Abstract
Tubulin-targeting agents attract undiminished attention as promising compounds for the design of anti-cancer drugs. Verubulin is a potent tubulin polymerization inhibitor, binding to colchicine-binding sites. In the present work, a series of verubulin analogues containing a cyclohexane or cycloheptane ring 1,2-annulated with pyrimidine moiety and various substituents in positions 2 and 4 of pyrimidine were obtained and their cytotoxicity towards cancer and non-cancerous cell lines was estimated. The investigated compounds revealed activity against various cancer cell lines with IC50 down to 1–4 nM. According to fluorescent microscopy data, compounds that showed cytotoxicity in the MTT test disrupt the normal cytoskeleton of the cell in a pattern similar to that for combretastatin A-4. The hit compound (N-(4-methoxyphenyl)-N,2-dimethyl-5,6,7,8-tetrahydroquinazolin-4-amine) was encapsulated in biocompatible nanocontainers based on Ca2+ or Mg2+ cross-linked alginate and it was demonstrated that its cytotoxic activity was preserved after encapsulation.
1. Introduction
Tubulin is a globular protein essential for the functioning of all eukaryotic cells. Heterodimers composed of α- and β-tubulin molecules polymerize to form microtubules, which play a key role in the formation of cytoskeleton, cell division and intracellular transport [1,2,3]. Tubulin- and microtubule-targeting agents have been the subject of intensive research and some of them are now widely used both in chemotherapy and as research tools to study the details of microtubule functioning. Despite widespread clinical use of anti-microtubule drugs, research in the field is constantly ongoing and associated mainly with the search for new alternatives with lower toxicities, better bioavailability and effectivity against resistant tumors [4,5,6,7,8,9,10].
The binding of tubulin-targeting agents to different regions of α,β-tubulin dimer causes its conformation switch and regulates microtubule behavior [3,4]. The compounds interacting with the taxol-binding site and stabilizing microtubules [11], as well as microtubule-destabilizing agents that bind to the vinca domain of tubulin [12], are used in chemotherapy. Several representatives of a large group of colchicine-binding site ligands have also been introduced into clinical trials, the most successful of which was fosbretabulin (combretastatin A-4 prodrug) [13]. However, so far none of these compounds has received full and final approval for clinical use. Considering the fact that many ligands of the colchicine-binding site are both efficient and easily accessible synthetically, they attract significant interest of researchers, and extensive efforts are being made to develop new molecules with this type of action [13,14,15]. Note that the common limitations for the use of these compounds as drug candidates are their low selectivity towards non-tumor cells (resulting in high general toxicity) and, for certain molecules, poor bioavailability.
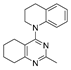
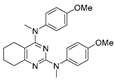
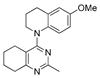
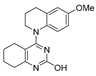
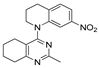
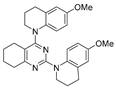
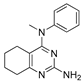
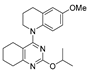
One of the colchicine site ligands that has successfully passed a phase II clinical trial is verubulin (Azixa, 1) [16,17,18,19] (Figure 1). Since the first reports of verubulin as a potent microtubule-disrupting agent and especially after the suspension of its further clinical promotion due to economic reasons [20], several research groups extensively studied the structure–activity relationship for the series of analogues of the lead molecule (Figure 1) [20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36]. In particular, the following modifications were carried out: the variations of the substituent in position 2 of quinazoline, conformational restriction of 4-methoxy-N-methylaniline moiety, replacement of quinazoline core with other aromatic heterocycles, etc.
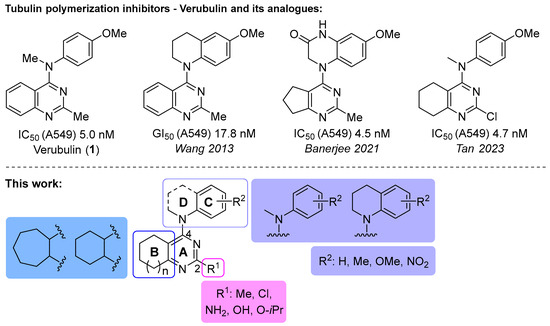
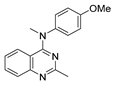
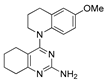
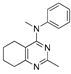
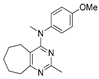
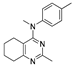
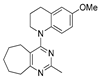
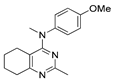
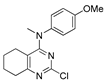
Figure 1.
Verubulin (1) [36], some of its previously described analogues [25,33,34] and compounds studied in the present work.
In 2021, Banerjee et al. [33] showed that replacement of the fused benzene ring in quinazoline for a saturated 5-membered ring can lead to compounds with high cytotoxicity (see example in Figure 1). Recently, J. Tan et al. [34] described a large series of verubulin analogues, in one of which the quinazoline core was replaced with tetrahydroquinazoline (Figure 1). This compound not only retained high activity but also demonstrated the best selectivity (by 4–5 times) for tumor cells, despite the fact that most of the active substances in the series were not selective at all [34]. The result obtained seemed interesting in the context of the strategy of designing pharmaceutically relevant molecules, including less toxic ones, by increasing their three-dimensionality (“escaping the flatland”), which has been widely discussed in recent decades [37,38,39,40]. Therefore, it seemed interesting to conduct a systematic study of verubulin analogues with a reduced degree of flatness due to the replacement of the benzene ring fused with pyrimidine for a six- or seven-membered saturated ring B (Figure 1). In the present work, we synthesized a series of such analogues, also varying their conformational rigidity and the substituents in positions 2 and 4 of heterocyclic core A (Figure 1), and studied them in cell viability assays on cell lines of cancer and non-cancerous etiology and other biotests.
Additionally, we explored the possibility to improve the toxicological profile of a selected compound via its encapsulation into nanoparticles formed from Ca2+ or Mg2+ cross-linked alginate biopolymers. Alginate is a biodegradable and biocompatible copolymer of guluronic and mannuronic acids, which has already received US Food and Drug Administration (FDA) approval for medical applications [41]. This natural anionic polymer (biomaterial) has been widely investigated and used in many areas of biomedical science and technology due to its biocompatibility, low toxicity, relatively low cost, softness and easy gelation upon addition of divalent cations (Ca2+ and Mg2+) [42]. The role of Ca2+ and Mg2+ ions is to form a framework structure of polysaccharide macromolecules, accompanied by a significant compactization of the initial macromolecules of alginate. Alginate has promising biopharmaceutical properties such as pH sensitivity, biocompatibility, biodegradability, mucoadhesiveness, lack of toxicity and immunogenicity, which make it attractive for the development of carriers for drug immobilization [43]. In our previous work, we have demonstrated that encapsulation of an isoxasole derivative with anti-cancer activity in alginate biopolymer can significantly increase its selectivity towards cancer cells [44].
2. Results and Discussion
2.1. Chemistry
4-Aminopyrimidines 2a–r were obtained via nucleophilic substitution of chlorine in the corresponding 4-cloropyrimidines upon the treatment with secondary amines in the presence of a catalytic amount of HCl (Scheme 1 and Scheme 2). In most cases, the reactions of 4-chloropyrimidines 3a–c with amines proceeded smoothly, affording target heterocycles in moderate to good yields.
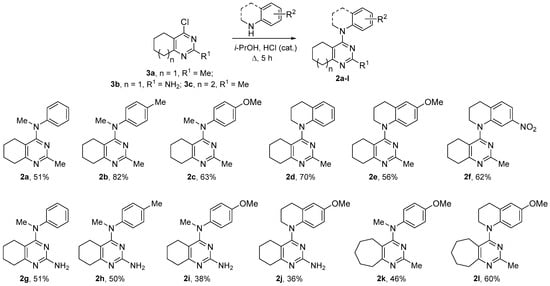
Scheme 1.
Synthesis of 4-aminopyrimidines 2a–l.
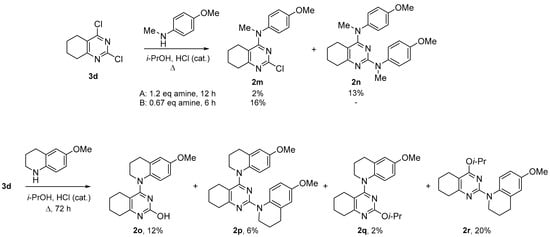
Scheme 2.
Synthesis of 4-aminopyrimidines 2m–r.
The reaction of 2,4-dichlorotetrahydroquinazoline (3d) with 4-methoxy-N-methylaniline led to a multicomponent mixture of product, from which only the prevailing product 2n resulted from two-fold substitution and compound 2m could be isolated in low yields (Scheme 2). By reducing the time and using the excess of dichloride 3d, we managed to obtain tetrahydroquinazoline 2m as the main product, though during the purification the yield of this compound decreased. The interaction of dichloride 3d and sterically hindered 6-methoxytetrahydroquinoline was even more complicated and required a long reflux, after which the compounds 2o–r could be isolated from the reaction mixture (Scheme 2). The structure of all the obtained compounds was unambiguously determined via NMR spectroscopy, using 2D techniques, when necessary (see Supplementary Materials).
2.2. Bioactivity Testing
2.2.1. Cytotoxicity of Verubulin Analogues and SAR Analysis
The obtained series of verubulin analogues 2a–r (which also included three by-products isolated in the reactions according to Scheme 2) were initially evaluated for cytotoxicity against commonly used human breast cancer cell line MCF7′ (fast-growth subclone), human lung epithelial carcinoma cell line A549 and etiologically non-cancer immortalized lung fibroblast cell line VA13 (WI38 subline 2RA). Cytotoxicity against immortalized human embryonic kidney cell line HEK293T, characterized by a high growth rate, was also studied regarding cytotoxicity against non-cancerous fast-growth cells. A standard MTT assay [45] was performed; verubulin (1) served as a positive reference. The data obtained are presented in Table 1 (for dose–response curves, see Supplementary Materials).

Table 1.
Cytotoxicity effects calculated as IC50 for 64 h treatment of the cell lines of different etiology.
Saturated verubulin derivative 2c retained high cytotoxicity of the parent molecule (1) similarly to its analogue with a chlorine substituent in position 2 of ring A (2m), described by Tan et al. [34]. Compounds 2c and 2m are more cytotoxic than their counterpart with a more polar amino group at C2 (2i), which is consistent with molecular dynamics modeling data (see below). Increasing the size of the alicycle B in the verubulin analogue 2c to a seven-membered ring (2k) reduces cytotoxicity by more than 10 times. A noticeable drop in activity during the expansion of ring B is most likely due to the steric hindrances it creates. Thus, this result denotes a limitation imposed on the volume of alicyclic ring B.
Saturated verubulin analogues (2c, 2k), regardless of the size of ring B, demonstrated no selectivity towards tumor cells in the pair A549/VA13. It should be noted that verubulin and its saturated analogue 2m which has been reported to be selective in the pair MCF7/MCF10A [34] demonstrated no selectivity in the pair A549/VA13 either.
Conformational restriction of 4-methoxy-N-methylaniline moiety in saturated verubulin derivative 2c led to compound 2e with slightly less activity, but with an IC50 still in the nanomolar concentration range. As seen in Figure 1, a similar change in activity was observed for the rigid analogue of verubulin obtained by Wang et al. [25]. The same modification for verubulin analogue 2k with a seven-membered ring B resulted in compound 2l, which was seven to twelve times less active. Thus, the cytotoxicity in the series of both “open” and “rigid” compounds, depending on ring B, changed as follows: benzene > cyclohexane > cycloheptane.
In a series of conformationally restricted tetrahydroquinazolines, the important role of a non-polar substituent at position 2 of ring A was also demonstrated: the replacement of the methyl group by an amino group slightly reduced cytotoxicity against cancer cells (2j vs. 2e). Similar replacement with a hydroxyl group led to a complete loss of cytotoxicity (2o vs. 2e), which was, however, largely restored if the hydroxyl was converted to non-polar ether (O-iPr) moiety (2q vs. 2o).
