Abstract
The Asteraceae are widely distributed throughout the world, with diverse functions and large genomes. Many of these genes remain undiscovered and unstudied. In this study, we discovered a new gene ClNUM1 in Chrysanthemum lavandulifolium and studied its function. In this study, bioinformatics, RT-qPCR, paraffin sectioning, and tobacco transgenics were utilized to bioinformatically analyze and functionally study the three variable splice variants of the unknown gene ClNUM1 cloned from C. lavandulifolium. The results showed that ClNUM1.1 and ClNUM1.2 had selective 3′ splicing and selective 5′ splicing, and ClNUM1.3 had selective 5′ splicing. When the corresponding transgenic tobacco plants were subjected to abiotic stress treatment, in the tobacco seedlings, the ClNUM1.1 gene and the ClNUM1.2 gene enhanced salt and low-temperature tolerance and the ClNUM1.3 gene enhanced low-temperature tolerance; in mature tobacco plants, the ClNUM1.1 gene was able to enhance salt and low-temperature tolerance, and the ClNUM1.2 and ClNUM1.3 genes were able to enhance low-temperature tolerance. In summary, there are differences in the functions of the different splice variants and the different seedling stages of transgenic tobacco, but all of them enhanced the resistance of tobacco to a certain extent. The analysis and functional characterization of the ClNUM1 gene provided new potential genes and research directions for abiotic resistance breeding in Chrysanthemum.
1. Introduction
Chrysanthemums are traditional Chinese flowers and among the most important ornamental flowers in the world, with high ornamental and application value [1]; however, at the same time, Chrysanthemums are very susceptible to abiotic stresses, which greatly limits their application, and their genetic background is relatively complex, so progress in Chrysanthemum resistance research has been relatively slow [2]. C. lavandulifolium is a diploid plant of the genus Chrysanthemum in the family Asteraceae with a relatively simple genetic background and is also one of the important parents of modern Chrysanthemum species, making it a model plant for the study of plants in the family Asteraceae. Therefore, genetics studies in C. lavandulifolium are important for resistance breeding in Chrysanthemum [3].
Studies on the resistance of Chrysanthemum plants have been carried out in many aspects, such as heat resistance and insect resistance, and the main bases are NAC, MYB, and WRKY transcription factors family [4,5,6,7,8,9,10,11,12]. For example, the SND1 and NST1 genes in the NAC family regulate secondary cell wall and lignin synthesis, increasing the broad-spectrum resistance of Leucanthemella linearis [13]. Chrysanthemum × morifolium CmWRKY15-1 enhanced resistance to chrysanthemum white rust by regulating the expression of CmNPR1 [14]. Chrysanthemum indicum var.aromaticum CiMYB32 responds to drought stress [15]. In the process of studying the NST1 gene of C. lavandulifolium, we accidentally cloned the unknown gene ClNUM1 and its alternative splicing and the gene has a conserved structural domain of Nuclease-associated DNA-binding domain 3 (NUMOD3).
NUMOD3 is a homing endonucleases and related proteins. Sitbon et al. identified four new short conserved sequence structural domains in homing endonucleases and related proteins [16]. NUMODs were named the new structural domains nuclear-associated modular DNA-binding structural domains. These domains are modular and occur in various combinations. One structural domain consists of a motif whose structure has been described as a new sequence-specific DNA-binding helix. Sequence similarity suggests that the other two structural domains are novel helix–turn–helix DNA-binding domains. Four families of homing endonucleases are known—HNH, GIY-YIG, His-Cys box, and LAGLIDADG—each characterized by and named after a short-conserved sequence motif in the structural domain of its nuclease [17]. Among them, NUMOD3 is found in single-stranded and tandem repeats of GIY-YIG and HNH proteins and includes the DNA-binding domain of the I-TevI homologous endonuclease [18,19]. It forms a unique extended structure that wraps around the DNA. This region binds to DNA in a sequence-specific manner, helically inserting and twisting the DNA groove [20].
Alternative splicing events, including alternative 5′ and 3′ splice site selection, exon skipping, and intron retention [21], are widespread mechanisms in eukaryotes [22]. In alternative splicing, a single gene produces different protein variants through different splice site combinations [23,24,25]. Selective splicing is involved in the regulation of various plant biological activities, including plant responses to biotic and abiotic stresses [26,27], regulation of plant flowering [28], and regulation of the plant biological clock [29]. Chaudhary et al. proposed that under stress conditions, plants buffer the level of normal protein synthesis by alternative splicing, reduce the translation of a large portion of the transcriptome, and produce protein isoforms that are adapted to the protein isoforms needed under stress [30]. Splice variants have been studied in plants such as maize, rice, citrus, and Arabidopsis thaliana [31,32,33,34], but there are still few studies on Chrysanthemum.
Since the conserved structural domain of these three splice variants is NUMOD3, we named these three splice variants ClNUM1.1, ClNUM1.2, and ClNUM1.3. The nucleotide sequences of the three variable splices were compared with the homologous gene CsNST1 in the NCBI database, and the homology of the nucleotide sequences was found to be as high as 83%. Afterward, the three splice variants of this gene were transferred into tobacco, and the biological functions of the different variants in the model plant tobacco were identified via RT-qPCR, morphometric indexes, physiological indexes, and paraffin sections, to study the function of ClNUM1 in plants under abiotic stress so as to explore potential functional genes for resistance breeding of Chrysanthemum.
2. Materials and Methods
2.1. Plant Materials, Vectors and Strains
The C. lavandulifolium histocultures and wild-type big-leaf tobacco (Nicotiana tabacum var. macrophylla), vector Super35S::GFP, E. coli strain DH5α, and Agrobacterium strain GV3101 used in this study were obtained from the group of Associate Prof. Xuebin Song at Qingdao Agricultural University.
2.2. Homologous Gene Cloning
Cynara scolymus is a plant in the family Asteraceae and is closely related to C. lavandulifolium. The coding region of the CsNST1 gene found in the NCBI database (accession number LOC112517473, NCBI) was used as a template for primer design, and the primer sequences were as follows (Table 1):

Table 1.
