRef]
- Zhong, J.; Huang, Y.; Mechref, Y. Derivatization of Sialylated Glycopeptides (DOSG) Enabling Site-Specific Isomeric Profiling Using LC-MS/MS. Anal. Chem. 2021, 93, 5763–5772. [Google Scholar] [CrossRef]
- Jiang, K.; Aloor, A.; Qu, J.; Xiao, C.; Wu, Z.; Ma, C.; Zhang, L.; Wang, P.G. Rapid and sensitive MALDI MS analysis of oligosaccharides by using 2-hydrazinopyrimidine as a derivative reagent and co-matrix. Anal. Bioanal. Chem. 2017, 409, 421–429. [Google Scholar] [CrossRef]
- Wu, Y.; Sha, Q.; Du, J.; Wang, C.; Zhang, L.; Liu, B.F.; Lin, Y.; Liu, X. Determination of N-glycans by high performance liquid chromatography using 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate as the glycosylamine labeling reagent. J. Chromatogr. A 2018, 1535, 114–122. [Google Scholar] [CrossRef]
- Ruhaak, L.R.; Zauner, G.; Huhn, C.; Bruggink, C.; Deelder, A.M.; Wuhrer, M. Glycan labeling strategies and their use in identification and quantification. Anal. Bioanal. Chem. 2010, 397, 3457–3481. [Google Scholar] [CrossRef]
- Mechref, Y.; Kang, P.; Novotny, M.V. Solid-phase permethylation for glycomic analysis. Methods Mol. Biol. 2009, 534, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Saraswat, M.; Mangalaparthi, K.K.; Garapati, K.; Pandey, A. TMT-Based Multiplexed Quantitation of N-Glycopeptides Reveals Glycoproteome Remodeling Induced by Oncogenic Mutations. ACS Omega 2022, 7, 11023–11032. [Google Scholar] [CrossRef] [PubMed]
- Dube, D.H.; Bertozzi, C.R. Glycans in cancer and inflammation—Potential for therapeutics and diagnostics. Nat. Rev. Drug Discov. 2005, 4, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, N.; Kizuka, Y. Glycans and cancer: Role of N-glycans in cancer biomarker, progression and metastasis, and therapeutics. Adv. Cancer Res. 2015, 126, 11–51. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez Reyes, C.D.; Huang, Y.; Atashi, M.; Zhang, J.; Zhu, J.; Liu, S.; Parikh, N.D.; Singal, A.G.; Dai, J.; Lubman, D.M.; et al. PRM-MS Quantitative Analysis of Isomeric N-Glycopeptides Derived from Human Serum Haptoglobin of Patients with Cirrhosis and Hepatocellular Carcinoma. Metabolites 2021, 11, 563. [Google Scholar] [CrossRef] [PubMed]
- Veillon, L.; Huang, Y.; Peng, W.; Dong, X.; Cho, B.G.; Mechref, Y. Characterization of isomeric glycan structures by LC-MS/MS. Electrophoresis 2017, 38, 2100–2114. [Google Scholar] [CrossRef]
- Kizuka, Y.; Kitazume, S.; Taniguchi, N. N-glycan and Alzheimer’s disease. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2447–2454. [Google Scholar] [CrossRef]
- Tena, J.; Tang, X.; Zhou, Q.; Harvey, D.; Barajas-Mendoza, M.; Jin, L.W.; Maezawa, I.; Zivkovic, A.M.; Lebrilla, C.B. Glycosylation alterations in serum of Alzheimer’s disease patients show widespread changes in N-glycosylation of proteins related to immune function, inflammation, and lipoprotein metabolism. Alzheimers Dement. 2022, 14, e12309. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Alves, I.; Gaifem, J.; Rabinovich, G.A. Immune regulatory networks coordinated by glycans and glycan-binding proteins in autoimmunity and infection. Cell Mol. Immunol. 2023, 20, 1101–1113. [Google Scholar] [CrossRef]
- Chien, M.W.; Fu, S.H.; Hsu, C.Y.; Liu, Y.W.; Sytwu, H.K. The Modulatory Roles of N-glycans in T-Cell-Mediated Autoimmune Diseases. Int. J. Mol. Sci. 2018, 19, 780. [Google Scholar] [CrossRef]
- Unione, L.; Gimeno, A.; Valverde, P.; Calloni, I.; Coelho, H.; Mirabella, S.; Poveda, A.; Arda, A.; Jimenez-Barbero, J. Glycans in Infectious Diseases. A Molecular Recognition Perspective. Curr. Med. Chem. 2017, 24, 4057–4080. [Google Scholar] [CrossRef] [PubMed]
- Reyes, C.D.G.; Onigbinde, S.; Sanni, A.; Bennett, A.I.; Jiang, P.; Daramola, O.; Ahmadi, P.; Fowowe, M.; Atashi, M.; Sandilya, V.; et al. N-Glycome Profile of the Spike Protein S1: Systemic and Comparative Analysis from Eleven Variants of SARS-CoV-2. Biomolecules 2023, 13, 1421. [Google Scholar] [CrossRef]