Finally, analysis of SAR in the series of obtained verubulin analogues shows the critical role of the para-methoxy substituent in ring C. Its removal reduced cytotoxicity by 2–3 orders of magnitude (2a vs. 2c, 2g vs. 2i), while its replacement with the methyl group was less detrimental, although it still reduced the activity by an order of magnitude (2b vs. 2c, 2j vs. 2h). Compound 2f containing a meta-nitro group in ring C was inactive. Miscellaneous compounds 2n, 2p and 2r obtained as by-products in the synthetic schemes were moderately cytotoxic.
2.2.2. Effect of Verubulin Analogues on Microtubule Dynamics
Since verubulin is known to stimulate microtubule disassociation, we performed an immunofluorescent microscopy study to determine the response of microtubule dynamics in carcinoma A549 cells to the treatment with representative compounds 2b,c,e. As shown in Figure 2, exposure to 5–10 μM of the compounds for 24 h leads to a pronounced or complete disruption of microtubules (Figure 2A–C), and the pattern of a diffuse distribution of the stained microtubule subunits corresponds to that caused by combretastatin CA-4 (Figure 2E). Aberrant cell morphology—contraction and rounding (Figure 2A,B) or contraction and elongation (Figure 2C)—was also observed. As expected, immunofluorescent staining of microtubules in the cells treated with compound 2a, characterized by low cytotoxicity, revealed no effect on the microtubule cytoskeleton even at very high concentrations of this compound (Figure 2D).
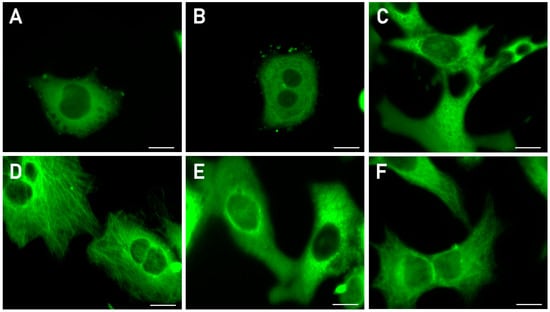
Figure 2.
Immunofluorescence microscopy images of the microtubules (MTs) in A549 cells treated with tested compounds, combretastatin (CA-4) as positive control or DMSO as negative control. (A) 2b, 10 μM (depolymerization of MTs). (B) 2e, 10 μM (full depolymerization of MTs). (C) 2c, 10 μM (full depolymerization of MTs). (D) 2a, 100 μM (intact MTs). (E) 0.03 μM CA-4 (full depolymerization of MTs). (F) 0.5% DMSO (intact MTs). Scale bar 10 μm. Images illustrate representative examples of two to four independent experiments.
An additional study of the effect of compounds 2c and 2e on microtubule assembly in vitro was monitored by recording the change in optical density at 355 nm during the GTP-dependent polymerization on crude preparation of tubulin (Tb) and microtubule-associated proteins (MAPs) from mouse brains. The obtained graphs of the change in optical density versus time are shown in Figure 3. It was shown that both 2c and 2e inhibited the initial rate of tubulin polymerization in a dose-dependent manner similarly to verubulin. The concentrations of the compounds were the same as in Figure 2. On the basis of initial rates (from 6 to 16 min for control and from 12 to 32 min for others) of the linear change in optical density at 355 nm vs. compound concentrations, where each point is the average of two different experiments with three repeats in each, the IC50 were calculated for 2c, 2e and verubulin: 7.2 ± 1.9 μM; 26 ± 6 μM and 3.1 ± 1.1 μM, respectively (Figure S2). It should be mentioned that unlike control probe (DMSO) in the presence of 2c, 2e and verubulin there are no features of depolymerization which causes a plateau in the control curve (Figure 3).
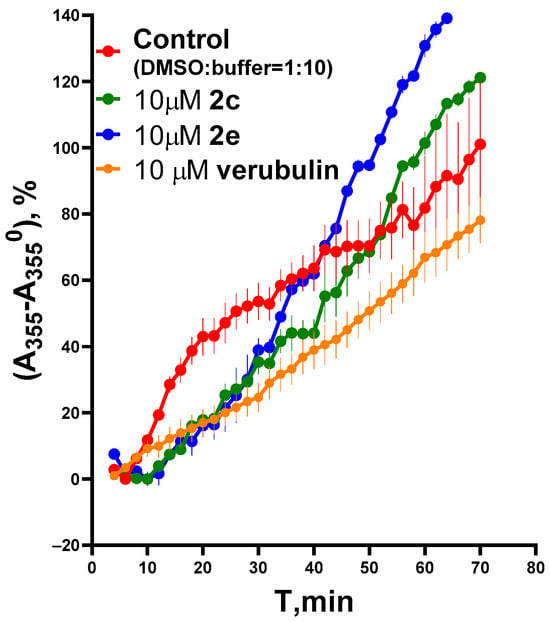
Figure 3.
Influence of compounds 2c and 2e in comparison with verubulin (positive control) on Tb + MAP polymerization as a time-dependent increase in absorbance at 355 nm. Background-subtracted data were normalized to control probe. This figure shows the results of the representative experiment. Each curve is the average of three repeats; each point is mean ± SEM.
2.2.3. Molecular Dynamics Simulations for Compound 2c
The obtained data (Figure 2 and Figure 3) confirm that new compounds significantly disturb the assembly of microtubules and exhibit characteristics similar to tubulin polymerization inhibitors, particularly such as verubulin. As was mentioned above, the latter binds to the colchicine site of tubulin, which is located in the β-subunit at the interface with the α-subunit of the α,β-heterodimer. The data of X-ray diffraction analysis of tubulin complexes with verubulin (PDB ID: 5XKF) [46] and several of its analogues [29,30,33,34] have been collected recently. To get an idea of how the saturated analogue of verubulin 2c (the most active in the series) binds to the protein, we performed molecular dynamics simulations (Figure 4). The plot of the root mean square deviations (RMSDs, Figure 4A) for the protein, GDP, GTP and ligand non-hydrogen atoms and the visual analysis of the trajectory confirm that system stability is retained from about the fiftieth nanosecond until the end of the course of the production simulation (100 ns), although the position of the ligand is slightly adjusted compared to the docking pose (by a shift of tetrahydroquinazoline residue).
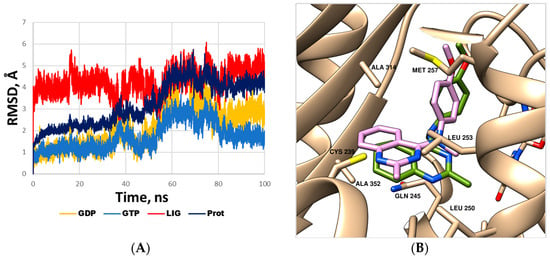
Figure 4.
(A) Root mean square deviation of the protein, GDP, GTP and ligand (2c) non-hydrogen atoms during molecular dynamics simulation of the complex with the α,β-tubulin heterodimer. (B) Binding mode of compound 2c refined using molecular dynamics simulation (detailed view of the binding site in the β-subunit). Ligand 2c is represented by a green-colored stick model; the amino acid residues located within 3 Å of the ligand are represented by beige stick models. The position of verubulin (PDB ID: 5XKF, matched to the β-subunit) is shown by a mauve-colored stick model. The guanosine triphosphate molecule bound in the α-subunits is shown by stick models on the right in the background. The hydrogen atoms are omitted for clarity.
As shown in Figure 4B, the saturated analogue of verubulin, compound 2c, is oriented in the colchicine-binding site similar to the parent molecule. Ring B in 2c is located in the hydrophobic pocket within the side chains of Ala252β, Ala314β, Ala315β, Ile316β. Both the methyl group attached to tetrahydroquinazoline and the methoxy group of 4-methoxyphenyl residue also form important hydrophobic interactions with the β-tubulin subunit (the former is exposed in the hydrophobic subpocket formed by the side chain of Leu250β, Leu253β and the aliphatic chain of Gln245, while the latter is located close to the aliphatic chains of Met257β and Lys350β). The N1 atom of ring A of verubulin analogues, as was shown by Banerjee et al. [29], can form a key water-mediated hydrogen bond with the main chain of Cys239β, and since such an interaction is water-mediated it is theoretically possible for compound 2c, despite some differences in the location of ring A compared to verubulin.
The binding mode of compound 2c corresponds to the observed SAR patterns, in particular, to a decrease in activity when the non-polar group at position 2 of ring A was replaced by a polar one, as well as a loss of activity upon removal of the methoxy group in ring C (see above, Table 1). The dramatic difference in cytotoxicity of compounds 2o and 2j (both with polar substituents at position 2 of ring A) is most likely due to the disruption of the above-mentioned water-mediated hydrogen bonds of the N1 with the key amino acid residue Cys239β caused by the hydroxyl group of 2o, capable of forming stronger hydrogen bonds than the amino group of its counterpart 2j. Overall, molecular modeling and SAR data further confirm that saturated verubulin analogue 2c retains the mode of action of the parent molecule.
2.2.4. Apoptosis Induction by Verubulin Analogues
Apoptosis induction by verubulin and hit compounds 2c,e,i,j,m was studied by flow cytometry to reveal the mechanism of HCT116 tumor cell death. Apoptosis is an important and active regulatory pathway of cell growth and proliferation. Annexin V is a calcium-dependent phospholipid-binding protein with a high affinity for phosphatidylserine (PS), a membrane component normally localized to the internal face of the cell membrane. Early in the apoptotic pathway, molecules of PS are translocated to the outer surface of the cell membrane where Annexin V can readily bind them [47,48].
All tested compounds were found to significantly induce apoptosis of HCT116 cells after 48 h of exposure (Figure 5). Nearly 77.5% of untreated HCT116 cells were viable (Annexin V (−) and 7-AAD (−)). For verubulin (1), the fraction of viable cells was reduced compared to the control, and almost half of the studied cells (49.3%) were found at the early (Annexin V (+) and 7-AAD (−) 16.3%) or late (Annexin V (+) and 7-AAD (+) 33%) apoptosis stages. The number of viable cells was about 50% of the entire studied population of HCT116 cells. Saturation of ring B (2c) was shown to affect the activation of apoptosis: the population of apoptotic cells decreased down to 30% when treated with 2c (Figure 5D). Further conformational restriction (2e) led to a decrease in the number of viable cells down to 30% and a significant increase in the number of cells at the late apoptosis stage (Figure 5E). The replacement of the methyl group in position 2 of the pyrimidine ring in compound 2c by chlorine (2m) had no effect on the apoptotic pattern compared to verubulin, while its replacement by a NH2 group (2i) changed it: the percentage of early apoptotic cells increased significantly (46.3%) and the population of cells in late apoptosis decreased. Thus, the introduction of an amino group into the pyrimidine ring affects the rate of cell entry into apoptosis. For compound 2j, the population of cells in early apoptosis increased up to 56.4%.

Figure 5.
Apoptotic profile of HCT116 cells treated with selected compounds after 48 h. Concentration of compounds 2 × IC50 (see Supplementary Materials).
The obtained data demonstrate that saturation of ring B in the verubulin molecule, though preserving both cytotoxicity and the effect on microtubules, does not lead to an increase in the selectivity. Therefore, we studied the opportunity to improve the toxicological profile for compound 2c via its encapsulation in Ca2+/Mg2+ cross-linked alginate nanoparticles as a drug delivery system.
2.3. Encapsulation of Compound 2c in Biocompatible Nanocontainers Based on Ca2+/Mg2+ Cross-Linked Alginate and Investigation of Cytotoxicity of Formulations via MTT Assay
Compound 2c was encapsulated in alginate (Alg)-based nanocontainers cross-linked with Ca2+ and Mg2+ ions synthesized following the procedure described in [44,49]. The molar ratio both for [Alginate monomer units]/[Ca2+] and [Alginate monomer units]/[Mg2+] was 10 ÷ 1. After purification and lyophilization, white complexes were obtained which were dissolved in water that resulted in (Alg)–Ca2+ and (Alg)–Mg2+ nanocontainer solutions, respectively.
Aminopyrimidine 2c was transformed into a protonated form 2c∙HCl for better interaction with the carboxyl groups of anionic nanocontainers (see Supplementary Materials).
The formation of polysaccharide nanocontainers as well as their interaction with aminopyrimidine 2c∙HCl was controlled by means of dynamic light scattering (DLS), laser microelectrophoresis and UV spectrophotometry (Figure S4).