The primer sequence for CsNST1 gene.
Then, the gene was cloned using C. lavandulifolium as a template according to the instructions of 2 × Phanta Flash Master Mix (Dye Plus) (Vazyme Biotech, Nanjing, China), and the obtained products were subjected to agarose gel electrophoresis migration experiments and then recovered by gel recovery using a FastPure Gel DNA Extraction Mini Kit (Vazyme Biotech, Nanjing, China), and the obtained products were sent to the company for sequencing to obtain different splice variants of ClNUM1 gene: ClNUM1.1, ClNUM1.2 and ClNUM1.3.
2.3. Stress Treatment of Chrysanthemum lavandulifolium
We transplanted C. lavandulifolium seedlings of 5 cm in height and good growth condition from sterile planting bottles (1/2 MS + 30 g/L sucrose + 6 g/LAGAR) and planted them in an artificial climate chamber at 25 °C, 16 h of light, 8 h of darkness, 3000 lux of light and 70% air humidity. Different treatments were applied to C. lavandulifolium after one week of incubation. The plants exposed to salt treatment were watered with 200 mmol/L NaCl, the drought treatment simulates drought conditions by not watering for a specified period of time, and the plants exposed to low-temperature treatment were placed in an incubator at 4 °C and sampled at 0 h, 1 h, 4 h, 8 h, 12 h, 24 h, and 36 h. We carried out at least three replicates per treatment.
2.4. RNA Extraction and RT-qPCR
We carried out RT-qPCR using wild-type C. lavandulifolium under different treatments to verify the expression patterns of the three transcripts under stress conditions. A certain amount of C. lavandulifolium and tobacco tissues was quick-frozen in liquid nitrogen and then quickly ground into powder and transferred to RNase-free 2 mL centrifuge tubes. Then, total RNA was extracted according to the instructions for the FastPure Plant Total RNA Isolation Kit (Vazyme Biotech, Nanjing, China). cDNA synthesis was performed according to the instructions for the HiScript III RT SuperMix for qPCR (+gDNA wiper) kit (Vazyme Biotech, Nanjing, China). RT-qPCR was performed on a StepONE Plus system (Applied Biosystems, Waltham, MA, USA) using SYBR qPCR Master Mix (Vazyme Biotech, Nanjing, China) and specific primers. The data obtained were analyzed by the 2−ΔΔCt method, and the internal reference gene was Actin7 (Table 2).

Table 2.
The primer sequence for Actin7 gene.
2.5. Agrobacterium Transformation Method
Three splice variants were recombined with the overexpression vector Super35S::GFP using the homologous recombination kit from Vazyme (ClonExpress® Ultra One Step Cloning Kit). The recombinant plasmids were transferred into the receptor cells of Agrobacterium tumefaciens GV3101. The transformed A. tumefaciens colonies were selected on LB-agar plates containing 50 mg L−1 kanamycin, 50 mg L−1 rifampicin, and 50 mg L−1 gentamicin. Positive colonies were picked for PCR amplification and identification, after which the colonies were subjected to the configuration of infiltration solution and transformed tobacco as described [35]. After screening and characterization, the transgenic tobaccos were obtained.
2.6. Transgenic Tobacco Resistance Screening and Management and Maintenance Methods
In this study, Super35S::GFP was used as the control group (CK), and Super35S::GFP, Super35S::ClNUM1.1, Super35S::ClNUM1.2, and Super35S::ClNUM1.3 tobacco seeds were sown in screening medium (MS + 50 mg/L Hgy + 30 g/L sucrose + 6 g/L agar) and incubated at 25 °C with 16 h of light and 8 h of darkness and 3000 lux light intensity. After the growth of true leaves, tobacco seedlings with similar growth status were picked and transplanted to the stress treatment medium in an aseptic environment as well as to an artificial climate chamber for subsequent treatment experiments.
2.7. Abiotic Stress Treatments and Morphometric Measurements
2.7.1. Stress Treatment of Transgenic Tobacco at the Seedling Stage
Tobacco seedlings with the same growth status in the screening medium were transferred to MS, MS + 200 mmol/L NaCl, and MS + 200 µmol/L ABA(Abscisic-Acid) media, ABA is used to simulate drought stress [13]. The incubation conditions of the control, salt, and ABA treatments were at 25 °C, 16 h of light and 8 h of darkness, and 3000 lux light intensity, and that of the low-temperature treatment was at 4 °C, 16 h of light and 8 h of darkness, and 3000 lux light intensity. After 24 days of treatment in the medium, the tobacco plants were carefully removed, and the roots were carefully cleaned from the medium. After cleaning, the control, salt, ABA, and low-temperature treated tobacco plants were subjected to fresh weight and root length determination (Table S1).
2.7.2. Stress Treatment of Transgenic Tobacco at the Mature Stage
Tobacco seedlings with the same growth status in the screening medium were selected and transferred to a charcoal substrate for two to three weeks and were subjected to control, salt, drought, and low-temperature treatments when they reached maturity. The control treatments were watered every other week, the salt treatments were watered with 200 mmol/L NaCl every other week, the drought treatments were subjected to simulated drought conditions by artificially controlling the water content of the soil, and the low-temperature treatments were placed in a 4 °C incubator. The control, drought, and salt treatments were all incubated at 25 °C. After 15 days of treatment in the artificial climate chamber, the growth status of the tobacco was photographed, and the height of the stalks was measured. Afterward, the stress-treated tobacco was rehydrated, and the height of the stalks was measured and photographed after seven days (Table S2).
2.8. Measurement of Physiological Parameters
To better illustrate the function of the splice variants, the chlorophyll content was determined in control and treated tobacco seedlings grown in an artificial climate chamber. Leaf samples were rinsed with distilled water and blotted dry with filter paper. The sides of the main veins of the leaves were cut into filaments <1 mm in width, and a 0.2 g sample was weighed out. The leaves were then immersed in 25 mL of 95% ethanol and left in the dark for 3 days. Absorbance was measured at wavelengths of 470 mm, 649 mm, and 665 mm using a UV spectrophotometer (Hitachi; Tokyo, Japan), and, finally, the total chlorophyll content was calculated.