- Freitas, D.; Campos, D.; Gomes, J.; Pinto, F.; Macedo, J.A.; Matos, R.; Mereiter, S.; Pinto, M.T.; Polónia, A.; Gartner, F.; et al. O-glycans truncation modulates gastric cancer cell signaling and transcription leading to a more aggressive phenotype. EBioMedicine 2019, 40, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Amon, R.; Reuven, E.M.; Leviatan Ben-Arye, S.; Padler-Karavani, V. Glycans in immune recognition and response. Carbohydr. Res. 2014, 389, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Liwosz, A.; Lei, T.; Kukuruzinska, M.A. N-glycosylation affects the molecular organization and stability of E-cadherin junctions. J. Biol. Chem. 2006, 281, 23138–23149. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.G.; Balmaña, M.; Macedo, J.A.; Poças, J.; Fernandes, Â.; de-Freitas-Junior, J.C.M.; Pinho, S.S.; Gomes, J.; Magalhães, A.; Gomes, C.; et al. Glycosylation in cancer: Selected roles in tumour progression, immune modulation and metastasis. Cell Immunol. 2018, 333, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ma, C.; Chin, L.S.; Li, L. Integrative glycoproteomics reveals protein N-glycosylation aberrations and glycoproteomic network alterations in Alzheimer’s disease. Sci. Adv. 2020, 6, eabc5802. [Google Scholar] [CrossRef] [PubMed]
- Bennun, S.V.; Hizal, D.B.; Heffner, K.; Can, O.; Zhang, H.; Betenbaugh, M.J. Systems Glycobiology: Integrating Glycogenomics, Glycoproteomics, Glycomics, and Other ‘Omics Data Sets to Characterize Cellular Glycosylation Processes. J. Mol. Biol. 2016, 428, 3337–3352. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, A.; Relvas-Santos, M.; Azevedo, R.; Santos, L.L.; Ferreira, J.A. Protein Glycosylation and Tumor Microenvironment Alterations Driving Cancer Hallmarks. Front. Oncol. 2019, 9, 380. [Google Scholar] [CrossRef]
- Guo, S.; Zhang, C.; Le, A. The limitless applications of single-cell metabolomics. Curr. Opin. Biotechnol. 2021, 71, 115–122. [Google Scholar] [CrossRef]
- Blanco, A.; Blanco, G. Chapter 5—Lipids. In Medical Biochemistry, 2nd ed.; Blanco, A., Blanco, G., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 105–129. [Google Scholar]
- Li, Z.; Cheng, S.; Lin, Q.; Cao, W.; Yang, J.; Zhang, M.; Shen, A.; Zhang, W.; Xia, Y.; Ma, X.; et al. Single-cell lipidomics with high structural specificity by mass spectrometry. Nat. Commun. 2021, 12, 2869. [Google Scholar] [CrossRef]
- Yang, K.; Han, X. Lipidomics: Techniques, Applications, and Outcomes Related to Biomedical Sciences. Trends Biochem. Sci. 2016, 41, 954–969. [Google Scholar] [CrossRef] [PubMed]
- Hoang, G.; Udupa, S.; Le, A. Chapter Six—Application of metabolomics technologies toward cancer prognosis and therapy. In International Review of Cell and Molecular Biology; Montrose, D.C., Galluzzi, L., Eds.; Academic Press: Cambridge, MA, USA, 2019; Volume 347, pp. 191–223. [Google Scholar]
- Wang, Z.; Zhu, H.; Xiong, W. Advances in mass spectrometry-based multi-scale metabolomic methodologies and their applications in biological and clinical investigations. Sci. Bull. 2023, 68, 2268–2284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Bao, C.; Jiang, L.; Wang, S.; Wang, K.; Lu, C.; Fang, H. When cancer drug resistance meets metabolomics (bulk, single-cell and/or spatial): Progress, potential, and perspective. Front. Oncol. 2022, 12, 1054233. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Bhat, G.R.; Sethi, I.; Sadida, H.Q.; Rah, B.; Mir, R.; Algehainy, N.; Albalawi, I.A.; Masoodi, T.; Subbaraj, G.K.; Jamal, F.; et al. Cancer cell plasticity: From cellular, molecular, and genetic mechanisms to tumor heterogeneity and drug resistance. Cancer Metastasis Rev. 2024, 43, 197–228. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xu, F.; Yao, J.; Mao, C.; Zhu, M.; Qian, M.; Hu, J.; Zhong, H.; Zhou, J.; Shi, X.; et al. Single-cell metabolic fingerprints discover a cluster of circulating tumor cells with distinct metastatic potential. Nat. Commun. 2023, 14, 2485. [Google Scholar] [CrossRef]
- Zhu, G.; Zhang, W.; Zhao, Y.; Chen, T.; Yuan, H.; Liu, Y.; Guo, G.; Liu, Z.; Wang, X. Single-Cell Metabolomics-Based Strategy for Studying the Mechanisms of Drug Action. Anal. Chem. 2023, 95, 4712–4720. [Google Scholar] [CrossRef]
- Zeng, Q.; Mousa, M.; Nadukkandy, A.S.; Franssens, L.; Alnaqbi, H.; Alshamsi, F.Y.; Safar, H.A.; Carmeliet, P. Understanding tumour endothelial cell heterogeneity and function from single-cell omics. Nat. Rev. Cancer 2023, 23, 544–564. [Google Scholar] [CrossRef]
- Chen, X.; Peng, Z.; Yang, Z. Metabolomics studies of cell–cell interactions using single cell mass spectrometry combined with fluorescence microscopy. Chem. Sci. 2022, 13, 6687–6695. [Google Scholar] [CrossRef]