The synthesized nanocontainers as well as nanocontainers filled with 2c were easily dispersed in bi-distilled water. In all cases, dynamic light scattering found one type of composite microgel with narrow particle size distribution (see Figure S5 for typical distribution functions). It was established that the effective hydrodynamic diameter (Dh) of the initial alginate macromolecules in an aqueous solution is 680 nm. The interaction of linear alginate both with Ca2+ and Mg2+ ions leads to a significant contraction in particle size: for samples (Alg)–Ca2+ and (Alg)–Mg2+, the particle diameters are 150 nm and 205 nm, respectively (Table 2). The effective diameters of 2c∙HCl–(Alg)–Ca2+ and 2c∙HCl–(Alg)–Mg2+ decrease down to 130 nm and 106 nm, respectively, that confirms the formation of the formulations. The polydispersity of alginate-based nanocarriers was evaluated using PDI [50]. It was shown that the synthesized nanocontainers are characterized by PDI values not exceeding 0.05. Negative EPM values indicate that both 2c∙HCl–(Alg)–Ca2+ and 2c∙HCl–(Alg)–Mg2+ are characterized by high aggregative stability.

Table 2.
Characteristics of aqueous dispersions of Ca2+ and Mg2+ cross-linked nanocontainers filled with immobilized compound 2c and their components.
The incorporation of aminopyrimidine 2c into microgels (entries 1 and 3) was also confirmed using UV spectroscopy (Figure S4). The absorption spectrum of protonated aminopyrimidine 2c shows two pronounced electronic transitions at wavelengths of 225 nm and 300 nm. In the absorption spectra of the interaction products of both microgels with 2c, there is a sharp decrease in the peak intensity at 225 nm. The peak intensity at 300 nm does not change significantly. The obtained result indicates the electrostatic interaction of the protonated form of 2c with the carboxyl groups of the microgels [51,52]. In turn, the preservation of the peak at 300 nm can be used for the quantitative determination of the 2c content in microgels. A calibration curve (Figure S4) was plotted to quantify the 2c content in the nanocontainers. It was established that both (Alg)–Ca2+ and (Alg)–Mg2+ bound 16 wt.% aminopyrimidine 2c that corresponds to the quantitative binding of 2c. In addition, the nature of the cross-linking ions does not affect the binding capacity of nanocontainers towards aminopyrimidine 2c.
To evaluate the ability of the nanocontainers to retain the loaded aminopyrimidine 2c, external dialysis solutions after purification of the microgels were analyzed by measuring the optical density at two wavelengths of 225 nm (D225) and 300 nm (D300), corresponding to the two maxima in the spectrum of the control aminopyrimidine solution (Figure S4). The D225 and D300 in external dialysis solutions after purification of systems 2c∙HCl–(Alg)–Ca2+ and 2c∙HCl–(Alg)–Mg2+ were found to be 0.18 and 0.012, respectively, which is comparable to the corresponding value for the solvent (bi-distilled water). That confirms that the loaded compound is retained in the nanocontainers.
The MTT assay was carried out on the A549 cell line and non-tumor hTERT-immortalized human fibroblasts for 48 h (Table 2; for cell survival curves, see Supplementary Materials). While (Alg)–Ca2+ and (Alg)–Mg2+ nanocontainers demonstrated no or low toxicity against cell lines of different etiology (entries 1 and 3, respectively), their formulations with compound 2c were found to preserve cytotoxic activity at the level of free amine 2c.
The differences in cell survival were demonstrated on non-tumor human hTERT-immortalized fibroblasts. While for the Mg2+ formulation (entry 3) IC50 values obtained for A549 and hTERT-immortalized fibroblast cell lines were the same, the Ca2+ formulation (entry 1) demonstrated two-fold selectivity towards tumor cells.
The data obtained allow us to conclude that the cytotoxic activity against the A549 cell line is preserved under the action of the compound 2c in alginate formulations compared to its free form. The encapsulation of the amine in (Alg)–Ca2+ nanoparticles is more promising from the point of view of selectivity to cancer cells.
3. Materials and Methods
3.1. Chemistry
3.1.1. General Remarks
1H and 13C NMR spectra were recorded on a 400 MHz Agilent 400-MR spectrometer (400.0 and 100.6 MHz for 1H and 13C respectively; Agilent Technologies, Santa Clara, CA, USA); chemical shift δ were measured with reference to the solvent (CDCl3, δH = 7.26 ppm, δC = 77.16 ppm). When necessary, assignments of signals in NMR spectra were made using 2D techniques. Fourier-transform infrared (FT-IR) spectra were obtained on a Bruker ALPHA FT-IR spectrometer (Bruker Daltonik GmbH, Bremen, Germany). Accurate mass measurements (HRMS) were performed on a Bruker micrOTOF II mass spectrometer (Bruker Daltonik GmbH, Bremen, Germany) with electrospray ionization (ESI). Mass spectra (MS) were obtained using a GC-MS Thermo ISQ instrument (Thermo Fisher Scientific, Waltham, MA, USA) with electron ionization (EI). Analytical thin layer chromatography was carried out with silica gel plates supported on aluminum (ALUGRAM® Xtra SIL G/UV254, Macherey-Nagel, Duren, Germany); the detection was carried out by a UV lamp (254 and 365 nm). Column chromatography was performed on silica gel (Silica 60, 0.015–0.04 mm, Macherey-Nagel, Duren, Germany). 4-Chloropyrimidines 3a [53], 3b [54], 3c [55], 3d [56] were obtained via described methods. All other starting materials were commercially available. All reagents except commercial products of satisfactory quality were purified according to literature procedures prior to use.
3.1.2. SNAr Reactions of 4-Chloropyrimidines 2a–d with Amines (General Method)
To the solution of chloropyrimidine 2a–d (0.55 mmol) and corresponding amine (0.66 mmol) in isopropanol (5 mL), two drops of HCl were added. The reaction mixture was refluxed for 5 h. When chloropyrimidine 2d was used as starting compound, reagent ratios and reaction time were modified, see below. Isopropanol was evaporated under reduced pressure and, to the residue, water (15 mL) was added and the resulting mixture was extracted with chloroform (3 × 15 mL). Combined organic layers were dried over MgSO4. The solvent was evaporated under reduced pressure and the product was isolated via preparative column chromatography.
N,2-Dimethyl-N-phenyl-5,6,7,8-tetrahydroquinazolin-4-amine (2a)
Yield 71 mg (51%). Yellowish solid, m.p. 84–85 °C; Rf = 0.61 (CHCl3–MeOH 10:1).
1H NMR (CDCl3, δ, ppm): 1.42–1.53 m (2H, CH2), 1.49–1.63 m (2H, CH2), 1.72–1.80 m (2H, CH2), 2.58 s (3H, CH3), 2.69–2.80 m (2H, CH2), 3.45 s (3H, CH3N), 6.95–7.03 m (2H, 2CH), 7.05–7.13 m (1H, CH), 7.24–7.34 m (2H, 2CH).
13C NMR (CDCl3, δ, ppm): 22.3 (CH2), 22.8 (CH2), 25.9 (CH3), 26.1 (CH2), 32.3 (CH2), 41.5 (CH3N), 116.2 (C(4a)), 124.2 (2CH), 124.4 (CH), 129.3 (2CH), 148.1 (C, Ph), 163.4 (C(4)), 163.9 (C(2)), 164.8 (C(8a)).
FT-IR (KBr tablet, ν, cm−1): 3440, 3055, 3034, 2932, 2862, 2657, 2436, 2360, 2335, 1976, 1938, 1910, 1865, 1834, 1734, 1596, 1558, 1495, 1468, 1438, 1423, 1384, 1333, 1278, 1218, 1186, 1158, 1119, 1100, 1074, 1028, 993, 904, 889, 861, 833, 759, 741, 709, 698, 662, 613, 580, 547, 497, 449.
MS (EI+, m/z (%)): 255 (1), 254 (14), 253 (87) [M]+, 252 (100), 238 (14) [M-CH3]+, 224 (11), 183 (5), 176 (11) [M-C6H5]+, 163 (5), 162 (55), 161 (12), 148 (8), 107 (7), 106 (20), 105 (5), 104 (7), 91 (6), 79 (11), 78 (5), 77 (21).
HRMS (ESI+, m/z): calculated for C16H19N3 [M + H]+ 254.1652, found 254.1651.
N,2-Dimethyl-N-(4-methylphenyl)-5,6,7,8-tetrahydroquinazolin-4-amine (2b)
Yield 121 mg (82%). Yellowish solid, m.p. 91–92 °C; Rf = 0.38 (CHCl3–MeOH 10:1).
1H NMR (CDCl3, δ, ppm): 1.42–1.53 m (2H, CH2), 1.62–1.78 m (4H, 2CH2), 2.33 s (3H, CH3Ar), 2.57 s (3H, CH3), 2.67–2.76 br.t (2H, CH2, 3.41 c (3H, CH3N), 6.87–6.93 m (2H, 2CH), 7.06–7.12 m (2H, 2CH).
13C NMR (CDCl3, δ, ppm): 21.1 (CH3Ar), 22.4 (CH2), 22.8 (CH2), 25.9 (CH3), 26.1 (CH2), 32.4 (CH2), 41.8 (CH3N), 115.8 (C(4a)), 124.6 (2CH), 130.0 (2CH), 134.3 (C, Ar), 145.7 (C, Ar), 163.3 (C(4)), 163.8 (C(2)), 164.5 (C(8a)).
FT-IR (KBr tablet, ν, cm−1): 3434, 3020, 2969, 2929, 2859, 2651, 2435, 2370, 2329, 1949, 1925, 1816, 1729, 1559, 1542, 1512, 1472, 1447, 1412, 1390, 1282, 1257, 1216, 1182, 1157, 1118, 1102, 1069, 1023, 994, 949, 906, 892, 863, 829, 765, 734, 722, 654, 630, 597, 573, 542, 501, 473, 444.
MS (EI+, m/z (%)): 269 (1), 268 (17), 267 (93) [M]+, 266 (100), 252 (15) [M-CH3]+, 250 (5), 238 (9), 176 (9), 163 (6), 162 (59), 161 (10), 148 (7), 147 (5), 121 (5), 120 (20), 119 (5), 118 (6), 107 (11), 106 (7), 105 (7), 91 (13), 79 (10), 77 (10), 65 (5).
HRMS (ESI+, m/z): calculated for C17H21N3 [M + H]+ 268.1808, found 268.1811.
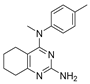
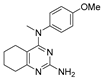
N-(4-methoxyphenyl)-N,2-dimethyl-5,6,7,8-tetrahydroquinazolin-4-amine (2c)
Yield 98 mg (63%). Beige solid, m.p. 87–88 °C; Rf = 0.68 (CHCl3–MeOH 10:1).
1H NMR (CDCl3, δ, ppm): 1.40–1.52 m (2H, CH2), 1.59–1.75 m (4H, 2CH2), 2.56 s (3H, CH3), 2.64–2.76 m (2H, CH2), 3.38 s (3H, CH3N), 3.80 s (3H, CH3O), 6.79–6.88 m (2H, 2CH), 6.92–7.02 m (2H, 2CH).
13C NMR (CDCl3, δ, ppm): 22.3 (CH2), 22.9 (CH2), 25.9 (CH3), 26.1 (CH2), 32.3 (CH2), 42.2 (CH3N), 55.6 (CH3O), 114.5 (2CH), 115.2 (C(4a)), 126.6 (2CH), 141.5 (C, Ar), 156.9 (C-O, Ar), 163.5 (C(4)), 163.6 (C(2)), 164.3 (C(8a)).
FT-IR (KBr tablet, ν, cm−1): 3444, 3054, 3010, 2934, 2916, 2859, 2839, 2432, 2065, 1947, 1870, 1610, 1558, 1544, 1512, 1463, 1443, 1413, 1385, 1297, 1251, 1183, 1172, 1156, 1117, 1099, 1037, 993, 949, 904, 891, 841, 816, 768, 734, 656, 631, 596, 573, 553, 506, 468, 441.