2.9. Paraffin Sectioning
We selected prime-aged tobacco with the same growth state and cut a 1.5 cm long part from 1/3 of the stem of the transgenic tobacco. The selected stalks were treated in FAA fixative (70% alcohol/formalin/acetic acid 18:1:1) for 24 h. The material was soaked in a mixture of hydrogen peroxide and glacial acetic acid (1:1) for 48 h to soften the material and then dehydrated with ethanol and embedded in paraffin wax. The samples were divided into 10 μm sections by a slicer. Afterward, fenugreek solid green staining was performed, and the thickness of the cell wall and the size of the cells were observed using a light microscope.
2.10. Statistical Analysis
All experiments were set up with three or more biological replicates. Data are expressed as the mean ± SD (standard deviation) and were analyzed by a t-test (* p ≤ 0.05 and ** p ≤ 0.01).
3. Results
3.1. Bioinformatics Analysis of the ClNUM1 Splice Variants
Three alternative splicing variants of ClNUM1 were cloned from C. lavandulifolium, named ClNUM1.1, ClNUM1.2, and ClNUM1.3 (Figure S1). ClNUM1.1 was 1164 bp in total length and encoded 370 amino acids, while ClNUM1.2 was 1176 bp and encoded 373 amino acids; CLNUM1.3 was 1134 bp and encoded 362 amino acids (Figure 1a). ClNUM1.1 and ClNUM1.2 had both selective 3′ splicing and selective 5′ splicing, and ClNUM1.3 had selective 5′ splicing (Figure 1b).

Figure 1.
Bioinformatics analysis of three different splice variants. (a) Comparison of amino acid sequences. Black is the same part of the amino acid, gray is the different part of the amino acid. (* represents the length sequence number of the omitted amino acid.) (b) Schematic structure of ClNUM1.1, ClNUM1.2, and ClNUM1.3. The gray parts represent exons, and the white parts represent introns.
However, three variable splices of this gene were found in the cloning results. Alternative splicing generates multiple mRNA transcripts from a single gene by assembling exons differently using selective splice sites in precursor mRNAs.
Further analysis of the three variables found that the conserved structural domains of these three variable splices were different from those of the NST gene, and the three variable splices belonged to the conserved structural domain of NUMOD3 after comparison.
3.2. ClNUM1.1, ClNUM1.2, and ClNUM1.3 in C. lavandulifolium Show Different Responses to Stress Treatments
To further investigate the response of ClNUM1.1, ClNUM1.2, and ClNUM1.3 in C. lavandulifolium under abiotic stress conditions, we comparatively analyzed the expression levels of the ClNUM1.1, ClNUM1.2, and ClNUM1.3 genes in wild-type C. lavandulifolium under different stress treatments (Figure 2).
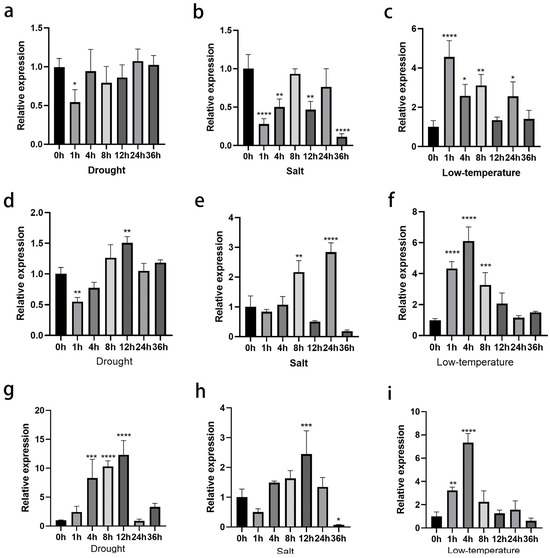
Figure 2.
Expression levels of ClNUM1.1, ClNUM1.2, and ClNUM1.3 in C. lavandulifolium under stress treatments. (a–c) Expression levels of ClNUM1.1 in C. lavandulifolium under drought, salt, and low-temperature treatments. (d–f) Expression levels of ClNUM1.2 in C. lavandulifolium under drought, salt, and low-temperature treatments. (g–i) Expression levels of ClNUM1.3 in C. lavandulifolium under drought, salt, and low-temperature treatments. All experiments were set up with three or more biological replicates (independent sample t-test; * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001).
The expression of ClNUM1.1 under drought and salt treatments was lower than that before the treatments, with a minimum decrease of 45.5% at 1 h in drought treatment and a minimum decrease of 88.7% at 36 h in salt treatment, while the expression of ClNUM1.1 under low-temperature treatment showed an overall trend higher than that before the treatments and reached a peak at 1 h, with a 3.56-fold increase in expression. Under drought treatment, the expression of ClNUM1.2 at 1 h and 12 h was significantly different from that before treatment, with a decrease of 45% and an increase of 50.7%, respectively. Under salt treatment, the expression increased significantly at 8 h and 24 h, with an increase of 1.16-fold and 1.84-fold, respectively, and the expression increased under low-temperature treatment compared with that before treatment, with increases of 3.34-fold, 5.13-fold, and 2.29-fold at 1 h, 4 h, and 8 h, respectively. The expression of ClNUM1.3 under drought treatment was elevated compared with that before treatment, increasing 7.30-fold, 9.30-fold, and 11.33-fold at 4 h, 8 h, and 12 h. Under salt treatment the expression of ClNUM1.3 was highest at 12 h, increasing 1.44-fold, and there was a significant increase in the expression of ClNUM1.3 under low-temperature treatment at 1 h and 4 h, increasing 2.22-fold and 6.33-fold, respectively, compared with that before treatment.
These results clearly show that ClNUM1.1, ClNUM1.2, and ClNUM1.3 in C. lavandulifolium all responded under the stress treatments, but the different splice variants responded differently to each treatment.