- León-Letelier, R.A.; Dou, R.; Vykoukal, J.; Yip-Schneider, M.T.; Maitra, A.; Irajizad, E.; Wu, R.; Dennison, J.B.; Do, K.-A.; Zhang, J.; et al. Contributions of the Microbiome-Derived Metabolome for Risk Assessment and Prognostication of Pancreatic Cancer. Clin. Chem. 2024, 70, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Zahra, W.; Rai, S.N.; Birla, H.; Singh, S.S.; Dilnashin, H.; Rathore, A.S.; Singh, S.P. The Global Economic Impact of Neurodegenerative Diseases: Opportunities and Challenges. In Bioeconomy for Sustainable Development; Keswani, C., Ed.; Springer: Singapore, 2020; pp. 333–345. [Google Scholar]
- Zheng, Y.; Liu, Z.; Xing, J.; Zheng, Z.; Pi, Z.; Song, F.; Liu, S. In situ analysis of single cell and biological samples with rGO-Cu functional probe ESI-MS spectrometry. Talanta 2020, 211, 120751. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Wu, B.; Yang, C.; Xu, X.; Shi, Z.; Jiang, D. Targeted Metabolomics Analysis of Serum Amino Acids in T2DM Patients. Diabetes Metab. Syndr. Obes. 2024, 17, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Li, G.; Zhang, X.; Zhang, C.; Li, M.; Qiu, Y.; Sun, W.; Dong, Y.; Li, S.; Li, J. Targeted metabolomics-based understanding of the sleep disturbances in drug-naïve patients with schizophrenia. BMC Psychiatry 2024, 24, 355. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.-J.; Kim, D.J.; Kang, D.-W.; Yang, W.; Jeong, H.-Y.; Kim, J.-M.; Ko, S.-B.; Lee, S.-H.; Yoon, B.-W.; Cho, J.-Y.; et al. Targeted Metabolomic Biomarkers for Stroke Subtyping. Transl. Stroke Res. 2024, 15, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; He, T.; Wang, H. Non-targeted metabolomics study for discovery of hepatocellular carcinoma serum diagnostic biomarker. J. Pharm. Biomed. Anal. 2024, 239, 115869. [Google Scholar] [CrossRef]
- Sun, S.; Chen, M.; Zhang, T.; Wang, Y.; Shen, W.; Zhang, T.; Liu, J.; Lan, H.; Zhao, J.; Lin, F.; et al. Identification of Key Factors in Cartilage Tissue During the Progression of Osteoarthritis Using a Non-targeted Metabolomics Strategy. Phenomics 2024. [Google Scholar] [CrossRef]
- Chen, W.; Guo, W.; Li, Y.; Chen, M. Integrative analysis of metabolomics and transcriptomics to uncover biomarkers in sepsis. Sci. Rep. 2024, 14, 9676. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, G.; Xie, C.; Zhou, Y.; Yang, R.; Wu, J.; Xu, J.; Tu, K. Metabolomics Analysis of Different Quinoa Cultivars Based on UPLC-ZenoTOF-MS/MS and Investigation into Their Antioxidant Characteristics. Plants 2024, 13, 240. [Google Scholar] [CrossRef]
- Windarsih, A.; Arifah, M.F.; Utami, I.D.; Suratno; Rohman, A. Detection of goat milk adulteration in horse milk using LC-HRMS-based non-targeted metabolomics and chemometrics. Chem. Pap. 2024, 78, 809–821. [Google Scholar] [CrossRef]
- Tian, Y.; Wang, W.; Jiang, W.; Zhang, G.; He, J.; Dong, S.; Zhou, Y.; Yang, W.; Tang, Q.; Yu, Y.; et al. Non-targeted metabolomics provides insights into the distinct amino acid and lipid metabolism in liver tissues of rainbow trout (Oncorhynchus mykiss) cultured in seawater at different temperatures. Aquaculture 2024, 579, 740188. [Google Scholar] [CrossRef]
- Yang, S.; Pathak, S.; Tang, H.; Zhang, D.; Chen, Y.; Ntezimana, B.; Ni, D.; Yu, Z. Non-Targeted Metabolomics Reveals the Effects of Different Rolling Methods on Black Tea Quality. Foods 2024, 13, 325. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Ma, R.; Wang, Y.; Tong, L.; Wen, W.; Mu, T.; Tian, J.; Yu, B.; Gu, Y.; Zhang, J. Non-targeted metabolomics identifies biomarkers in milk with high and low milk fat percentage. Food Res. Int. 2024, 179, 113989. [Google Scholar] [CrossRef] [PubMed]
- Mo, L.; Fang, L.; Yao, W.; Nie, J.; Dai, J.; Liang, Y.; Qin, L. LC-QTOF/MS-based non-targeted metabolomics to explore the toxic effects of di(2-ethylhexyl) phthalate (DEHP) on Brassica chinensis L. Sci. Total Environ. 2024, 918, 170817. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Wu, H.; Fang, X.; Feng, X.; Zhou, L. The cardiovascular toxicity of polystyrene microplastics in rats: Based on untargeted metabolomics analysis. Front. Pharmacol. 2024, 15, 1336369. [Google Scholar] [CrossRef]