MS (EI+, m/z (%)): 285 (2), 284 (17), 283 (100) [M]+, 282 (60), 268 (25) [M-CH3]+, 254 (5), 176 (6), 163 (8), 162 (69), 148 (9), 147 (11), 142 (5), 136 (13), 123 (9), 121 (19), 120 (8), 107 (8), 106 (8), 92 (5), 79 (15), 78 (6), 77 (15).
HRMS (ESI+, m/z): calculated for C17H21N3O [M + H]+ 284.1757, found 284.1757.
4-(3,4-Dihydroquinolin-1(2H)-yl)-2-methyl-5,6,7,8-tetrahydroquinazoline (2d)
Yield 108 mg (70%). Beige solid, m.p. 118–119 °C; Rf = 0.80 (CHCl3–MeOH 10:1).
1H NMR (CDCl3, δ, ppm): 1.48–1.65 m (2H, CH2, THQ), 1.72–1.85 m (2H, CH2, THQ), 1.96–2.13 m (4H, CH2, THQ + CH2, DHQ), 2.58 (3H, CH3), 2.72–2.90 m (4H, CH2, THQ + CH2, DHQ), 3.76–3.85 m (2H, CH2N, DHQ), 6.36–6.46 m (1H, CH), 6.80–6.88 m (1H, CH), 6.92–7.01 m (1H, CH), 7.04–7.13 m (1H, CH).
13C NMR (CDCl3, δ, ppm): 22.5 (CH2, THQ), 22.6 (CH2, THQ), 24.1 (CH2, DHQ), 25.8 (CH3), 25.9 (CH2, THQ), 27.1 (CH2, DHQ), 32.4 (CH2, THQ), 47.3 (CH2N, DHQ), 117.9 (CH), 118.7 (C(4a)), 121.2 (CH), 126.0 (CH), 127.8 (C, DHQ), 128.9 (CH), 141.7 (C, DHQ), 162.5 (C(4)), 164.7 (C(2)), 166.0 (C(8a)).
FT-IR (KBr tablet, ν, cm−1): 3445, 3062, 3034, 2945, 2924, 2875, 2843, 2430, 1916, 1733, 1643, 1603, 1551, 1491, 1423, 1405, 1337, 1292, 1254, 1216, 1193, 1155, 1133, 1111, 1051, 1034, 994, 974, 945, 903, 886, 832, 759, 659, 622, 580, 547, 498, 426.
MS (EI+, m/z (%)): 281 (2), 280 (17), 279 (94) [M]+, 278 (100), 264 (10) [M-CH3]+, 251 (7), 250 (18), 237 (6), 236 (7), 209 (7), 162 (14), 149 (15), 148 (26), 147 (6), 133 (9), 132 (75), 131 (11), 130 (15), 126 (8), 117 (11), 115 (8), 107 (11), 106 (9), 104 (6), 91 (7), 80 (5), 79 (12), 78 (5), 77 (13).
HRMS (ESI+, m/z): calculated for C18H21N3 [M + H]+ 280.1808, found 280.1809.
4-(6-Methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-methyl-5,6,7,8-tetrahydroquinazoline (2e)
Yield 95 mg (56%). Yellowish solid, m.p. 116–117 °C; Rf = 0.46 (CHCl3–MeOH 20:1).
1H NMR (CDCl3, δ, ppm): 1.41–1.57 m (2H, CH2, THQ), 1.66–1.78 m (2H, CH2, THQ), 1.82–1.91 m (2H, CH2, THQ), 1.92–2.03 m (2H, CH2, DHQ), 2.57 s (3H, CH3), 2.67–2.77 m (2H, CH2, DHQ), 2.78–2.88 m (2H, CH2, THQ), 3.72 s (3H, CH3O), 3.75–3.85 m (2H, CH2N, DHQ), 6.45 d (1H, 3J = 8.7 Hz, CH), 6.57 dd (1H, 3J = 8.7 Hz, 4J = 2.7 Hz, CH), 6.65 d (1H, 4J = 2.7 Hz, CH).
13C NMR (CDCl3, δ, ppm): 21.8 (CH2, THQ), 22.4 (CH2, THQ), 24.1 (CH2, DHQ), 24.6 (CH3), 26.3 (CH2, THQ), 27.0 (CH2, DHQ), 30.9 (CH2, THQ), 47.0 (CH2N, DHQ), 55.4 (CH3O), 111.2 (CH), 113.8 (CH), 116.7 (C(4a)), 120.6 (CH), 131.5 (C, DHQ), 134.3 (C, DHQ), 155.4 (C-O, DHQ), 162.1 (C(4)), 162.9 (C(2), C(8a)).
FT-IR (KBr tablet, ν, cm−1): 3470, 3428, 3239, 3055, 2936, 2858, 2747, 2688, 2583, 2361,1777, 1614, 1552, 1502, 1467, 1438, 1404, 1320, 1296, 1262, 1236, 1204, 1163, 1136, 1114, 1054, 1038, 1024, 995, 931, 882, 864, 833, 781, 758, 738, 714, 655, 630, 604, 575, 548, 451, 429.
MS (EI+, m/z (%)): 311 (2), 310 (20), 309 (100) [M]+, 308 (55), 281 (5), 280 (9), 266 (7), 163 (9), 162 (73), 161 (11), 160 (6), 155 (8), 149 (9), 148 (20), 147 (20), 146 (5), 141 (5), 131 (5), 130 (5), 107 (8), 106 (9), 79 (19), 77 (13).
HRMS (ESI+, m/z): calculated for C19H23N3O [M + H]+ 310.1914, found 310.1916.
2-Methyl-4-(7-nitro-3,4-dihydroquinolin-1(2H)-yl)-5,6,7,8-tetrahydroquinazoline (2f)
Yield 110 mg (62%). Yellow solid, m.p. 184–185 °C; Rf = 0.14 (light petrol–EtOAc–MeOH 3:1:0.01).
1H NMR (CDCl3, δ, ppm): 1.59–1.72 m (2H, CH2, THQ), 1.82–1.91 m (2H, CH2, THQ), 2.05–2.14 m (2H, CH2, DHQ), 2.14–2.22 m (2H, CH2, THQ), 2.62 s (3H, CH3), 2.84–2.98 m (4H, CH2, THQ + CH2, DHQ), 3.76–3.85 m (2H, CH2N, DHQ), 7.18 d (1H, 4J = 2.2 Hz, CH), 7.20 d (1H, 3J = 8.3 Hz, CH), 7.65 dd (1H, 3J = 8.3 Hz, 4J = 2.2 Hz, CH).
13C NMR (CDCl3, δ, ppm): 22.44 (CH2, THQ), 22.46 (CH2, THQ), 22.8 (CH2, DHQ), 25.7 (CH2, THQ), 25.8 (CH3), 27.7 (CH2, DHQ), 32.5 (CH2, THQ), 47.7 (CH2, DHQ), 111.3 (CH), 114.9 (CH), 119.9 (C(4a)), 129.8 (CH), 133.3 (C, DHQ), 142.5 (C, DHQ), 146.9 (CNO2, DHQ), 162.1 (C(4)), 165.5 (C(2)), 168.0 (C(8a)).
FT-IR (KBr tablet, ν, cm−1): 3446, 2949, 2911, 2862, 2423, 1919, 1775, 1617, 1550, 1518, 1494, 1458, 1421, 1394, 1342, 1275, 1248, 1213, 1154, 1135, 1119, 1097, 1050, 996, 923, 884, 862, 821, 803, 758, 736, 683, 659, 627, 584, 523, 475.
MS (EI+, m/z (%)): 326 (1), 325 (12), 324 (57) [M]+, 323 (12), 309 (6) [M-CH3]+, 308 (6), 307 (26), 296 (5), 290 (12), 282 (6), 277 (5), 249 (5), 177 (22), 173 (5), 163 (5), 162 (22), 160 (6), 149 (19), 148 (100), 147 (13), 131 (7), 130 (10), 121 (7), 107 (31), 106 (15), 104 (9), 103 (5), 80 (9), 79 (17), 78 (7), 77 (17), 42 (5).
HRMS (ESI+, m/z): calculated for C18H20N4O2 [M + H]+ 325.1659, found 325.1655.
N4-Methyl-N4-phenyl-5,6,7,8-tetrahydroquinazolinyl-2,4-diamine (2g)
Yield 71 mg (51%). Beige solid, m.p. 128–129 °C; Rf = 0.38 (CHCl3–MeOH 10:1).
1H NMR (CDCl3, δ, ppm): 1.35–1.48 m (2H, CH2), 1.53–1.69 m (4H, 2CH2), 2.57–2.71 m (2H, CH2), 3.39 s (3H, CH3), 5.31 br.s (2H, NH2), 6.98–7.07 m (2H, 2CH), 7.12–7.21 m (1H, CH), 7.27–7.36 m (2H, 2CH).
13C NMR (CDCl3, δ, ppm): 21.8 (CH2), 23.0 (CH2), 25.7 (CH2), 30.9 (CH2), 41.9 (CH3N), 109.3 (C(4a)), 125.0 (2CH), 125.2 (CH), 129.4 (2CH), 147.3 (C, Ph), 158.6 (C(2)), 162.0 (C(8a)), 164.5 (C(4)).
HRMS (ESI+, m/z): calculated for C15H18N4 [M + H]+ 255.1604, found 255.1599.
N4-Methyl-N4-(4-methylphenyl)-5,6,7,8-tetrahydroquinazolinyl-2,4-diamine (2h)
Yield 74 mg (50%). Beige solid, m.p. 135–136 °C; Rf = 0.31 (CHCl3–MeOH 10:1).
1H NMR (CDCl3, δ, ppm): 1.30–1.45 m (2H, CH2), 1.45–1.54 m (2H, CH2), 1.54–1.64 m (2H, CH2), 2.35 s (3H, CH3), 2.62–2.74 m (2H, CH2), 3.39 s (3H, CH3N), 6.24 br.s (2H, NH2), 6.91–7.03 m (2H, 2CH), 7.11–7.20 m (2H, 2CH).
13C NMR (CDCl3, δ, ppm): 20.7 (CH2), 21.1 (CH3, Ar), 22.6 (CH2), 25.4 (CH2), 28.4 (CH2), 42.9 (CH3N), 108.0 (C(4a)), 126.0 (2CH), 130.2 (2CH), 137.1 (C, Ar), 143.1 (C, Ar), 155.16 (C(2)), 155.21 (C(8a)), 164.3 (C(4)).
HRMS (ESI+, m/z): calculated for C16H20N4 [M + H]+ 269.1761, found 269.1755.
N4-(4-Methoxyphenyl)-N4-methyl-5,6,7,8-tetrahydroquinazolinyl-2,4-diamine (2i)
Yield 59 mg (38%). Beige solid, m.p. 132–133 °C; Rf = 0.4 (CHCl3–MeOH 10:1).
1H NMR (CDCl3, δ, ppm): 1.39–1.48 m (2H, CH2), 1.57–1.67 m (4H, 2CH2), 2.54–2.62 m (2H, CH2), 3.31 s (3H, CH3N), 3.81 s (3H, CH3O), 4.69 br.s (2H, NH2), 6.78–6.86 m (2H, 2CH), 6.95–7.01 m (2H, 2CH).
13C NMR (CDCl3, δ, ppm): 22.4 (CH2), 23.2 (CH2), 25.9 (CH2), 32.3 (CH2), 42.2 (CH3N), 55.6 (CH3O), 109.0 (C(4a)), 114.5 (2CH), 126.7 (2CH), 141.5 (C, Ar), 156.8 (C, Ar), 160.1 (C), 164.7 (C), 164.9 (C).
HRMS (ESI+, m/z): calculated for C16H20N4O [M + H]+ 285.1710, found 285.1707.
2-Amino-4-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-5,6,7,8-tetrahydroquinazoline (2j)
Yield 61 mg (36%). Beige solid, m.p. 145–146 °C; Rf = 0.65 (light petrol–EtOAc 2:3).