3.3. Phenotypic Analysis of Young ClNUM1 Transgenic Tobacco under Abiotic Stress
To investigate the function of the ClNUM1.1, ClNUM1.2, and ClNUM1.3 splice variants, the present study was carried out to determine the root length, fresh weight, and chlorophyll content of the treated transgenic tobacco by abiotic stress treatment. After 24 days of treatment, we found that compared with before treatment, tobacco grew significantly under CK, NaCl, and low-temperature treatments, but the growth signs were not obvious under ABA conditions (Figure 3). The measurements showed that there was no significant difference in root length among the transgenic tobacco plants at 25 °C with 16 h of light and 8 h of darkness and 3000 lux, and the fresh weight of ClNUM1.1 and ClNUM1.3 was not significantly different from that of CK; the fresh weight of ClNUM1.2 increased by 47.5% compared with that of CK; under ABA treatment, ClNUM1.2 and ClNUM1.3 showed an increase in root length and fresh weight compared to CK, their root length increased by 32.7% and 18.2%, respectively, and their fresh weight increased by 53.4%, showing a significant difference, while ClNUM1.1 showed an increase in root length by 106.1% compared to CK, and its fresh weight was not significantly different; the root length and fresh weight of all three transgenic tobacco lines were increased under low-temperature treatment compared with CK, with a 30.6%, 35.2%, and 16.5% increase in root length and 251%, 183.8%, and 87.4% increase in fresh weight for ClNUM1.1, ClNUM1.2, and ClNUM1.3, respectively, showing significant differences; under salt treatment, the fresh weight of all three transgenic tobacco lines was increased and significantly different from CK, and the root length of ClNUM1.1 and ClNUM1.2 was increased by 10.8% and 39%, respectively, compared to that of CK, but the root length of ClNUM1.3 showed little change compared to that of CK (Figure 4). We found that the growth of CK was inhibited under all the stress treatments, as well as the growth of the ClNUM1.1, ClNUM1.2, and ClNUM1.3 transgenic tobacco lines under ABA treatment.
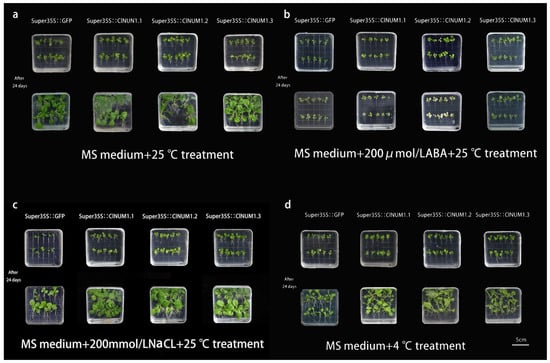
Figure 3.
Growth status of tobacco under different treatments in the medium: (a) growth status under control treatment, (b) growth status under ABA treatment, (c) growth status under salt treatment, and (d) growth status under low-temperature treatment.
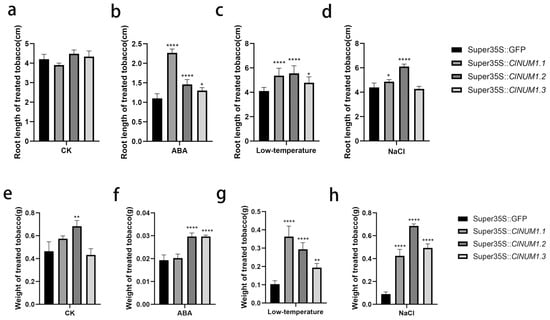
Figure 4.
Root length and fresh weight of tobacco under different treatments after 24 days. (a–d) Root length of transgenic tobacco under control, drought, low-temperature, and salt treatments. (e–h) Fresh weight of transgenic tobacco under control, drought, low-temperature, and salt treatments All experiments were set up with three or more biological replicates (independent sample t-test; * p < 0.05, ** p < 0.01, **** p < 0.0001).
The experimental results showed that ClNUM1.1 transgenic tobacco had a strong ability to tolerate salt and low temperature at the seedling stage; ClNUM1.2 transgenic tobacco had a strong ability to tolerate salt and low temperature; and ClNUM1.3 transgenic tobacco had a strong ability to tolerate low temperature.
3.4. Phenotypic Study of ClNUM1 Transgenic Tobacco at Maturity under Abiotic Stress and Determination of Physiological Indexes
To further illustrate the function of the splice variants under abiotic stress treatments, we observed the growth status of mature transgenic tobacco seedlings under stress treatments and statistically compiled the changes in the stem height of tobacco before and after rehydration under the stress treatments, as well as the chlorophyll content of the transgenic tobacco after the treatments.
In this study, it was found that under the control treatment, transgenic tobacco did not show a significant difference in stem height change after treatment with the same growth rate, but after rewatering, ClNUM1.1, ClNUM1.2, and ClNUM1.3 showed significantly slower growth rates compared to CK (Figure 5).

Figure 5.
Growth states of mature tobacco seedlings under different treatments. (a) Growth state under the control treatment. (b) Growth state under drought treatment. (c) Growth state under salt treatment. (d) Growth state under low-temperature treatment.
Under drought treatment, the stem height of ClNUM1.2 and ClNUM1.3 increased by 1.5 cm and 9.1 cm compared with that of CK, but the growth recovery rate of ClNUM1 transgenic tobacco was significantly slower than that of CK after rehydration; the stem height of ClNUM1.1 exhibited a slower growth rate than that of CK after both treatment and rehydration. Under salt treatment, the stem height of ClNUM1.1 increased by 0.27 cm and 0.05 cm after both treatment and rehydration compared with that of CK; the stem height of ClNUM1.2 grew at a slower rate than that of CK after treatment but recovered very quickly after rehydration; and the average stem height of ClNUM1.3 was 5.5 cm higher than that of CK after treatment but recovered very slowly after rehydration. The average stalk heights of ClNUM1.1, ClNUM1.2, and ClNUM1.3 were 0.13 cm, 0.54 cm, and 1.1 cm higher than that of CK under the low-temperature treatment, and they recovered better than CK after rewatering (Figure 6a,b).
