Abstract
The development of novel agents to combat COVID-19 is of high importance. SARS-CoV-2 main protease (Mpro) is a highly attractive target for the development of novel antivirals and a variety of inhibitors have already been developed. Accumulating evidence on the pathobiology of COVID-19 has shown that lipids and lipid metabolizing enzymes are critically involved in the severity of the infection. The purpose of the present study was to identify an inhibitor able to simultaneously inhibit both SARS-CoV-2 Mpro and phospholipase A2 (PLA2), an enzyme which plays a significant role in inflammatory diseases. Evaluating several PLA2 inhibitors, we demonstrate that the previously known potent inhibitor of Group IIA secretory PLA2, GK241, may also weakly inhibit SARS-CoV-2 Mpro. Molecular mechanics docking and molecular dynamics calculations shed light on the interactions between GK241 and SARS-CoV-2 Mpro. 2-Oxoamide GK241 may represent a lead molecular structure for the development of dual PLA2 and SARS-CoV-2 Mpro inhibitors.
1. Introduction
With more than 450 million cases of infected people and 6 million casualties globally, the discovery of efficient agents to treat COVID-19, which is caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), is an unmet need [1]. Pioneering studies showed that enzymes, such as RNA-dependent RNA polymerase (RdRp) and SARS-CoV-2 main protease (Mpro), are attractive targets for the development of novel antiviral agents [2,3].
Studies aiming to understand the pathobiology of COVID-19 have also demonstrated the involvement of lipids and lipid metabolizing enzymes in this potentially lethal infection [4,5]. Phospholipases A2 (PLA2s) are enzymes which catalyze the hydrolysis of membrane glycerophospholipids, releasing free fatty acids (FFAs) and lysophospholipids and initiating arachidonic acid (AA) cascade and promotion of inflammation [6,7,8]. Proteomics studies on SARS-CoV-2 infected cells have revealed alterations of proteins linked to the inflammatory response due to the viral infection [9]. The expression of two PLA2s, namely cytosolic PLA2 (GIVA cPLA2) and secreted PLA2 (GIIA sPLA2) was notably differentiated after 24 h of infection [9]. Plasma metabolomic and lipidomic studies associated with COVID-19 showed elevated levels of FFAs and reduction in phosphatidylcholines (PCs), which indicated increased enzymatic activity of PLA2s [10]. Large-scale plasma analysis has revealed that lipids are strongly involved in the response to infection [11]. The concentrations of oleic acid (OA, C18:1) and AA (C20:4) were directly correlated to the severity of the disease in COVID-19 patients who required admission to an intensive care unit [11]. Most recently, an independent cohort study has demonstrated elevated levels of GIIA sPLA2 in the plasma of deceased patients in comparison to patients with severe or mild COVID-19, indicating that GIIA sPLA2 is associated with increased mortality due to COVID-19 [12]. This study highlights the importance of GIIA sPLA2, establishing this enzyme as a factor that leads to severe COVID-19 morbidity and mortality, and suggesting it as a therapeutic target to prevent COVID-19 mortality.
Previously described data [4,5,9,10,11,12], as well as data reported in recent review articles summarizing the role of PLA2s in inflammatory diseases [13,14], prompted us to explore if we could identify a small-molecule inhibitor able to simultaneously inhibit PLA2 and SARS-CoV-2 Mpro. It would be advantageous, if we could target two enzymes with a dual inhibitor able to simultaneously block virus replication by inhibiting SARS-CoV-2 Mpro and regulate the inflammatory response by inhibiting PLA2. In the present study, we focus on 2-oxoamide (also known as α-ketoamide) small molecules as appropriate agents to inhibit both PLA2 and SARS-CoV-2 Mpro and we describe the first dual inhibitor of GIIA sPLA2 and SARS-CoV-2 Mpro.
2. Results
2.1. Design of Inhibitors
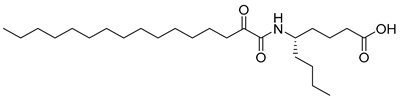
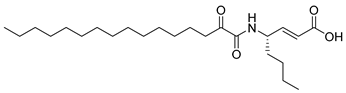
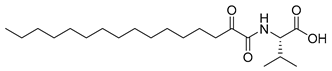
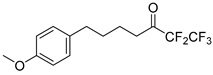
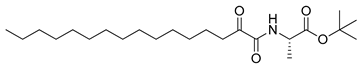
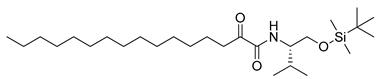
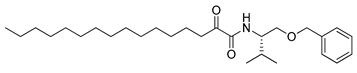
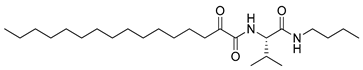
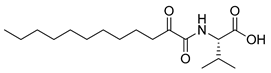
In 2020, Hilgenfeld and coworkers demonstrated that 2-oxoamides are potent inhibitors of SARS-CoV-2 Mpro [15] and reported the X-ray structure of SARS-CoV-2 Mpro in complex with the 2-oxoamide inhibitor 1 (Figure 1) [15]. SARS-CoV-2 Mpro is a cysteine protease and its key cysteine residue may attack small-molecule inhibitors containing either a reactive carbonyl group or a Michael acceptor functionality [15,16,17,18,19,20,21]. In previous years, we have designed and synthesized a variety of 2-oxoamides as inhibitors of PLA2s. More specifically, we have developed 2-oxoamides (2), which are based on non-natural δ- or γ-amino acids and selectively inhibit GIVA cPLA2 [22,23], while 2-oxoamides (3) based on natural α-amino acids selectively inhibit GIIA sPLA2 [24].