1H NMR (CDCl3, δ, ppm): 1.44–1.59 m (2H, CH2, THQ), 1.67–1.80 m (2H, CH2, THQ), 1.83–1.94 m (2H, CH2, THQ), 1.95–2.06 m (2H, CH2, DHQ), 2.60–2.72 m (2H, CH2, THQ), 2.72–2.81 m (2H, CH2, DHQ), 3.66–3.80 m (2H, CH2, DHQ), 3.78 s (3H, CH3O), 5.06 br.s (2H, NH2), 6.55 d (1H, 3J = 8.7 Hz, CH), 6.62 dd (1H, 3J = 8.7 Hz, 4J = 2.7 Hz, CH), 6.70 d (1H, 4J = 2.7 Hz, CH).
13C NMR (CDCl3, δ, ppm): 22.3 (CH2), 23.0 (CH2), 24.2 (CH2), 26.1 (CH2), 27.3 (CH2), 31.6 (CH2), 47.0 (CH2N), 55.6 (CH3O), 111.0 (C(4a), THQ), 111.5 (CH), 113.8 (CH), 121.0 (CH), 131.0 (C, DHQ), 135.0 (C, DHQ), 155.1 (C, DHQ), 155.1 (C, THQ), 159.8 (C, THQ), 163.7 (C, THQ).
HRMS (ESI+, m/z): calculated for C18H22N4O [M + H]+ 311.1866, found 311.1863.
N-(4-Methoxyphenyl)-N,2-dimethyl-6,7,8,9-tetrahydro-5H-cyclohepta[d]pyrimidin-4-amine (2k)
Yield 75 mg (46%). Beige solid, m.p. 92–93 °C; Rf = 0.66 (CHCl3–MeOH 10:1).
1H NMR (CDCl3, δ, ppm): 1.07–1.18 m (2H, CH2), 1.53–1.71 m (4H, 2CH2), 2.16–2.29 m (2H, CH2), 2.58 s (3H, CH3), 2.76–2.88 m (2H, CH2), 3.35 s (3H, CH3N), 3.78 s (3H, CH3O), 6.77–6.85 m (2H, 2CH), 6.87–6.96 m (2H, 2CH).
13C NMR (CDCl3, δ, ppm): 25.7 (CH3), 25.8 (CH2), 26.3 (CH2), 27.7 (CH2), 31.9 (CH2), 38.6 (CH2), 42.1 (CH3N), 55.6 (CH3O), 114.8 (2CH, Ar), 120.5 (C(4a)), 125.1 (2CH, Ar), 142.9 (C, Ar), 156.5 (C, Ar), 163.4 (C(2)), 163.5 (C(4)), 171.0 (C(9a)).
HRMS (ESI+, m/z): calculated for C18H23N3O [M + H]+ 298.1914, found 298.1917.
4-(6-Methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-methyl-6,7,8,9-tetrahydro-5H-cyclohepta[d]pyrimidine (2l)
Yield 107 mg (60%). Orange solid, m.p. 122–123 °C; Rf = 0.58 (CHCl3–MeOH 10:1).
1H NMR (CDCl3, δ, ppm): 1.37–1.50 m (2H, CH2), 1.65–1.84 m (4H, 2CH2), 1.98–2.03 m (2H, CH2, DHQ), 2.31–2.43 m (2H, CH2), 2.60 s (3H, CH3), 2.77–2.86 m (2H, CH2, DHQ), 2.90–3.00 m (2H, CH2), 3.69–3.82 m (2H, CH2N, DHQ), 3.76 s (3H, CH3O), 6.44 d (1H, 3J = 8.8 Hz, CH), 6.57 dd (1H, 3J = 8.8 Hz, 4J = 2.8 Hz, CH), 6.68 d (1H, 4J = 2.8 Hz, CH).
13C NMR (CDCl3, δ, ppm): 23.7 (CH2, DHQ), 25.3 (CH3), 25.8 (CH2), 27.2 (CH2), 27.5 (CH2, DHQ), 28.2 (CH2), 32.1 (CH2), 38.4 (CH2), 48.0 (CH2N, DHQ), 55.6 (CH3O), 112.0 (CH), 114.3 (CH), 118.6 (CH), 123.1 (C(4a)), 128.7 (C, Ar), 136.4 (C, Ar), 154.5 (C-O, Ar), 162.4 (C), 164.1 (2C).
HRMS (ESI+, m/z): calculated for C20H25N3O [M + H]+ 324.2070, found 324.2068.
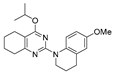
2-Chloro-N-(4-methoxyphenyl)-N-methyl-5,6,7,8-tetrahydroquinazolin-4-amine (2m)
Yield 27 mg (16%) from chloropyrimidine 2d (0.83 mmol, 168 mg) and 4-methoxy-N-methylaniline (0.55 mmol, 75 mg), the reaction mixture was refluxed for 6 h. White solid, m.p. 138–139 °C; Rf = 0.82 (CHCl3–MeOH 20:1).
1H NMR (CDCl3, δ, ppm): 1.38–1.51 m (2H, CH2), 1.58–1.71 m (4H, 2CH2), 2.67–2.76 m (2H, CH2), 3.38 s (3H, CH3N), 3.81 s (3H, CH3O), 6.79–6.90 m (2H, 2CH), 6.97–7.03 m (2H, 2CH).
13C NMR (CDCl3, δ, ppm): 21.9 (CH2), 22.6 (CH2), 26.2 (CH2), 32.4 (CH2), 42.7 (CH3N), 55.6 (CH3O), 114.7 (2CH), 116.1 (C(4a)), 127.1 (2CH), 140.2 (C, Ar), 156.9 (C(2)), 157.7 (C, Ar), 164.7 (C(4)), 167.1 (C(8a)).
HRMS (ESI+, m/z): calculated for C16H18ClN3O [M + H]+ 304.1211, 306.1183; found 304.1206, 306.1181.
N,N′-bis(4-methoxyphenyl)-N,N′-dimethyl-5,6,7,8-tetrahydroquinazolin-2,4-diamine (2n)
Yield 17 mg (13%), from chloropyrimidine 2d (0.55 mmol, 112 mg) and 4-methoxy-N-methylaniline (0.66 mmol, 90 mg), the reaction mixture was refluxed for 12 h. Brown amorphous solid, m.p. 35–40 °C; Rf = 0.18 (CH2Cl2–MeOH 20:1).
1H NMR (CDCl3, δ, ppm): 1.35–1.50 m (2H, CH2), 1.55–1.73 m (4H, 2CH2), 2.55–2.67 m (2H, CH2), 3.17 s (3H, CH3N), 3.53 s (3H, CH3N), 3.80 s (3H, CH3O), 3.82 s (3H, CH3O), 6.78–6.85 m (2H, 2CH), 6.84–6.93 m (2H, 2CH), 6.93–7.00 m (2H, 2CH), 7.28–7.35 m (2H, 2CH).
13C NMR (CDCl3, δ, ppm): 22.6 (CH2), 23.4 (CH2), 25.9 (CH2), 32.9 (CH2), 38.4 (CH3N), 42.7 (CH3N), 55.54 (CH3O), 55.58 (CH3O), 108.0 (C(4a)), 113.6 (2CH), 114.3 (2CH), 126.7 (2CH), 127.4 (2CH), 139.8 (C, Ar), 141.8 (C, Ar), 156.3 (C, Ar), 156.6 (C, Ar), 159.5 (C(2)), 163.6 (C(4)), 165.0 (C(8a)).
HRMS (ESI+, m/z): calculated for C24H28N4O2 [M + H]+ 405.2285, found 405.2284.
2-Hydroxy-4-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-5,6,7,8-tetrahydroquinazoline (2o)
Yield 18 mg (12%), from chloropyrimidine 2d (0.96 mmol, 194 mg) and 4-methoxy-N-methylaniline (0.48 mmol, 78 mg), the reaction mixture was refluxed for 60 h. Yellowish solid, m.p. 185–187 °C; Rf = 0.67 (CHCl3–MeOH 20:1).
1H NMR (CDCl3, δ, ppm): 1.66–1.83 m (4H, 2CH2, THQ), 1.89–2.02 m (2H, CH2, DHQ), 2.37–2.49 m (2H, CH2, THQ), 2.49–2.60 m (2H, CH2, THQ), 2.67–2.79 m (2H, CH2, DHQ), 3.81 s (3H, CH3), 3.88–3.97 m (2H, CH2, DHQ), 6.69–6.84 m (2H, 2CH), 7.07–7.15 m (1H, CH), 8.73 br.s (1H, OH).
13C NMR (CDCl3, δ, ppm): 21.8 (CH2, THQ), 22.4 (CH2, THQ), 22.7 (CH2, THQ), 23.8 (CH2, DHQ), 27.1 (CH2, DHQ), 32.5 (CH2, THQ), 45.1 (CH2N, DHQ), 55.7 (CH3O), 111.9 (C(4a)), 112.9 (CH), 115.5 (CH), 123.4 (CH), 130.1 (C, Ar), 135.2 (C, Ar), 150.0 (C(4)), 157.4 (C, Ar), 162.89 (C), 162.93 (C).
HRMS (ESI+, m/z): calculated for C18H21N3O2 [M + H]+ 312.1707, found 312.1695.
2,4-Bis(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-5,6,7,8-tetrahydroquinazoline (2p)
Yield 6 mg (6%), from chloropyrimidine 2d (0.96 mmol, 194 mg) and 4-methoxy-N-methylaniline (0.48 mmol, 78 mg), the reaction mixture was refluxed for 60 h. Yellow solid, m.p. 150–152 °C; Rf = 0.46 (light petrol–EtOAc 4:1).
1H NMR (CDCl3, δ, ppm): 1.49–1.61 m (2H, CH2, THQ), 1.71–1.82 m (2H, CH2, THQ), 1.88–2.08 m (6H, CH2, THQ + 2CH2, DHQ), 2.64–2.88 m (6H, CH2, THQ + 2CH2, DHQ), 3.68–3.75 m (2H, CH2, DHQ), 3.77 s (3H, CH3), 3.78 s (3H, CH3), 3.97–4.10 m (2H, CH2, DHQ), 6.51 d (1H, 3J = 8.8 Hz, CH), 6.59 dd (1H, 3J = 8.8 Hz, 4J = 2.8 Hz, CH), 6.62–6.73 m (3H, 3CH), 7.7d0 d (1H, 3J = 9.0 Hz, CH).
13C NMR (CDCl3, δ, ppm): 22.9 (CH2, THQ), 23.2 (CH2, THQ), 24.0 (CH2, DHQ), 24.5 (CH2, DHQ), 26.2 (CH2, THQ), 27.5 (CH2, DHQ), 28.2 (CH2, DHQ), 32.9 (CH2, THQ), 45.0 (CH2N, DHQ), 46.7 (CH2N, DHQ), 55.6 (2CH3O), 111.1 (C(4a)), 111.3 (CH), 111.4 (CH), 113.0 (CH), 113.6 (CH), 120.3 (CH), 125.6 (CH), 130.7 (C, Ar), 131.1 (C, Ar), 133.9 (C, Ar), 135.7 (C, Ar), 154.4 (C-O, Ar), 154.7 (C-O, Ar), 159.1 (C(2)), 162.5 (C(4)), 166.1 (C(8a)).
HRMS (ESI+, m/z): calculated for C28H32N4O2 [M + H]+ 457.2598, found 457.2586.
2-Isopropoxy-4-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-5,6,7,8-tetrahydroquinazoline (2q)
Yield 4 mg (2%), from chloropyrimidine 2d (0.96 mmol, 194 mg) and 4-methoxy-N-methylaniline (0.48 mmol, 78 mg), the reaction mixture was refluxed for 60 h. Yellowish solid, m.p. 135–137 °C; Rf = 0.33 (light petrol–EtOAc 4:1).
1H NMR (CDCl3, δ, ppm): 1.37 d (6H, 3J = 6.2 Hz, 2CH3, i-Pr), 1.49–1.57 m (2H, CH2, THQ), 1.71–1.80 m (2H, CH2, THQ), 1.88–1.96 m (2H, CH2, THQ), 1.96–2.07 m (2H, CH2, DHQ), 2.70–2.80 m (4H, CH2, THQ + CH2, DHQ), 3.78 s (3H, CH3), 3.75–3.83 m (2H, CH2, DHQ), 5.23 quint (1H, 3J = 6.2 Hz, CH, i-Pr), 6.51 d (1H, 3J = 8.8 Hz, CH), 6.61 dd (1H, 3J = 8.8 Hz, 4J = 2.9 Hz, CH), 6.70 d (1H, 4J = 2.9 Hz, CH).