- Xiao, X.; Zhou, Y.; Li, X.; Jin, J.; Durham, J.; Ye, Z.; Wang, Y.; Hennig, B.; Deng, P. 13C-Stable isotope resolved metabolomics uncovers dynamic biochemical landscape of gut microbiome-host organ communications in mice. Microbiome 2024, 12, 90. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Tu, X.; Zang, Q.; Zhu, Y.; Li, L.; Zhang, R.; Abliz, Z. Liquid chromatography–mass spectrometry-based metabolomics and fluxomics reveals the metabolic alterations in glioma U87MG multicellular tumor spheroids versus two-dimensional cell cultures. Rapid Commun. Mass. Spectrom. 2024, 38, e9670. [Google Scholar] [CrossRef]
- Peper, C.J.; Kilgore, M.D.; Jiang, Y.; Xiu, Y.; Xia, W.; Wang, Y.; Shi, M.; Zhou, D.; Dumont, A.S.; Wang, X.; et al. Tracing the path of disruption: 13C isotope applications in traumatic brain injury-induced metabolic dysfunction. CNS Neurosci. Ther. 2024, 30, e14693. [Google Scholar] [CrossRef]
- Osipenko, S.; Bashilov, A.; Vishnevskaya, A.; Rumiantseva, L.; Levashova, A.; Kovalenko, A.; Tupertsev, B.; Kireev, A.; Nikolaev, E.; Kostyukevich, Y. Investigating the Metabolism of Plants Germinated in Heavy Water, D2O, and H218O-Enriched Media Using High-Resolution Mass Spectrometry. Int. J. Mol. Sci. 2023, 24, 15396. [Google Scholar] [CrossRef]
- Brorsen, L.F.; McKenzie, J.S.; Tullin, M.F.; Bendtsen, K.M.S.; Pinto, F.E.; Jensen, H.E.; Haedersdal, M.; Takats, Z.; Janfelt, C.; Lerche, C.M. Cutaneous squamous cell carcinoma characterized by MALDI mass spectrometry imaging in combination with machine learning. Sci. Rep. 2024, 14, 11091. [Google Scholar] [CrossRef]
- Bag, S.; Oetjen, J.; Shaikh, S.; Chaudhary, A.; Arun, P.; Mukherjee, G. Impact of spatial metabolomics on immune-microenvironment in oral cancer prognosis: A clinical report. Mol. Cell. Biochem. 2024, 479, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, I.; Rodriguez-Alvarez, N.; François, L.; Viot, D.; Poosti, F.; Aronica, E.; Dedeurwaerdere, S.; Barton, P.; Cillero-Pastor, B.; Heeren, R.M.A. Spatial omics reveals molecular changes in focal cortical dysplasia type II. Neurobiol. Dis. 2024, 195, 106491. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, N.; Kunzke, T.; Shen, J.; Zens, P.; Prade, V.M.; Feuchtinger, A.; Berezowska, S.; Walch, A. Spatial metabolomics identifies distinct tumor-specific and stroma-specific subtypes in patients with lung squamous cell carcinoma. Npj Precis. Oncol. 2023, 7, 114. [Google Scholar] [CrossRef] [PubMed]
- Conroy, L.R.; Clarke, H.A.; Allison, D.B.; Valenca, S.S.; Sun, Q.; Hawkinson, T.R.; Young, L.E.A.; Ferreira, J.E.; Hammonds, A.V.; Dunne, J.B.; et al. Spatial metabolomics reveals glycogen as an actionable target for pulmonary fibrosis. Nat. Commun. 2023, 14, 2759. [Google Scholar] [CrossRef] [PubMed]
- Züllig, T.; Trötzmüller, M.; Köfeler, H.C. Lipidomics from sample preparation to data analysis: A primer. Anal. Bioanal. Chem. 2020, 412, 2191–2209. [Google Scholar] [CrossRef]
- Kenwood, B.M.; Merrill, A.H. Lipidomics. In Encyclopedia of Cell Biology; Bradshaw, R.A., Stahl, P.D., Eds.; Academic Press: Waltham, MA, USA, 2016; pp. 147–159. [Google Scholar]
- Zhao, Y.-Y.; Cheng, X.-l.; Lin, R.-C. Chapter One—Lipidomics Applications for Discovering Biomarkers of Diseases in Clinical Chemistry. In International Review of Cell and Molecular Biology; Jeon, K.W., Ed.; Academic Press: Cambridge, MA, USA, 2014; Volume 313, pp. 1–26. [Google Scholar]
- Costa, A.C.; Riça, L.B.; van de Bilt, M.; Zandonadi, F.S.; Gattaz, W.F.; Talib, L.L.; Sussulini, A. Application of Lipidomics in Psychiatry: Plasma-Based Potential Biomarkers in Schizophrenia and Bipolar Disorder. Metabolites 2023, 13, 600. [Google Scholar] [CrossRef]
- Chiurchiù, V.; Tiberi, M.; Matteocci, A.; Fazio, F.; Siffeti, H.; Saracini, S.; Mercuri, N.B.; Sancesario, G. Lipidomics of Bioactive Lipids in Alzheimer’s and Parkinson’s Diseases: Where Are We? Int. J. Mol. Sci. 2022, 23, 6235. [Google Scholar] [CrossRef]
- Graham, Z.A.; Siedlik, J.A.; Toro, C.A.; Harlow, L.; Cardozo, C.P. Boldine Alters Serum Lipidomic Signatures after Acute Spinal Cord Transection in Male Mice. Int. J. Environ. Res. Public Health 2023, 20, 6591. [Google Scholar] [CrossRef]