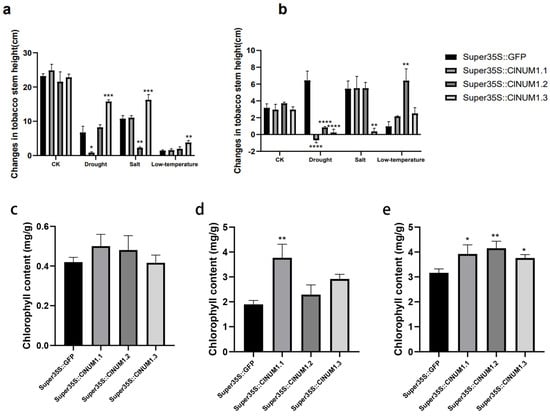
Figure 6.
Changes in stem height of mature tobacco seedlings after different treatments and after rehydration and chlorophyll content after treatments. (a) Changes in stem height of tobacco after stress treatment. (b) Changes in stem height after 7 days of restoration to normal growth conditions under salt, drought, and low-temperature stresses. (c) Chlorophyll content after drought treatment. (d) Chlorophyll content after salt treatment. (e) Chlorophyll content after low-temperature treatment. All experiments were set up with three or more biological replicates (independent sample t-test; * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001).
Under drought treatment, the chlorophyll content in ClNUM1 transgenic tobacco was not significantly different from that in CK; under salt treatment, the chlorophyll content of ClNUM1.1 transgenic tobacco was twice as high as that of CK, with a significant difference, and the chlorophyll content of ClNUM1.2 and ClNUM1.3 transgenic tobacco was 1.2 and 1.5 times as high as that of CK, with an increase from CK but no significant difference; the chlorophyll contents of ClNUM1.1, ClNUM1.2, and ClNUM1.3 transgenic tobacco were 1.24, 1.31, and 1.2 times higher than those of CK, respectively, under low-temperature treatment, all of which were significant differences (Figure 6c–e).
The test results showed that ClNUM1.1 transgenic tobacco had strong salt and low-temperature tolerance at the mature seedling stage; ClNUM1.2 transgenic tobacco had strong low-temperature tolerance; and ClNUM1.3 transgenic tobacco had strong low-temperature tolerance.
3.5. Anatomical Analysis of Transgenic Tobacco Stalks
Paraffin sections were taken from one-third of 12-week-old transgenic tobacco stalks and stained with Senka solid green, and the structures of the epidermis, cortex, bast, xylem, and pith could be observed under a light microscope (Figure 7a–h). Both saffron and solid green stain the cell walls of plants; saffron stains the lignified and corky cell walls as well as the ducts of plants red, while solid green stains the cellulose cell walls as well as the sieve tubes of plants green. We measured cell wall thickness and cell size of twenty each in the xylem of transgenic tobacco stalks (Figure 7i,j). Through observation, we found that the cell wall thickness of ClNUM1.1 and ClNUM1.3 transgenic tobacco increased compared to that of CK by 1.08-fold and 1.16-fold, respectively, and the cell size also increased compared to that of CK by 1.06 and 1.11 times, respectively. The cell wall thickness and cell size of ClNUM1.3 transgenic tobacco were significantly different; the cell wall thickness and cell size of ClNUM1.2 transgenic tobacco were both reduced compared to those of CK with no significant differences.
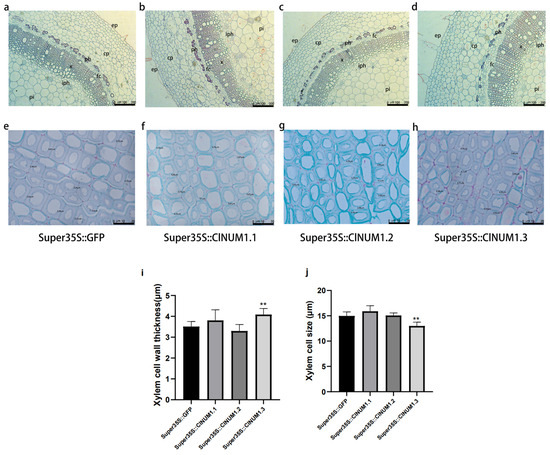
Figure 7.
Paraffin sections of transgenic tobacco stems as well as xylem cell wall thickness and xylem cell size of different genotypes of tobacco. (a–h) Stem cross-sections of CK, ClNUM1.1, ClNUM1.2, and ClNUM1.3 transgenic tobacco under normal growth conditions. (i) Xylem cell wall thickness of different genotypes of tobacco. (j) Xylem cell size of different genotypes of tobacco. Twenty cell sizes and cell wall thicknesses were measured for each transgenic plant. ep = epidermis, cp = cortical parenchyma, ph = phloem, iph = inner phloem, fc = osteocortex, x = xylem, pi = pith. (independent sample t-test; ** p < 0.01.)
4. Discussion
Alternative splicing is an important pathway for eukaryotes to generate significant regulatory and proteomic complexity. Alternative splicing has two main outcomes—proteome diversification and gene expression regulation [36]—and alternative splicing can increase transcriptome variability and complexity. It is considered to be one of the possible sources of large phenotypic differences between species. In the clipping type, alternative splicing of 5′ and 3′ splice sites is performed by selecting either a 5′ splice donor or a 3′ splice acceptor to retain or splice all or part of the exon sequence [37]. In this paper, the splicing variants we obtained were A5SS and A3SS, which enriches the chrysanthemum variable splicing types. In many studies, alternative splicing has been shown to play an important role in the growth and development of plants and animals, as well as in the response to adversity.
The ClNUM1 gene was discovered by chance during the cloning of the homologous gene CcNST1 in C. lavandulifolium, and a NUMOD3 conserved domain was found in this gene. Since the gene is a new and unknown gene, we named it ClNUM1. In addition, during the cloning process, we found three splicing variants of the ClNUM1 gene, named ClNUM1.1, ClNUM1.2, and ClNUM1.3.