13C NMR (CDCl3, δ, ppm): 22.2 (2CH3, i-Pr), 22.6 (CH2, THQ), 23.0 (CH2, THQ), 24.4 (CH2, DHQ), 26.3 (CH2, THQ), 27.4 (CH2, DHQ), 32.7 (CH2, THQ), 46.7 (CH2, DHQ), 55.6 (CH3O), 69.1 (CH-O, i-Pr), 111.3 (CH), 113.1 (C(4a)), 113.7 (CH), 120.5 (CH), 131.3 (C, Ar), 135.3 (C, Ar), 154.9 (C-O, Ar), 162.7 (C), 163.9 (C), 167.6 (C).
HRMS (ESI+, m/z): calculated for C21H27N3O2 [M + H]+ 354.2176, found 354.2166.
4-Isopropoxy-2-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-5,6,7,8-tetrahydroquinazoline (2r)
Yield 34 mg (20%), from chloropyrimidine 2d (0.96 mmol, 194 mg) and 4-methoxy-N-methylaniline (0.48 mmol, 78 mg), the reaction mixture was refluxed for 60 h. White solid, m.p. 94–95 °C; Rf = 0.75 (light petrol–EtOAc 2:1).
1H NMR (CDCl3, δ, ppm): 1.30 d (6H, 3J = 6.2 Hz, 2CH3, i-Pr), 1.69–1.83 m (4H, 2CH2, THQ), 1.88–2.01 m (2H, CH2, DHQ), 2.36–2.49 m (2H, CH2, THQ), 2.57–2.69 m (2H, CH2, THQ), 2.70–2.80 m (2H, CH2, DHQ), 3.79 s (3H, CH3), 3.93–4.04 m (2H, CH2, DHQ), 5.22 quint (1H, 3J = 6.2 Hz, CH-O, i-Pr), 6.64 d (1H, 4J = 2.9 Hz, CH), 6.70 dd (1H, 3J = 9.0 Hz, 4J = 2.9 Hz, CH), 7.73 d (1H, 3J = 9.0 Hz, CH).
13C NMR (CDCl3, δ, ppm): 21.6 (CH2, THQ), 22.2 (2CH3, i-Pr), 22.7 (CH2, THQ), 22.8 (CH2, THQ), 24.0 (CH2, DHQ), 28.1 (CH2, DHQ), 32.4 (CH2, THQ), 45.1 (CH2, DHQ), 55.6 (CH3O), 68.2 (CH-O, i-Pr), 106.9 (C(4a)), 111.3 (CH), 112.8 (CH), 125.9 (CH), 131.2 (C, Ar), 133.9 (C, Ar), 154.7 (C-O, Ar), 158.5 (C(2)), 165.2 (C(8a)), 166.9 (C(4)).
HRMS (ESI+, m/z): calculated for C21H27N3O2 [M + H]+ 354.2176, found 354.2169.
3.2. Biology
3.2.1. Cell Cultures
Human breast cancer cell line MCF7′ and human lung adenocarcinoma cell line A549 were kindly provided by Dr. S. Dmitriev (Lomonosov Moscow State University), immortalized human fibroblast cell line VA13 (WI38 subline 2RA) was kindly provided by Dr. M. Rubtsova (Lomonosov Moscow State University), human embryonic kidney HEK293T cell line was kindly provided by Dr. E. Knyazhanskaya (Lomonosov Moscow State University). MCF7′, VA13, A549 and HEK293T cell lines were maintained in DMEM/F-12 (Thermo Fisher Scientific, Waltham, MA, USA) culture medium containing 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA) and 50 μg/mL penicillin and 0.05 mg/mL streptomycin at 37 °C (Thermo Fisher Scientific, Waltham, MA, USA) in 5% CO2. Cells were maintained at 37 °C in a humidified MCO-18AC incubator (Sanyo, Tokyo, Japan) supplied with 5% CO2. Cell cultures were tested for the absence of mycoplasma and validated by STR.
3.2.2. Cell Viability Assay (MTT Assay)
The cytotoxicity of the substances was tested using the 3-(4,5-dimethylthiazol-2-yl)2,5-diphenyl tetrazolium bromide (MTT) assay [41] with some modifications. A total of 2500 cells per well for the MCF7′, HEK293T, A549 cell lines or 4000 cells per well for the VA-13 cell line were plated out in 135 μL of DMEM-F12 media (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) in a 96-well plate and incubated in the 5% CO2 incubator for the first 20 h without treatment. Then, 15.8 μL of media–DMSO solutions of tested substances (final DMSO concentrations in the media were 0.5% or less) was added to the cells (triplicate each) and cells were treated for 64 h with 0.023 mg/L–50 mg/L (eight dilutions) of compounds 2f,n,o,r; 1.14 μg/L–2.5 mg/L (eight dilutions) of compounds 2a,b,d,g,h,k,l,p,q; 57.2 ng/L–125 μg/L (eight dilutions) of compounds 2e,i,j; 22.9 ng/L–50 μg/L (eight dilutions) of compounds 1,2c,m. Doxorubicin was used as a control substance. Then, the MTT reagent (Paneco LLC, Moscow, Russia) was added to cells up to a final concentration of 0.5 g/L (10 × stock solution in PBS was used) and incubated for 2 h at 37 °C in the incubator, under an atmosphere of 5% CO2. Then, the MTT solution was discarded, and 140 μL of DMSO (PharmaMed LLC, Moscow, Russia) was added. The plates were swayed on a shaker (120 rpm) to dissolve the formazan. The absorbance was measured using a microplate reader (VICTOR X5 Light Plate Reader, PerkinElmer, Waltham, MA, USA) at a wavelength of 555 nm (in order to measure formazan concentration). The results were used to construct dose–response graphs and to estimate IC50 values (IC50 is the concentration resulting in half of the maximal cytotoxic effect) with GraphPad Prism V, GraphPadSoftware, Inc., San Diego, CA, USA.
3.2.3. Immunofluorescent Microscopy
A549 cells were seeded on coverslips (treated with poly-l-lysine at a concentration of 33 mg/L for an hour). Compounds were added up to the corresponding concentration after 24 h attachment and growth of the cells. Then, cells were incubated 24 h.
The cells were fixed with methanol (precooled in liquid nitrogen for 20 min), no additional permeabilization was performed. To prevent non-specific binding, cells were incubated with 4% BSA in PBS for one hour. Then, a solution of mouse monoclonal antibodies against alpha-tubulin cross-linked with Alexa 488 fluoroform (Thermo Fisher Scientific, Waltham, MA, USA, 32-2588) at a concentration of 5 μg/mL in 1% BSA solution in PBS was added to the cells, and the slides were incubated at +5 °C in a water chamber for 24 h. After the end of the incubation time, the cells were washed several times with PBS buffer solution and fixed on a glass slide with Moviol. Next day, cells were imaged with a ZEISS LSM 900 (ZEISS Microscopy, GmbH, Germany) and images processed with ImageJ.
3.2.4. Tubulin + MAP Polymerization
A tubulin + MAP polymerization assay was conducted as previously described in [57].
3.2.5. Cell Death
HCT116 (colon carcinoma) cells (3 × 105 cells in 2 mL of DMEM) were seeded in a 6-well plate and incubated with compounds 1,2c,e,i,j,m and cisplatin at 2 × IC50 (values based on MTT assay, see Supplementary Materials) for 48 h. After incubation, the cells were harvested by trypsinization, precipitated by centrifugation (3000 rpm), washed with cold PBS and recentrifuged. Aliquots of cells were processed as recommended in the Muse Annexin V& Dead Cell Kit (Luminex Corporation, Austin, TX, USA). The results were recorded on a Muse Cell Analyzer flow cytometer (Merck, Rahway, NJ, USA).
3.3. MTT Study of Compound 2c, Encapsulated in Alginate-Based Nanocontainers
3.3.1. General Remarks
The following reagents were used: sodium alginate (Na-Alg), calcium chloride (CaCl2), magnesium sulfate (MgSO4); all reagents from Sigma (Merck, Rahway, NJ, USA).
Dynamic light scattering (DLS) measurements were carried out in 0.15 M NaCl aqueous solution using a Photocor Complex photometer (Photocor Instruments, Moscow, Russia) equipped with a He-Ne 10 mW laser (λ = 633 nm) as the light source. The data were processed using DynaLS (DynaLS Version 2.7.1) software. Mean hydrodynamic sizes of particles were determined by DLS at the fixed scattering angle (90°). Software provided by the manufacturer was employed to calculate diameter values. Electrophoretic mobility (EPM) of particles was measured by laser microelectrophoresis in a thermostatic cell using a Brookhaven Zeta Plus (Brookhaven Instruments, Holtsville, NY, USA) device with the corresponding software. UV/vis spectroscopy measurements were performed with a UV-PC PE-5400UV spectrophotometer (Ecros, Moscow, Russia).
3.3.2. Preparation of Nanocontainers
All nanocontainers were synthesized at room temperature. A dilute solution of CaCl2 (1 mL) containing 0.56 mg of the compound was added dropwise to 50 mL of a 0.1% Na-Alg solution. A dilute solution of MgSO4 (1 mL) containing 0.60 mg of the compound was added dropwise to 50 mL of a 0.1% Na-Alg solution.
The resulting mixtures was intensely stirred for 24 h and placed in dialysis bags (MWCO~12 kDa, «Sigma», Merck, Rahway, NJ, USA) and dialyzed for 24 h against deionized water. After dialysis, the solutions were lyophilized.
3.3.3. Preparation of Nanocontainers Filled with 2c
A solution containing 1 mg of aminopyrimidine in 50 μL HCl was added dropwise to 5 mL of an aqueous solution of the Alg–Ca2+ (and Alg–Mg2+) nanocontainer containing 5 mg of the compound. The reaction mixture was intensely stirred for 24 h and neutralized by NaOH solution to pH~7. High-molecular products were purified by flow dialysis. Then, lyophilization was used.
3.3.4. Cytotoxicity Studies of Nanocontainers Filled with 2c
Lung adenocarcinoma A549, breast adenocarcinoma MDA-MB-231 and non-tumor fibroblasts immortalized with h-TERT cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were routinely propagated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (HyClone, Logan, UT, USA), 2 mM L-glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin (PanEco, Moscow, Russia) at 37 °C, 5% CO2 in a humidified atmosphere. Cells in the logarithmic phase of growth were used in the experiments.
The cytotoxicity of compounds for human cell lines (A549, MDA-MB-231, non-tumor fibroblasts) was assessed in MTT assays. Cells were plated in 96-well plates (NUNC, Thermo Fisher Scientific, Waltham, MA, USA, 5∙103 cells in 190 μL culture medium per well). After 24 h at 37 °C, 5% CO2 in a humidified atmosphere, cells were treated with compounds dissolved with DMEM from stocks: 2c∙HCl–(Alg)–Ca2+ (C2c 100 μM, CCOO-groups 1000 μM), (Alg)–Ca2+ (CCOO-groups 1000 μM), 2c∙HCl–(Alg)–Mg2+ (C2c 100 μM, CCOO-groups 1000 μM), (Alg)–Mg2+ (CCOO-groups 1000 μM), 2c (C2c 200μM in DMSO) to final amine concentrations of 0.8–25 μM or equivalent amounts of samples (Alg)–Ca2+ and (Alg)–Mg2+ without amine. Doxorubicin (Teva, Amsterdam, Netherlands) was used as control compound.
Cytotoxicity was assessed in a formazan conversion assay (MTT test) after a 48 h exposure. After the completion of drug exposure, 0.5 mg/mL 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT reagent) was added to cells for 2 h, the culture medium was removed, cells were resuspended in 100 μL DMSO and the optical densities were measured on a Multiscan FC plate spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) at a wavelength of 571 nm. The percentage of surviving cells for each dose was calculated as the quotient of the average optical density in the wells after incubation with this dose to the average optical density of the control wells (the values of the latter are taken as 100%). Five independent experiments were performed for each concentration. Standard deviations did not exceed 10%.