- Senko, D.; Gorovaya, A.; Stekolshchikova, E.; Anikanov, N.; Fedianin, A.; Baltin, M.; Efimova, O.; Petrova, D.; Baltina, T.; Lebedev, M.A.; et al. Time-Dependent Effect of Sciatic Nerve Injury on Rat Plasma Lipidome. Int. J. Mol. Sci. 2022, 23, 15544. [Google Scholar] [CrossRef]
- Meikle, P.J.; Wong, G.; Barlow, C.K.; Kingwell, B.A. Lipidomics: Potential role in risk prediction and therapeutic monitoring for diabetes and cardiovascular disease. Pharmacol. Ther. 2014, 143, 12–23. [Google Scholar] [CrossRef]
- Rasmiena, A.A.; Ng, T.W.; Meikle, P.J. Metabolomics and ischaemic heart disease. Clin. Sci. 2012, 124, 289–306. [Google Scholar] [CrossRef] [PubMed]
- Perrotti, F.; Rosa, C.; Cicalini, I.; Sacchetta, P.; Del Boccio, P.; Genovesi, D.; Pieragostino, D. Advances in Lipidomics for Cancer Biomarkers Discovery. Int. J. Mol. Sci. 2016, 17, 1992. [Google Scholar] [CrossRef] [PubMed]
- Horgusluoglu, E.; Neff, R.; Song, W.-M.; Wang, M.; Wang, Q.; Arnold, M.; Krumsiek, J.; Galindo-Prieto, B.; Ming, C.; Nho, K.; et al. Integrative metabolomics-genomics approach reveals key metabolic pathways and regulators of Alzheimer’s disease. Alzheimer’s Dement. 2022, 18, 1260–1278. [Google Scholar] [CrossRef] [PubMed]
- Bernardes, J.P.; Mishra, N.; Tran, F.; Bahmer, T.; Best, L.; Blase, J.I.; Bordoni, D.; Franzenburg, J.; Geisen, U.; Josephs-Spaulding, J.; et al. Longitudinal Multi-omics Analyses Identify Responses of Megakaryocytes, Erythroid Cells, and Plasmablasts as Hallmarks of Severe COVID-19. Immunity 2020, 53, 1296–1314.e1299. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Hu, E.; Ouyang, J.; Zhong, X.; Wang, F.; Liu, K.; Cai, L.; Zhou, Y.; Wang, Y.; Chen, G.; et al. Integrated omics landscape of hepatocellular carcinoma suggests proteomic subtypes for precision therapy. Cell Rep. Med. 2023, 4, 101315. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Wu, Y.; Cheng, W.; Wu, Y.; Wang, L.; Zhuang, L. Identification of novel prognostic biomarkers by integrating multi-omics data in gastric cancer. BMC Cancer 2021, 21, 460. [Google Scholar] [CrossRef] [PubMed]
- Clark, C.; Dayon, L.; Masoodi, M.; Bowman, G.L.; Popp, J. An integrative multi-omics approach reveals new central nervous system pathway alterations in Alzheimer’s disease. Alzheimer’s Res. Ther. 2021, 13, 71. [Google Scholar] [CrossRef] [PubMed]
- Strefeler, A.; Jan, M.; Quadroni, M.; Teav, T.; Rosenberg, N.; Chatton, J.-Y.; Guex, N.; Gallart-Ayala, H.; Ivanisevic, J. Molecular insights into sex-specific metabolic alterations in Alzheimer’s mouse brain using multi-omics approach. Alzheimer’s Res. Ther. 2023, 15, 8. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.E.; Kemp, M.L. Integration of machine learning and genome-scale metabolic modeling identifies multi-omics biomarkers for radiation resistance. Nat. Commun. 2021, 12, 2700. [Google Scholar] [CrossRef]
- Nativio, R.; Lan, Y.; Donahue, G.; Sidoli, S.; Berson, A.; Srinivasan, A.R.; Shcherbakova, O.; Amlie-Wolf, A.; Nie, J.; Cui, X.; et al. An integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer’s disease. Nat. Genet. 2020, 52, 1024–1035. [Google Scholar] [CrossRef]
- Heintz-Buschart, A.; May, P.; Laczny, C.C.; Lebrun, L.A.; Bellora, C.; Krishna, A.; Wampach, L.; Schneider, J.G.; Hogan, A.; de Beaufort, C.; et al. Integrated multi-omics of the human gut microbiome in a case study of familial type 1 diabetes. Nat. Microbiol. 2016, 2, 16180. [Google Scholar] [CrossRef] [PubMed]
- Lupberger, J.; Croonenborghs, T.; Roca Suarez, A.A.; Van Renne, N.; Jühling, F.; Oudot, M.A.; Virzì, A.; Bandiera, S.; Jamey, C.; Meszaros, G.; et al. Combined Analysis of Metabolomes, Proteomes, and Transcriptomes of Hepatitis C Virus–Infected Cells and Liver to Identify Pathways Associated With Disease Development. Gastroenterology 2019, 157, 537–551.e539. [Google Scholar] [CrossRef]
- Hudson, K.M.; Shiver, E.; Yu, J.; Mehta, S.; Jima, D.D.; Kane, M.A.; Patisaul, H.B.; Cowley, M. Transcriptomic, proteomic, and metabolomic analyses identify candidate pathways linking maternal cadmium exposure to altered neurodevelopment and behavior. Sci. Rep. 2021, 11, 16302. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Zhang, N.; Ren, D.; Yu, C.; Zhao, B.; Zhang, Y. Integrated spatial transcriptome and metabolism study reveals metabolic heterogeneity in human injured brain. Cell Rep. Med. 2023, 4, 101057. [Google Scholar] [CrossRef]