We characterized the functions of the different splice variants by analyzing morphological and physiological indices of seedling and mature lines of transgenic tobacco under abiotic stress treatments. It was found that the three splice variants enhanced the stress resistance of tobacco (seedling and maturity) and promoted plant growth to some extent, which was consistent with the function of the NAC transcription factor family [38,39].
In the seedling stress experiment, we found that the root length and fresh weight of ClNUM1 transgenic tobacco were not positively correlated under stress conditions, and sometimes the root length increased but fresh weight decreased or vice versa. In the stress experiment at maturity, we found two interesting phenomena: one is that the growth potential of ClNUM1 transgenic tobacco under stress conditions was significantly higher than that of CK after treatment, but the recovery rate of the growth potential after rewatering was slower than that of CK. The other is that the growth potential of ClNUM1 transgenic tobacco after treatment was not significantly different from that of CK, but the recovery rate of the growth potential after rewatering significantly differed from that of CK. We speculate that this may be a response of the plant to resist external environmental changes.
Previous studies have found that NST can thicken the secondary cell wall of Populus and Arabidopsis [40,41], and the change of the secondary cell wall can enhance grapevine and Populus resistance to abiotic stress [42,43]. By observing the cross-section of transgenic tobacco, it was found that compared with CK, the secondary cell wall thickness of transgenic tobacco has changed to a certain extent and the resistance to abiotic stress has been enhanced. We speculated that this gene can regulate the change of plant cell wall thickness and thus regulate plant resistance; this is consistent with previous studies. Abiotic stress will affect the chlorophyll metabolism of plants and thus affect the chlorophyll content of plants [44]. The strength of plant photosynthesis can be reflected by measuring the chlorophyll content so as to evaluate the growth status of plants under abiotic stress. Under the treatment, the chlorophyll content of transgenic tobacco was higher than CK, indicating that the growth status of transgenic tobacco was better than CK, and the resistance was higher than CK.
We have made a preliminary validation for ClNUM1 function in tobacco based on this experiment; however, how ClNUM1 in C. lavandulifolium regulates plant stress resistance more precisely by affecting secondary cell walls remains to be further studied. Subsequent studies on genes related to secondary cell wall synthesis will reveal the regulatory network and resistance mechanism of ClNUM1 in C. lavandulifolium under abiotic stress.
5. Conclusions
In this study, by analyzing the amino acid sequences of the three splice variants of the ClNUM1 gene of C. lavandulifolium, we found that ClNUM1.1 and ClNUM1.2 had two selective splicing types and ClNUM1.3 had selective 5′ splicing. The function of the splice variants of the unknown C. lavandulifolium gene ClNUM1 was verified in tobacco. It was found that ClNUM1.1 enhanced salt and low-temperature tolerance, ClNUM1.2 enhanced salt and low-temperature tolerance, and ClNUM1.3 enhanced low-temperature tolerance in tobacco. By observing the paraffin sections, we found that the thickness of the xylem cells was related to the growth rate of the plant, with thick cell walls in fast-growing plants and thin cell walls in slow-growing plants. However, the molecular mechanism underlying the role of the ClNUM1 gene in secondary cell wall synthesis and the synthesis of secondary cell wall components such as lignin, cellulose, and polysaccharides is not clear, which is something we need to focus on in future studies. In this study, we discovered a new unknown gene, ClNUM1, in C. lavandulifolium and verified the function of the splice variants of this gene in tobacco, thus providing a new potential gene for resistance breeding in Chrysanthemum and enriching the diversity of resistance breeding in Chrysanthemum.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cimb46060314/s1, Figure S1: The figure shows a comparison of the nucleotide sequences of ClNUM1 with its three spliced variants. Table S1: Mean and SD of root length and fresh weight of ClNUM1 transgenic tobacco at seedling stage after treatment. Table S2: Mean and SD of stem height change in mature ClNUM1 transgenic tobacco after treatment and after recovery.
Author Contributions
W.Z., H.W., X.S., and Y.G. carried out the majority of the research and the data analysis and completed the draft preparation and revision. X.H., Y.L., W.H., S.C., and X.Z. completed the experimental design, the data analysis, and the writing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation Youth Fund Project (32101580), the Survey and Collection Project of Herbal Plant Germplasm Resources at Shandong Provincial Forest and Grass Germplasm Resources Center (6602423134), and the Qingdao Agricultural University Doctoral Start-Up Fund (6631122019).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Yu, L.; Dang, J.; Liu, X.; Jiang, H. Investigation and Research on the Applied Value of Chrysanthemum. Contemp. Hortic. 2022, 13, 21–23. [Google Scholar] [CrossRef]