3.4. Molecular Dynamics Simulation
Molecular dynamics simulation was performed according to a previously published detailed protocol [57] using the model obtained from the tetramer structure of α,β-tubulin dimer PDB ID: 6GJ4 [58] and CHARMM36/CGenFF 4.4 force field [59,60] in GROMACS 2021.2 software [61] (the starting structure of the protein–ligand complex was obtained by means of molecular docking using AutoDock Vina 1.1.2 software [62]). For the visualization of the results, CPPTRAJ software 5.1.0 [63] in the Amber-Tools 21 package [64] and UCSF Chimera software 1.16 [65] were used; the position of verubulin (PDB ID: 5XKF [46]) was matched to the β-subunit for comparison.
4. Conclusions
In summary, a series of verubulin analogues, where the quinazoline core was replaced by tetrahydroquinazoline or cyclohepta[d]pyrimidine, with varying substituents at positions 2 and 4 of pyrimidine were obtained. The estimation of their cytotoxicity via MTT assay showed a spectrum of cytotoxic action from close to that of verubuliN′s to almost non-toxicity, offering useful information for SAR analysis and further research. 2-Methyl- and 2-chlorotetrahydroquinazolines, containing 4-methoxy-N-methylaniline moiety at position 4, were found to be the most potent: they revealed cytotoxicity with IC50 down to 1–4 nM, depending on the cancer (or non-cancer) cell line. According to fluorescent microscopy data, compounds that showed cytotoxicity in the MTT assay disrupt the normal cytoskeleton of the cell, and the pattern of destruction is similar to that for combretastatin A-4. Encapsulation of 4-aminotetrahydroquinazoline 2c into alginate-based nanocontainers was performed to show that the cytotoxic action of the encapsulated compound 2c is preserved compared to its free form, that opens the way to the further search for drug delivery systems for verubulin analogues.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ph16101499/s1, cells survival curves for compounds 1,2a–r, cytotoxicity against HCT116 cell line for selected compounds, details for preparation of nanocontainers filled with compound 2c, cells survival curves for nanocontainers filled with compound 2c, copies of NMR spectra.
Author Contributions
Conceptualization, O.N.Z. and E.B.A.; software, N.A.Z.; investigation, K.N.S., D.N.L., Y.K.G., N.A.Z., Y.A.G., D.A.S., Y.S.H., L.A.V., E.A.S., P.N.S., E.F.S., A.R.L., V.V.S., A.A.M., M.T.N. and A.A.S.; data curation, K.N.S., D.A.S., E.F.S., V.V.S. and Y.A.G.; writing—original draft preparation, D.N.L. and K.N.S.; writing—review and editing, D.A.S., E.F.S., O.N.Z., E.R.M. and E.B.A.; visualization, Y.K.G., N.A.Z., D.A.S., E.F.S. and Y.A.G.; supervision, K.N.S., D.A.S., E.F.S., A.A.Y. and O.N.Z.; project administration, E.R.M. and E.B.A.; funding acquisition, E.B.A. and D.A.S. All authors have read and agreed to the published version of the manuscript.
Funding
Synthetic work and encapsulation experiments for nanocontainers were supported by the Ministry of Science and Higher Education of the Russian Federation (Agreement with Zelinsky Institute of Organic Chemistry RAS No. 075-15-2020-803). Cytotoxicity tests and mechanistical investigations by fluorescent microscopy were supported by RSF 22-14-00099. Cell cultivation and MTT assay for alginate formulations were carried out in the Core Facility of the Institute of Biochemical Physics RAS “New Materials and Technologies” and supported by the RF State Program for IBCP RAS (Project №122041400114-2).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and supplementary material.
Acknowledgments
The research was carried out using the NMR Agilent 400-MR spectrometer purchased under the program of MSU development. The study of tubulin polymerization in vitro was carried out using equipment of the Center for Collective Use of the IPAC RAS, funded within the framework of state assignment (subject No. FFSN-2021-0005).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Pellegrini, F.; Budman, D.R. Review: Tubulin Function, Action of Antitubulin Drugs, and New Drug Development. Cancer Investig. 2005, 23, 264–273. [Google Scholar] [CrossRef]
- Borisy, G.; Heald, R.; Howard, J.; Janke, C. Microtubules: 50 years on from the discovery of tubulin. Nat. Rev. Mol. Cell Biol. 2016, 17, 322–328. [Google Scholar] [CrossRef]
- Janke, C.; Magiera, M.M. The tubulin code and its role in controlling microtubule properties and functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 307–326. [Google Scholar] [CrossRef]
- Naaz, F.; Haidera, M.R.; Shafi, S.; Yar, M.S. Anti-tubulin agents of natural origin: Targeting taxol, vinca, and colchicine binding domains. Eur. J. Med. Chem. 2019, 171, 310–331. [Google Scholar] [CrossRef] [PubMed]
- Arnst, K.E.; Banerjee, S.; Chen, H.; Deng, S.; Hwang, D.-J.; Li, W.; Miller, D.D. Current advances of tubulin inhibitors as dual acting small molecules for cancer therapy. Med. Res. Rev. 2019, 39, 1398–1426. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Ren, Y.; Pan, W.; Liu, J.; Chen, J. Discovery of Novel Acridane-Based Tubulin Polymerization Inhibitors with Anticancer and Potential Immunomodulatory Effects. J. Med. Chem. 2023, 66, 627–640. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.-Y.; Song, C.-H.; Liu, X.-J.; Wang, X.; Jia, M.-Q.; Wang, W.; Liu, W.-B.; Fu, X.-J.; Jin, C.-Y.; Song, J.; et al. Discovery of novel N-benzylarylamide-dithiocarbamate based derivatives as dual inhibitors of tubulin polymerization and LSD1 that inhibit gastric cancers. Eur. J. Med. Chem. 2023, 252, 115281. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Wu, J.-Q.; Cai, X.; Guan, W.; Zeng, Z.; Ou, Y.; Wu, X.; Li, J.; Fang, X.; Liu, J.; et al. Design, synthesis, and biological evaluation of diaryl heterocyclic derivatives targeting tubulin polymerization with potent anticancer activities. Eur. J. Med. Chem. 2023, 252, 115284. [Google Scholar] [CrossRef]
- Tang, M.; Liu, Y.-H.; Liu, H.; Mao, Q.; Yu, Q.; Kitagishi, H.; Zhang, Y.-M.; Xiao, L.; Liu, Y. Supramolecular Dual Polypeptides Induced Tubulin Aggregation for Synergistic Cancer Theranostics. J. Med. Chem. 2022, 65, 13473–13481. [Google Scholar] [CrossRef]
- Lin, S.; Du, T.; Zhang, J.; Wu, D.; Tian, H.; Zhang, K.; Jiang, L.; Lu, D.; Sheng, L.; Li, Y.; et al. Optimization of Benzamide Derivatives as Potent and Orally Active Tubulin Inhibitors Targeting the Colchicine Binding Site. J. Med. Chem. 2022, 65, 16372–16391. [Google Scholar] [CrossRef]
- Gallego-Jara, J.; Lozano-Terol, G.; Sola-Martínez, R.A.; Cánovas-Díaz, M.; de Diego Puente, T. A Compressive Review about Taxol®: History and Future Challenges. Molecules 2020, 25, 5986. [Google Scholar] [CrossRef]
- Sottomayor, M.; Ros Barceló, A. The vinca alkaloids: From biosynthesis and accumulation in plant cells, to uptake, activity and metabolism in animal cells. In Studies in Natural Products Chemistry (Bioactive Natural Products); Rahman, A., Ed.; Elsevier Science: Amsterdam, The Netherland, 2005; Volume 33, pp. 813–857. [Google Scholar]
- McLoughlin, E.C.; O’Boyle, N.M. Colchicine-Binding Site Inhibitors from Chemistry to Clinic: A Review. Pharmaceuticals 2020, 13, 8. [Google Scholar] [CrossRef]
- Li, W.; Sun, H.; Xu, S.; Zhu, Z.; Xu, J. Tubulin inhibitors targeting the colchicine binding site: A perspective of privileged structures. Future Med. Chem. 2017, 9, 1765–1794. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Miller, D.D.; Li, W. Molecular interactions at the colchicines binding site in tubulin: An X-ray crystallography perspective. Drug Discov. Today 2022, 27, 759–776. [Google Scholar] [CrossRef] [PubMed]
- Kasibhatla, S.; Baichwal, V.; Cai, S.X.; Roth, B.; Skvortsova, I.; Skvortsov, S.; Lukas, P.; English, N.M.; Sirisoma, N.; Drewe, J.; et al. MPC-6827: A Small-Molecule Inhibitor of Microtubule Formation That Is Not a Substrate for Multidrug Resistance Pumps. Cancer Res. 2007, 67, 5865–5871. [Google Scholar] [CrossRef]
- Alvarez, R.; Aramburu, L.; Puebla, P.; Caballero, E.; Gonzalez, M.; Vicente, A.; Medarde, M.; Pelaez, R. Pyridine Based Antitumour Compounds Acting at the Colchicine Site. Curr. Med. Chem. 2016, 23, 1100–1130. [Google Scholar] [CrossRef]
- Bansal, R.; Malhotra, A. Therapeutic progression of quinazolines as targeted chemotherapeutic agents. Eur. J. Med. Chem. 2021, 211, 113016. [Google Scholar] [CrossRef]
- Chamberlain, M.C.; Grimm, S.; Recht, S.P.L.; Zhu, J.Z.; Kim, L.; Rosenfeld, S.; Fadul, C.E. Brain Tumor Investigational Consortium (BTIC). A phase 2 trial of verubulin for recurrent glioblastoma: A prospective study by the brain tumor investigational consortium (BTIC). J. Neurooncol. 2014, 118, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Mahal, K.; Resch, M.; Ficner, R.; Schobert, R.; Biersack, B.; Mueller, T. Effects of the Tumor-Vasculature-Disrupting Agent Verubulin and Two Heteroaryl Analogues on Cancer Cells, Endothelial Cells, and Blood Vessels. ChemMedChem 2014, 9, 847–854. [Google Scholar] [CrossRef]
- Sirisoma, N.; Kasibhatla, S.; Pervin, A.; Zhang, H.; Jiang, S.; Willardsen, J.A.; Anderson, M.B.; Baichwal, V.; Mather, G.G.; Jessing, K.; et al. Discovery of 2-Chloro-N-(4-methoxyphenyl)-N-methylquinazolin-4-amine (EP128265, MPI-0441138) as a Potent Inducer of Apoptosis with High In Vivo Activity. J. Med. Chem. 2008, 51, 4771–4779. [Google Scholar] [CrossRef] [PubMed]
- Sirisoma, N.; Pervin, A.; Zhang, H.; Jiang, S.; Willardsen, J.A.; Anderson, M.B.; Mather, G.; Pleiman, C.M.; Kasibhatla, S.; Tseng, B.; et al. Discovery of N-(4-Methoxyphenyl)-N,2-dimethylquinazolin-4-amine, a Potent Apoptosis Inducer and Efficacious Anticancer Agent with High Blood Brain Barrier Penetration. J. Med. Chem. 2009, 52, 2341–2351. [Google Scholar] [CrossRef] [PubMed]
- Sirisoma, N.; Pervin, A.; Zhang, H.; Jiang, S.; Willardsen, J.A.; Anderson, M.B.; Mather, G.; Pleiman, C.M.; Kasibhatla, S.; Tseng, B.; et al. Discovery of N-methyl-4-(4-methoxyanilino)quinazolines as potent apoptosis inducers. Structure–activity relationship of the quinazoline ring. Bioorg. Med. Chem. Lett. 2010, 20, 2330–2334. [Google Scholar] [CrossRef]