- Vicari, M.; Mirzazadeh, R.; Nilsson, A.; Shariatgorji, R.; Bjärterot, P.; Larsson, L.; Lee, H.; Nilsson, M.; Foyer, J.; Ekvall, M.; et al. Spatial multimodal analysis of transcriptomes and metabolomes in tissues. Nat. Biotechnol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Lopez Soriano, V.; Dueñas Rey, A.; Mukherjee, R.; Inglehearn, C.F.; Coppieters, F.; Bauwens, M.; Willaert, A.; De Baere, E.; Genomics England Research, C. Multi-omics analysis in human retina uncovers ultraconserved cis-regulatory elements at rare eye disease loci. Nat. Commun. 2024, 15, 1600. [Google Scholar] [CrossRef]
- Pattaroni, C.; Begka, C.; Cardwell, B.; Jaffar, J.; Macowan, M.; Harris, N.L.; Westall, G.P.; Marsland, B.J. Multi-omics integration reveals a nonlinear signature that precedes progression of lung fibrosis. Clin. Transl. Immunol. 2024, 13, e1485. [Google Scholar] [CrossRef]
- Garcia-Segura, M.E.; Durainayagam, B.R.; Liggi, S.; Graça, G.; Jimenez, B.; Dehghan, A.; Tzoulaki, I.; Karaman, I.; Elliott, P.; Griffin, J.L. Pathway-based integration of multi-omics data reveals lipidomics alterations validated in an Alzheimer’s disease mouse model and risk loci carriers. J. Neurochem. 2023, 164, 57–76. [Google Scholar] [CrossRef]
- Qiu, C.; Yu, F.; Su, K.; Zhao, Q.; Zhang, L.; Xu, C.; Hu, W.; Wang, Z.; Zhao, L.; Tian, Q.; et al. Multi-omics Data Integration for Identifying Osteoporosis Biomarkers and Their Biological Interaction and Causal Mechanisms. iScience 2020, 23, 100847. [Google Scholar] [CrossRef]
- Byeon, S.K.; Madugundu, A.K.; Garapati, K.; Ramarajan, M.G.; Saraswat, M.; Kumar, M.P.; Hughes, T.; Shah, R.; Patnaik, M.M.; Chia, N.; et al. Development of a multiomics model for identification of predictive biomarkers for COVID-19 severity: A retrospective cohort study. Lancet Digit. Health 2022, 4, e632–e645. [Google Scholar] [CrossRef]
- Huang, C.; You, Z.; He, Y.; Li, J.; Liu, Y.; Peng, C.; Liu, Z.; Liu, X.; Sun, J. Combined transcriptomics and proteomics forecast analysis for potential biomarker in the acute phase of temporal lobe epilepsy. Front. Neurosci. 2023, 17, 1145805. [Google Scholar] [CrossRef] [PubMed]
- Suryadevara, R.; Gregory, A.; Lu, R.; Xu, Z.; Masoomi, A.; Lutz, S.M.; Berman, S.; Yun, J.H.; Saferali, A.; Ryu, M.H.; et al. Blood-based Transcriptomic and Proteomic Biomarkers of Emphysema. Am. J. Respir. Crit. Care Med. 2024, 209, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Sivakumar, P.; Ammar, R.; Thompson, J.R.; Luo, Y.; Streltsov, D.; Porteous, M.; McCoubrey, C.; Cantu, E.; Beers, M.F.; Jarai, G.; et al. Integrated plasma proteomics and lung transcriptomics reveal novel biomarkers in idiopathic pulmonary fibrosis. Respir. Res. 2021, 22, 273. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Fang, J.; Bekris, L.M.; Kim, Y.H.; Pieper, A.A.; Leverenz, J.B.; Cummings, J.; Cheng, F. AlzGPS: A genome-wide positioning systems platform to catalyze multi-omics for Alzheimer’s drug discovery. Alzheimer’s Res. Ther. 2021, 13, 24. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, Y.; Zhao, W.; Jiang, S.; Huang, Y.; Hou, J.; Zhang, X.; Zhai, Z.; Yang, C.; Wang, J.; et al. Integrative multi-omics and drug–response characterization of patient-derived prostate cancer primary cells. Signal Transduct. Target. Ther. 2023, 8, 175. [Google Scholar] [CrossRef] [PubMed]
- Leo, I.R.; Aswad, L.; Stahl, M.; Kunold, E.; Post, F.; Erkers, T.; Struyf, N.; Mermelekas, G.; Joshi, R.N.; Gracia-Villacampa, E.; et al. Integrative multi-omics and drug response profiling of childhood acute lymphoblastic leukemia cell lines. Nat. Commun. 2022, 13, 1691. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Song, M.; Kim, B.; Shim, I.; Kim, D.S.; Natarajan, P.; Do, R.; Won, H.-H. Prioritization of therapeutic targets for dyslipidemia using integrative multi-omics and multi-trait analysis. Cell Rep. Med. 2023, 4, 101112. [Google Scholar] [CrossRef] [PubMed]
- Leinonen, R.; Sugawara, H.; Shumway, M.; International Nucleotide Sequence Database Collaboration. The Sequence Read Archive. Nucleic Acids Res. 2010, 39, D19–D21. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef]
- Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2015, 44, D67–D72. [Google Scholar] [CrossRef]