- Cheng, X. Interspecific Hybridization between Chrysanthemum Species and Research on Resistant Germplasm Innovation. Master’s Thesis, University of Nanjing Agriculture, Nanjing, China, 2012. [Google Scholar]
- Peng, Y. Salt-Tolerance Analysis of Chrysanthemum lavandulifolium and Transformation of DlBADH Promoters to Chrysanthemum. Master’s Thesis, Beijing Forestry University, Beijing, China, 2011. [Google Scholar]
- An, C.; Sheng, L.P.; Du, X.P.; Wang, Y.J.; Zhang, Y.; Song, A.P.; Jiang, J.F.; Guan, Z.Y.; Fang, W.M.; Chen, F.D.; et al. Overexpression of CmMYB15 provides chrysanthemum resistance to aphids by regulating the biosynthesis of lignin. Hortic. Res. 2019, 6, 84. [Google Scholar] [CrossRef]
- Huang, H.; Wang, Y.; Wang, S.L.; Wu, X.; Yang, K.; Niu, Y.J.; Dai, S.L. Transcriptome-wide survey and expression analysis of stress-responsive NAC genes in Chrysanthemum lavandulifolium. Plant Sci. 2012, 193, 18–27. [Google Scholar] [CrossRef]
- Wang, K.; Wu, Y.H.; Tian, X.Q.; Bai, Z.Y.; Liang, Q.Y.; Liu, Q.L.; Pan, Y.Z.; Zhang, L.; Jiang, B.B. Overexpression of DgWRKY4 Enhances Salt Tolerance in Chrysanthemum Seedlings. Front. Plant Sci. 2017, 8, 1592. [Google Scholar] [CrossRef]
- Jaffar, M.A.; Song, A.P.; Faheem, M.; Chen, S.M.; Jiang, J.F.; Liu, C.; Fan, Q.Q.; Chen, F.D. Involvement of CmWRKY10 in Drought Tolerance of Chrysanthemum through the ABA-Signaling Pathway. Int. J. Mol. Sci. 2016, 17, 693. [Google Scholar] [CrossRef] [PubMed]
- Li, C.N.; Ng, C.K.Y.; Fan, L.M. MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 2015, 114, 80–91. [Google Scholar] [CrossRef]
- Li, F.; Zhang, Y.; Tian, C.; Wang, X.H.; Zhou, L.J.; Jiang, J.F.; Wang, L.K.; Chen, F.D.; Chen, S.M. Molecular module of CmMYB15-like-Cm4CL2 regulating lignin biosynthesis of chrysanthemum (Chrysanthemum morifolium) in response to aphid (Macrosiphoniella sanborni) feeding. New Phytol. 2023, 237, 1776–1793. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhong, M.; Wu, Y.H.; Bai, Z.Y.; Liang, Q.Y.; Liu, Q.L.; Pan, Y.Z.; Zhang, L.; Jiang, B.B.; Jia, Y.; et al. Overexpression of a chrysanthemum transcription factor gene DgNAC1 improves the salinity tolerance in chrysanthemum. Plant Cell Rep. 2017, 36, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.J.; Sheng, L.P.; Zhang, H.R.; Du, X.P.; An, C.; Xia, X.L.; Chen, F.D.; Jiang, J.F.; Chen, S.M. CmMYB19 Over-Expression Improves Aphid Tolerance in Chrysanthemum by Promoting Lignin Synthesis. Int. J. Mol. Sci. 2017, 18, 619. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.J.; Geng, Z.Q.; Wang, Y.X.; Wang, Y.G.; Liu, S.H.; Chen, C.W.; Song, A.P.; Jiang, J.F.; Chen, S.M.; Chen, F.D. A novel transcription factor CmMYB012 inhibits flavone and anthocyanin biosynthesis in response to high temperatures in chrysanthemum. Hortic. Res. 2021, 8, 248. [Google Scholar] [CrossRef]
- Wang, H.; Guo, Y.N.; Hao, X.Y.; Zhang, W.X.; Xu, Y.X.; He, W.T.; Li, Y.X.; Cai, S.Y.; Zhao, X.; Song, X.B. Alternative Splicing for Leucanthemella linearis NST1Contributes to Variable Abiotic Stress Resistance in Transgenic Tobacco. Genes 2023, 14, 1549. [Google Scholar] [CrossRef] [PubMed]
- Gao, G. Chrysanthemum CmWRKY15-1 Enhanced Resistance to Chrysanthemum White Rust by Regulating the Expression of CmNPR1. Master’s Thesis, Shenyang Agricultural University, Shenyang, China, 2022. [Google Scholar]
- Meng, R. Study on the Function of Shennong Chrysanthemum CiMYB32 in Response to Drought Stress. Master’s Thesis, Northeast Forestry University, Harbin, China, 2023. [Google Scholar]
- Sitbon, E.; Pietrokovski, S. New types of conserved sequence domains in DNA-binding regions of homing endonucleases. Trends Biochem. Sci. 2003, 28, 473–477. [Google Scholar] [CrossRef]
- Belfort, M.; Roberts, R.J. Homing endonucleases: Keeping the house in order. Nucleic Acids Res. 1997, 25, 3379–3388. [Google Scholar] [CrossRef]
- Lagerback, P.; Carlson, K. Amino Acid Residues in the GIY-YIG Endonuclease II of Phage T4 Affecting Sequence Recognition and Binding as Well as Catalysis. J. Bacteriol. 2008, 190, 5533–5544. [Google Scholar] [CrossRef][Green Version]
- Andersson, C.E.; Lagerback, P.; Carlson, K. Structure of Bacteriophage T4 Endonuclease II Mutant E118A, a Tetrameric GIY-YIG Enzyme. J. Mol. Biol. 2010, 379, 1003–1016. [Google Scholar] [CrossRef]
- Van Roey, P.; Waddling, C.A.; Fox, K.M.; Belfort, M.; Derbyshire, V. Intertwined structure of the DNA-binding domain of intron endonuclease I-TevI with its substrate. EMBO J. 2001, 20, 3631–3637. [Google Scholar] [CrossRef]
- Raczynska, K.D.; Simpson, C.G.; Ciesiolka, A.; Szewc, L.; Lewandowska, D.; McNicol, J.; Szweykowska-Kulinska, Z.; Brown, J.W.S.; Jarmolowski, A. Involvement of the nuclear cap-binding protein complex in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 2010, 38, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Jia, Z.; Pu, Q.; Tian, Y.; Zhu, F.; Liu, Y. ABA Mediates Plant Development and Abiotic Stress via Alternative Splicing. Int. J. Mol. Sci. 2022, 23, 3796. [Google Scholar] [CrossRef]