- Gangjee, A.; Zhao, Y.; Lin, L.; Raghavan, S.; Roberts, E.G.; Risinger, A.L.; Hamel, E.; Mooberry, S.L. Synthesis and Discovery of Water-Soluble Microtubule Targeting Agents that Bind to the Colchicine Site on Tubulin and Circumvent Pgp Mediated Resistance. J. Med. Chem. 2010, 53, 8116–8128. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-F.; Wang, S.-B.; Ohkoshi, E.; Wang, L.-T.; Hamel, E.; Qian, K.; Morris-Natschke, S.L.; Lee, K.-H.; Xie, L. N-Aryl-6-methoxy-1,2,3,4-tetrahydroquinolines: A novel class of antitumor agents targeting the colchicine site on tubulin. Eur. J. Med. Chem. 2013, 67, 196–207. [Google Scholar] [CrossRef]
- Gangjee, A.; Zhao, Y.; Raghavan, S.; Rohena, C.C.; Mooberry, S.L.; Hamel, E. Structure−Activity Relationship and in Vitro and in Vivo Evaluation of the Potent Cytotoxic Anti-microtubule Agent N-(4-Methoxyphenyl)-N,2,6-trimethyl-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-aminium Chloride and Its Analogues As Antitumor Agents. J. Med. Chem. 2013, 56, 6829–6844. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-F.; Guan, F.; Ohkoshi, E.; Guo, W.; Wang, L.; Zhu, D.-Q.; Wang, S.-B.; Wang, L.-T.; Hamel, E.; Yang, D.; et al. Optimization of 4-(N-Cycloamino)phenylquinazolines as a Novel Class of Tubulin-Polymerization Inhibitors Targeting the Colchicine Site. J. Med. Chem. 2014, 57, 1390–1402. [Google Scholar] [CrossRef]
- Devambatla, R.K.V.; Namjoshi, O.A.; Choudhary, S.; Hamel, E.; Shaffer, C.V.; Rohena, C.C.; Mooberry, S.L.; Gangjee, A. Design, Synthesis, and Preclinical Evaluation of 4-Substituted-5-methyl-furo[2,3-d]pyrimidines as Microtubule Targeting Agents That Are Effective against Multidrug Resistant Cancer Cells. J. Med. Chem. 2016, 59, 5752–5765. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Arnst, K.E.; Wang, Y.; Kumar, G.; Deng, S.; Yang, L.; Li, G.-B.; Yang, J.; White, S.W.; Li, W.; et al. Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy. J. Med. Chem. 2018, 61, 1704–1718. [Google Scholar] [CrossRef]
- Arnst, K.E.; Banerjee, S.; Wang, Y.; Chen, H.; Li, Y.; Yang, L.; Li, W.; Miller, D.D.; Li, W. X-ray Crystal Structure Guided Discovery and Antitumor Efficacy of Dihydroquinoxalinone as Potent Tubulin Polymerization Inhibitors. ACS Chem. Biol. 2019, 14, 2810–2821. [Google Scholar] [CrossRef]
- Lia, W.; Yina, Y.; Shuaia, W.; Xua, F.; Yaoa, H.; Liub, J.; Cheng, K.; Xua, J.; Zhud, Z.; Xu, S. Discovery of novel quinazolines as potential anti-tubulin agents occupying three zones of colchicine domain. Bioorg. Chem. 2019, 83, 380–390. [Google Scholar] [CrossRef]
- Loidreau, Y.; Nourrisson, M.-R.; Fruit, C.; Corbière, C.; Marchand, P.; Besson, T. Microwave-Assisted Synthesis of Potential Bioactive Benzo-, Pyrido- or Pyrazino-thieno[3,2-d]pyrimidin-4-amine Analogs of MPC-6827. Pharmaceuticals 2020, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Mahmud, F.; Deng, S.; Ma, L.; Yun, M.-K.; Fakayode, S.O.; Arnst, K.E.; Yang, L.; Chen, H.; Wu, Z.; et al. X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor. J. Med. Chem. 2021, 64, 13072–13095. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Wu, C.; Zhang, J.; Yu, Q.; Wang, X.; Zhang, L.; Ge, M.; Wang, Z.; Ouyang, L.; Wang, Y. Design, Synthesis, and Biological Evaluation of Heterocyclic-Fused Pyrimidine Chemotypes Guided by X-ray Crystal Structure with Potential Antitumor and Anti-multidrug Resistance Efficacy Targeting the Colchicine Binding Site. J. Med. Chem. 2023, 66, 3588–3620. [Google Scholar] [CrossRef] [PubMed]
- Evdokimova, A.V.; Alexeev, A.A.; Nurieva, E.V.; Milaeva, E.R.; Kuznetsov, S.A.; Zefirova, O.N. N-(4-Methoxyphenyl)-substituted bicyclic isothioureas: Effect on morphology of cancer cells. Mendeleev Commun. 2021, 31, 288–290. [Google Scholar] [CrossRef]
- Yang, H.; An, B.; Li, X.; Zeng, W. Evaluation of 4-phenylamino-substituted naphthalene-1,2-diones as tubulin polymerization inhibitors. Bioorg. Med. Chem. Lett. 2018, 28, 3057–3063. [Google Scholar] [CrossRef]
- Lovering, F.; Bikker, J.; Humblet, C. Escape from flatland: Increasing saturation as an approach to improving clinical success. J. Med. Chem. 2009, 52, 6752–6756. [Google Scholar] [CrossRef]
- Lovering, F. Escape from Flatland 2: Complexity and promiscuity. Med. Chem. Commun. 2013, 4, 515–519. [Google Scholar] [CrossRef]
- Cox, B.; Zdorichenko, V.; Cox, P.B.; Booker-Milburn, K.I.; Paumier, R.; Elliott, L.D.; Robertson-Ralph, M.; Bloomfield, G. Escaping from Flatland: Substituted Bridged Pyrrolidine Fragments with Inherent Three-Dimensional Character. ACS Med. Chem. Lett. 2020, 11, 1185–1190. [Google Scholar] [CrossRef]
- Klein, H.F.; Hamilton, D.J.; de Esch, I.J.P.; Wijtmans, M.; O’Brien, P. Escape from planarity in fragment-based drug discovery: A synthetic strategy analysis of synthetic 3D fragment libraries. Drug Discov. Today 2022, 27, 2484–2496. [Google Scholar] [CrossRef]
- Severino, P.; da Silva, C.F.; Andrade, L.N.; de Lima Oliveira, D.; Campos, J.; Souto, E.B. Alginate nanoparticles for drug delivery and targeting. Curr. Pharm. Des. 2019, 25, 1312–1334. [Google Scholar] [CrossRef]
- Lee, K.Y.; Mooney, D.J. Alginate: Properties and biomedical applications. Prog. Polym. Sci. 2012, 37, 106–126. [Google Scholar] [CrossRef]
- Shilpa, A.; Agrawal, S.; Ray, A.R. Controlled delivery of drugs from alginate matrix. J. Macromol. Sci. Part C Polym. Rev. 2003, 43, 187–221. [Google Scholar] [CrossRef]
- Spiridonov, V.V.; Sadovnikov, K.S.; Vasilenko, D.A.; Sedenkova, K.N.; Lukmanova, A.R.; Markova, A.A.; Shibaeva, A.V.; Bolshakova, A.V.; Karlov, S.S.; Averina, E.B.; et al. Synthesis and evaluation of the anticancer activity of the water-dispersible complexes of 4-acylaminoisoxazole derivative with biocompatible nanocontainers based on Ca2+ (Mg2+) cross-linked alginate. Mendeleev Commun. 2022, 32, 591–593. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J. Immunol. Methods. 1983, 65, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Yang, T.; Yang, J.; Wang, T.; Yu, Y.; Wang, Y.; Chen, Q.; Bai, P.; Li, D.; Ye, H.; et al. SKLB060 Reversibly Binds to Colchicine Site of Tubulin and Possesses Efficacy in Multidrug-Resistant Cell Lines. Cell. Physiol. Biochem. 2018, 47, 489–504. [Google Scholar] [CrossRef] [PubMed]
- Tait, J.F.; Gibson, D.; Fujikawa, K. Phospholipid binding properties of human placental anticoagulant protein-I, a member of the lipocortin family. J. Biol. Chem. 1989, 264, 7944–7949. [Google Scholar] [CrossRef]
- Andree, H.A.; Reutelingsperger, C.P.; Hauptmann, R.; Hemker, H.C.; Hermens, W.T.; Willems, G.M. Binding of vascular anticoagulant alpha (VAC alpha) to planar phospholipid bilayers. J. Biol. Chem. 1990, 265, 4923–4928. [Google Scholar] [CrossRef]
- Spiridonov, V.V.; Lukmanova, A.R.; Pozdyshev, D.V.; Markova, A.A.; Volodina, Y.L.; Golovina, G.V.; Shakhmatov, V.V.; Kuzmin, V.A.; Muronetz, V.I.; Yaroslavov, A.A. Ionically cross-linked micro-sized hydrogels with encapsulated drug: Structure, cell uptake kinetics and cytotoxicity. Mendeleev Commun. 2023, 33, 553–555. [Google Scholar] [CrossRef]
- Farkas, N.; Kramar, J.A. Dynamic light scattering distributions by any means. J. Nanopart. Res. 2021, 23, 120. [Google Scholar] [CrossRef]
- Spiridonov, V.; Orlova, M.; Ivanov, I.; Panova, I.; Orlov, A.; Antonova, Y.; Yaroslavov, A. Synthesis of microgels based on carboxymethylcellulose cross-linked with zinc(II) ions and heterocyclic effectors of NO-synthase. Colloids Surf. A Physicochem. Eng. Asp. 2020, 585, 124104. [Google Scholar] [CrossRef]
- Grante, I.; Actins, A.; Orola, L. Protonation effects on the UV/Vis absorption spectra of imatinib: A theoretical and experimental study. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2014, 129, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.W.; Rose, F.L. 1080. S-triazolopyrimidines. Part I. Synthesis as potential therapeutic agents. J. Chem. Soc. 1963, 5642–5659. [Google Scholar] [CrossRef]
- Ohno, S.; Mizukoshi, K.; Komatsu, O.; Kunoh, Y.; Nakamura, Y.; Katoh, E.; Nagasaka, M. Synthesis and Hypoglycemic Activity of 7,8-Dihydro-6H-thiopyrano[3,2-d]pyrimidine Derivatives and Related Compounds. Chem. Pharm. Bull. 1986, 34, 4150–4165. [Google Scholar] [CrossRef] [PubMed]
- Sedenkova, K.N.; Zverev, D.V.; Nazarova, A.A.; Lavrov, M.I.; Radchenko, E.V.; Grishin, Y.K.; Gabrel’yan, A.V.; Zamoyski, V.L.; Grigoriev, V.V.; Averina, E.B.; et al. Novel Nanomolar Allosteric Modulators of AMPA Receptor of Bis(pyrimidine) Series: Synthesis, Biotesting and SAR Analysis. Molecules 2022, 27, 8252. [Google Scholar] [CrossRef]
- Zhou, H.-J.; Wang, J.; Yao, B.; Wong, S.; Djakovic, S.; Kumar, B.; Rice, J.; Valle, E.; Soriano, F.; Menon, M.-K.; et al. Discovery of a First-in-Class, Potent, Selective, and Orally Bioavailable Inhibitor of the p97 AAA ATPase (CB-5083). J. Med. Chem. 2015, 58, 9480–9497. [Google Scholar] [CrossRef]
- Sadovnikov, K.S.; Vasilenko, D.A.; Gracheva, Y.A.; Zefirov, N.A.; Radchenko, E.V.; Palyulin, V.A.; Grishin, Y.K.; Vasilichin, V.A.; Shtil, A.A.; Shevtsov, P.N.; et al. Novel substituted 5-methyl-4-acylaminoisoxazoles as antimitotic agents: Evaluation of selectivity to LNCaP cancer cells. Arch. Pharm. 2022, 355, e2100425. [Google Scholar] [CrossRef] [PubMed]
- Brindisi, M.; Ulivieri, C.; Alfano, G.; Gemma, S.; de Asís Balaguer, F.; Khan, T.; Grillo, A.; Chemi, G.; Menchon, G.; Prota, A.E.; et al. Structure-activity relationships, biological evaluation and structural studies of novel pyrrolonaphthoxazepines as antitumor agents. Eur. J. Med. Chem. 2019, 162, 290–320. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; MacKerell, A.D. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Trott, O.A.; Olson, J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An overview of the Amber biomolecular simulation package. WIREs Comput. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).