- Wang, G.; Wu, S.; Xiong, Z.; Qu, H.; Fang, X.; Bao, Y. CROST: A comprehensive repository of spatial transcriptomics. Nucleic Acids Res. 2023, 52, D882–D890. [Google Scholar] [CrossRef] [PubMed]
- Vizcaíno, J.A.; Côté, R.; Reisinger, F.; Foster, J.M.; Mueller, M.; Rameseder, J.; Hermjakob, H.; Martens, L. A guide to the Proteomics Identifications Database proteomics data repository. Proteomics 2009, 9, 4276–4283. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W.; Bandeira, N.; Sharma, V.; Perez-Riverol, Y.; Carver, J.J.; Kundu, D.J.; García-Seisdedos, D.; Jarnuczak, A.F.; Hewapathirana, S.; Pullman, B.S.; et al. The ProteomeXchange consortium in 2020: Enabling ‘big data’ approaches in proteomics. Nucleic Acids Res 2020, 48, D1145–D1152. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Aoki-Kinoshita, K.F.; Ishihama, Y.; Okuda, S. GlycoPOST realizes FAIR principles for glycomics mass spectrometry data. Nucleic Acids Res. 2020, 49, D1523–D1528. [Google Scholar] [CrossRef] [PubMed]
- Haug, K.; Salek, R.M.; Conesa, P.; Hastings, J.; de Matos, P.; Rijnbeek, M.; Mahendraker, T.; Williams, M.; Neumann, S.; Rocca-Serra, P.; et al. MetaboLights—An open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res. 2013, 41, D781–D786. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Beck, S. Making multi-omics data accessible to researchers. Sci. Data 2019, 6, 251. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Chen, F.; Gao, F.; Li, L.; Liu, K.; You, L.; Hua, C.; Yang, F.; Liu, W.; Peng, C.; et al. CNSA: A data repository for archiving omics data. Database 2020, 2020, baaa055. [Google Scholar] [CrossRef] [PubMed]
- Athieniti, E.; Spyrou, G.M. A guide to multi-omics data collection and integration for translational medicine. Comput. Struct. Biotechnol. J. 2023, 21, 134–149. [Google Scholar] [CrossRef]
- Yamada, R.; Okada, D.; Wang, J.; Basak, T.; Koyama, S. Interpretation of omics data analyses. J. Hum. Genet. 2021, 66, 93–102. [Google Scholar] [CrossRef]
- Dunkler, D.; Sánchez-Cabo, F.; Heinze, G. Statistical analysis principles for Omics data. Methods Mol. Biol. 2011, 719, 113–131. [Google Scholar] [CrossRef] [PubMed]
- Rouam, S. False Discovery Rate (FDR). In Encyclopedia of Systems Biology; Dubitzky, W., Wolkenhauer, O., Cho, K.-H., Yokota, H., Eds.; Springer: New York, NY, USA, 2013; pp. 731–732. [Google Scholar]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, S.; Drăghici, S. Identifying Significantly Impacted Pathways and Putative Mechanisms with iPathwayGuide. Curr. Protoc. Bioinform. 2017, 57, 7.15.11–17.15.30. [Google Scholar] [CrossRef]
- Wu, L.; Liu, F.; Cai, H. IOAT: An interactive tool for statistical analysis of omics data and clinical data. BMC Bioinform. 2021, 22, 326. [Google Scholar] [CrossRef]
- Buy, M.; Digan, W.; Chen, X.; Husson, J.; Ménager, M.; Rieux-Laucat, F.; Garcelon, N. A Multi-Omics Common Data Model for Primary Immunodeficiencies. Stud. Health Technol. Inform. 2022, 290, 56–60. [Google Scholar] [CrossRef]
- Harbig, T.A.; Fratte, J.; Krone, M.; Nieselt, K. OmicsTIDE: Interactive exploration of trends in multi-omics data. Bioinform. Adv. 2023, 3, vbac093. [Google Scholar] [CrossRef]
- Fang, Z.; Ma, T.; Tang, G.; Zhu, L.; Yan, Q.; Wang, T.; Celedón, J.C.; Chen, W.; Tseng, G.C. Bayesian integrative model for multi-omics data with missingness. Bioinformatics 2018, 34, 3801–3808. [Google Scholar] [CrossRef]
- Ma, C.; Wu, M.; Ma, S. Analysis of cancer omics data: A selective review of statistical techniques. Brief. Bioinform. 2022, 23, bbab585. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Mukhopadhyay, I. TiMEG: An integrative statistical method for partially missing multi-omics data. Sci. Rep. 2021, 11, 24077. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes, A. Food analysis and Foodomics. J. Chromatogr. A 2009, 1216, 7109. [Google Scholar] [CrossRef]
- Picó, C.; Serra, F.; Rodríguez, A.M.; Keijer, J.; Palou, A. Biomarkers of Nutrition and Health: New Tools for New Approaches. Nutrients 2019, 11, 1092. [Google Scholar] [CrossRef] [PubMed]
- Hill, E.B.; Tang, M.; Long, J.M.; Kemp, J.F.; Westcott, J.L.; Hendricks, A.E.; Reisdorph, N.A.; Campbell, W.W.; Krebs, N.F.; Schaefer, C.; et al. mini-MED: Study protocol for a randomized, multi-intervention, semi-controlled feeding trial of a Mediterranean-amplified vs. habitual Western dietary pattern for the evaluation of food-specific compounds and cardiometabolic health. Trials 2024, 25, 101. [Google Scholar] [CrossRef] [PubMed]