- James, A.B.; Syed, N.H.; Bordage, S.; Marshall, J.; Nimmo, G.A.; Jenkins, G.I.; Herzyk, P.; Brown, J.W.S.; Nimmo, H.G. Alternative Splicing Mediates Responses of the Arabidopsis Circadian Clock to Temperature Changes. Plant Cell 2012, 24, 961–981. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Xu, Q.; Wang, X. Regulation of the circadian clock through pre-mRNA splicing in Arabidopsis. J. Exp. Bot. 2014, 65, 1973–1980. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, A.; Riegler, S.; Ayatollahi, Z.; Cavallari, N.; Giono, L.E.; Nimeth, B.A.; Mutanwad, K.V.; Schweighofer, A.; Lucyshyn, D.; Barta, A.; et al. Targeting alternative splicing by RNAi: From the differential impact on splice variants to triggering artificial pre-mRNA splicing. Nucleic Acids Res. 2021, 49, 1133–1151. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Liu, Y.; Kang, D.; Fan, K.; Wang, C.; Wang, G.; Liu, Y. Two alternative splicing variants of maize HKT1;1 confer salt tolerance in transgenic tobacco plants. Plant Cell Tissue Organ Cult. 2015, 123, 569–578. [Google Scholar] [CrossRef]
- Lv, B.; Hu, K.; Tian, T.; Wei, K.; Zhang, F.; Jia, Y.; Tian, H.; Ding, Z. The pre-mRNA splicing factor RDM16 regulates root stem cell maintenance in Arabidopsis. J. Integr. Plant Biol. 2021, 63, 662–675. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Deng, Q.; Wu, M.; Wang, Z.; Wei, D.; Wang, H.; Xiang, H.; Zhang, H.; Tang, Q. Mechanisms of alternative splicing in regulating plant flowering: A review. Chin. J. Biotechnol. 2021, 37, 2991–3004. [Google Scholar] [CrossRef]
- Simpson, C.G.; Fuller, J.; Calixto, C.P.G.; McNicol, J.; Booth, C.; Brown, J.W.S.; Staiger, D. Monitoring Alternative Splicing Changes in Arabidopsis Circadian Clock Genes. Methods Mol. Biol. 2016, 1398, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, S.; Jabre, I.; Reddy, A.S.N.; Staiger, D.; Syed, N.H. Perspective on Alternative Splicing and Proteome Complexity in Plants. Trends Plant Sci. 2019, 24, 496–506. [Google Scholar] [CrossRef]
- Thatcher, S.R.; Danilevskaya, O.N.; Meng, X.; Beatty, M.; Zastrow-Hayes, G.; Harris, C.; Van Allen, B.; Habben, J.; Li, B.L. Genome-Wide Analysis of Alternative Splicing during Development and Drought Stress in Maize. Plant Physiol. 2016, 170, 586–599. [Google Scholar] [CrossRef] [PubMed]
- Alhabsi, A.; Butt, H.; Kirschner, G.K.; Blilou, I.; Mahfouz, M.M.; Sunkar, R. SCR106 splicing factor modulates abiotic stress responses by maintaining RNA splicing in rice. J. Exp. Bot. 2023, 75, 802–818. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.-X.; Wu, Y.-M.; Zhang, J.-X.; Zhang, J.-X.; Zhou, H.; Zeng, R.-F.; Zheng, W.-X.; Qiu, M.-Q.; Zhou, J.-J.; Xie, Z.-Z.; et al. A bZIP transcription factor CiFD regulates drought- and low-temperature-induced flowering by alternative splicing in citrus. J. Integr. Plant Biol. 2023, 65, 674–691. [Google Scholar] [CrossRef]
- Albaqami, M.; Laluk, K.; Reddy, A.S.N. The Arabidopsis splicing regulator SR45 confers salt tolerance in a splice isoform-dependent manner. Plant Mol. Biol. 2019, 100, 379–390. [Google Scholar] [CrossRef]
- Horsch, R.B.; Fry, J.E.; Hoffmann, N.L.; Wallroth, M.; Eichholtz, D.; Rogers, S.G.; Fraley, R.T. A simple and general method for transferring genes into plants. Science 1985, 227, 1229–1231. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.C.R.; Tittes, S.; Mendieta, J.P.; Collier-zans, E.; Rowe, H.C.; Rieseberg, L.H.; Kane, N.C. Genetics of alternative splicing evolution during sunflower domestication. Proc. Natl. Acad. Sci. USA 2018, 115, 6768–6773. [Google Scholar] [CrossRef]
- Iñiguez, L.P.; Ramírez, M.; Barbazuk, W.B.; Hernández, G. Identification and analysis of alternative splicing events in Phaseolus vulgaris and Glycine max. BMC Genom. 2017, 18, 650. [Google Scholar] [CrossRef]
- Xu, Y.; Cheng, J.; Hu, H.; Yan, L.; Jia, J.; Wu, B. Genome-Wide Identification of NAC Family Genes in Oat and Functional Characterization of AsNAC109 in Abiotic Stress Tolerance. Plants 2024, 13, 1017. [Google Scholar] [CrossRef]
- Chen, N.; Shao, Q.; Lu, Q.; Li, X.; Gao, Y.; Xiao, Q. Research progress on function of NAC transcription factors in tomato (Solanum lycopersicum L.). Euphytica 2023, 219, 22. [Google Scholar] [CrossRef]
- Takata, N.; Awano, T.; Nakata, M.T.; Sano, Y.; Sakamoto, S.; Mitsuda, N.; Taniguchi, T. Populus NST/SND orthologs are key regulators of secondary cell wall formation in wood fibers, phloem fibers and xylem ray parenchyma cells. Tree Physiol. 2019, 39, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Luo, F.; Zhong, Y.; He, J.; Li, L. Modulation of NAC transcription factor NST1 activity by XYLEM NAC DOMAIN1 regulates secondary cell wall formation in Arabidopsis. J. Exp. Bot. 2020, 71, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhang, C.; Wang, Y.; Zhang, Y.; Zhang, Y.; Wang, Y.; Wang, C. Expression profiles of genes regulated by BplMYB46 in Betula platyphylla. J. For. Res. 2019, 30, 2267–2276. [Google Scholar] [CrossRef]
- Malacarne, G.; Lagreze, J.; Martin, B.R.S.; Malnoy, M.; Moretto, M.; Moser, C.; Costa, L.D. Insights into the cell-wall dynamics in grapevine berries during ripening and in response to biotic and abiotic stress. Plant Mol. Biol. 2024, 114, 38. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, W.; Niu, D.; Liu, X. Effects of abiotic stress on chlorophyll metabolism. Plant Sci. 2024, 342, 112030. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).