- Trimigno, A.; Khakimov, B.; Savorani, F.; Poulsen, S.K.; Astrup, A.; Dragsted, L.O.; Engelsen, S.B. Human urine 1H NMR metabolomics reveals alterations of protein and carbohydrate metabolism when comparing habitual Average Danish diet vs. healthy New Nordic diet. Nutrition 2020, 79–80, 110867. [Google Scholar] [CrossRef] [PubMed]
- Dilmore, A.H.; Martino, C.; Neth, B.J.; West, K.A.; Zemlin, J.; Rahman, G.; Panitchpakdi, M.; Meehan, M.J.; Weldon, K.C.; Blach, C.; et al. Effects of a ketogenic and low-fat diet on the human metabolome, microbiome, and foodome in adults at risk for Alzheimer’s disease. Alzheimer’s Dement. 2023, 19, 4805–4816. [Google Scholar] [CrossRef]
| Omics Spotlight | Omics Integration | Applications | References |
|---|---|---|---|
| Diagnosis and prognosis | G+T+P+M | Alzheimer’s disease diagnosis | [148] |
| T+Me+scT | Evaluation of Covid-19 prognosis | [149] | |
| G+T+P+pP | Hepatocellular carcinoma classification | [150] | |
| G+T | Identification of prognostic biomarkers for gastric cancer | [151] | |
| P+M+L | Search for pathway alterations in Alzheimer’s disease | [152,153] | |
| G+T+M | Identification of metabolite biomarkers for predicting radiation resistance | [154] | |
| T+P+E | Epigenetic alterations associated with Alzheimer’s disease | [155] | |
| G+T+P | Gastrointestinal microbiome and diabetes mellitus (type 1) | [156] | |
| G+P+M | Liver disease pathogenesis | [157] | |
| T+P+M | Effect of heavy-metal exposure in neurodevelopment | [158] | |
| sT+sM | Spatial resolution approaches to study brain injuries | [159] | |
| sT+sM | Multimodal spatial approach used in Parkinson’s disease | [160] | |
| scT+E | Identification of genomic variants associated with eye diseases | [161] | |
| Biomarker | T+M+L | Progression of lung fibrosis | [162] |
| G+T+P | Metabolic mapping of Alzheimer’s disease | [163] | |
| G+T+M+Me | Identification of osteoporosis biomarkers | [164] | |
| M+L | Predictive biomarkers for COVID-19 severity | [165] | |
| G+T+P+pP | Exploration for Hepatocellular carcinoma biomarkers | [150] | |
| P+Gly | Identification of mild cognitive impairment biomarkers | [53] | |
| T+P | Temporal lobe epilepsy biomarkers | [166] | |
| T+P | Emphysema biomarkers | [167] | |
| T+P | Idiopathic pulmonary fibrosis | [168] | |
| Drug targets and therapeutics | G+T+P | Alzheimer’s drug discovery | [169] |
| G+T+P | Prostate cancer diagnosis and therapies | [170] | |
| P+Gly | Glycosylation profile of the S1 protein of eleven SARS-CoV-2 variants | [92] | |
| G+T | Drug response profiling of childhood acute lymphoblastic leukemia cell lines | [171] | |
| G+T | Prioritization of therapeutic targets for dyslipidemia | [172] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gutierrez Reyes, C.D.; Alejo-Jacuinde, G.; Perez Sanchez, B.; Chavez Reyes, J.; Onigbinde, S.; Mogut, D.; Hernández-Jasso, I.; Calderón-Vallejo, D.; Quintanar, J.L.; Mechref, Y. Multi Omics Applications in Biological Systems. Curr. Issues Mol. Biol. 2024, 46, 5777-5793. https://doi.org/10.3390/cimb46060345
Gutierrez Reyes CD, Alejo-Jacuinde G, Perez Sanchez B, Chavez Reyes J, Onigbinde S, Mogut D, Hernández-Jasso I, Calderón-Vallejo D, Quintanar JL, Mechref Y. Multi Omics Applications in Biological Systems. Current Issues in Molecular Biology. 2024; 46(6):5777-5793. https://doi.org/10.3390/cimb46060345
Chicago/Turabian StyleGutierrez Reyes, Cristian D., Gerardo Alejo-Jacuinde, Benjamin Perez Sanchez, Jesus Chavez Reyes, Sherifdeen Onigbinde, Damir Mogut, Irma Hernández-Jasso, Denisse Calderón-Vallejo, J. Luis Quintanar, and Yehia Mechref. 2024. "Multi Omics Applications in Biological Systems" Current Issues in Molecular Biology 46, no. 6: 5777-5793. https://doi.org/10.3390/cimb46060345
APA StyleGutierrez Reyes, C. D., Alejo-Jacuinde, G., Perez Sanchez, B., Chavez Reyes, J., Onigbinde, S., Mogut, D., Hernández-Jasso, I., Calderón-Vallejo, D., Quintanar, J. L., & Mechref, Y. (2024). Multi Omics Applications in Biological Systems. Current Issues in Molecular Biology, 46(6), 5777-5793. https://doi.org/10.3390/cimb46060345