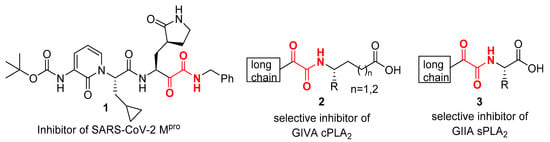
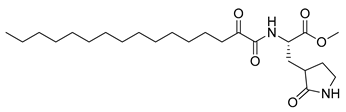
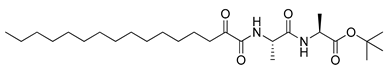
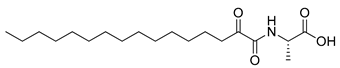
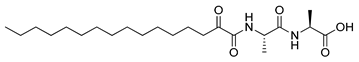
Figure 1.
Structures of 2-oxoamide inhibitor of SARS-CoV-2 Mpro and 2-oxoamide inhibitors of GIVA cPLA2 and GIIA sPLA2.
To explore if known inhibitors of PLA2s may inhibit SARS-CoV-2 Mpro, we selected three 2-oxoamides previously developed by us, which are selective inhibitors of either GIVA cPLA2 (AX109 and AX074) [22,23] or GIIA sPLA2 (GK241) [24], and pentafluoroethyl ketone GK187, which selectively inhibits calcium-independent GVIA iPLA2 [25]. Their structures are shown in Table 1. Since our initial in vitro studies have shown that GIIA sPLA2 inhibitor GK241 inhibits SARS-CoV-2 Mpro, we have also synthesized several GK241 analogs for SAR studies.

Table 1.
In vitro inhibition of SARS-CoV-2 Mpro by known PLA2 inhibitors.
2.2. Synthesis
2-Oxoamides 7a–g were synthesized in two steps from long chain α-hydroxycarboxylic acids 4a,b and amines 5a–g (see Scheme 1 below). The coupling was carried out employing 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC∙HCl) as the coupling agent in the presence of 1-hydroxybenzotriazole (HOBt) and was followed by an oxidation reaction using Dess–Martin periodinane (Scheme 1) [26].
Scheme 1.
Synthesis of 2-oxoamides from 2-hydroxycarboxylic acids 4a,b and amines 5a–g. (a) EDC∙HCl, HOBt, Et3N, dry CH2Cl2, 1 h at 0 °C, 16 h at r.t.; (b) Dess–Martin periodinane, dry CH2Cl2, 1 h, r.t.
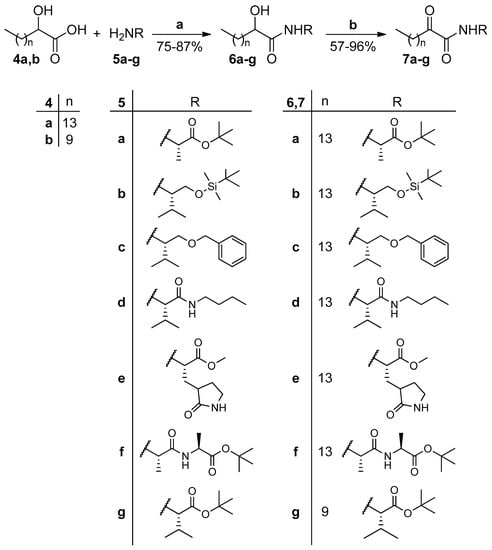
tert-Butyl esters 7a and 7f,g were deprotected by treatment with trifluoroacetic acid (TFA) to provide the corresponding carboxylic acids 8a–c (Scheme 2).
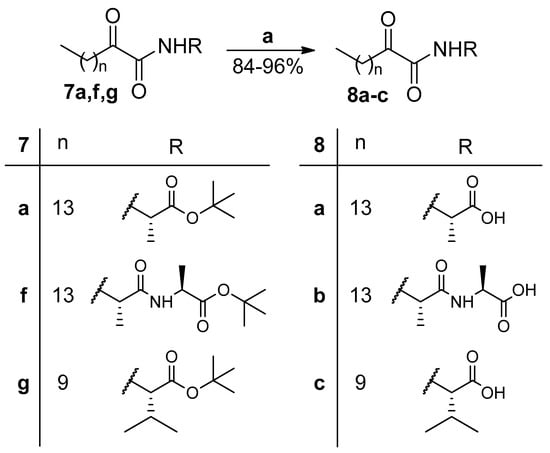
Scheme 2.
Removal of tert-butyl ester group. (a) 50% TFA in CH2Cl2 (0.5 M), 3 h, r.t.
Amine 5b was synthesized from carbobenzoxy-L-valinol by protection of the hydroxyl group, using tert-butyldimethylsilyl chloride, as described in [27], and then removal of the Cbz group. Amine 5c was synthesized from tert-butyloxycarbonyl-L-valinol (9) by protection of the hydroxyl group, followed by removal of the Boc group (Scheme 3).
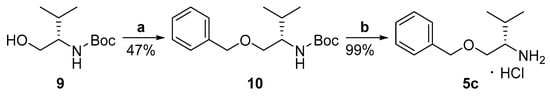
Scheme 3.
Synthesis of amine 5c. (a) BnBr, NaH, dry DMF, 16 h, 0 °C to r.t.; (b) 4N HCl/MeOH, 2 h, r.t.
2.3. Inhibition of SARS-CoV-2 Mpro by 2-Oxoamide PLA2 Inhibitors and Analogs
The inhibitory potency of the known PLA2 inhibitors and all of the new 2-oxoamides synthesized against SARS-CoV-2 Mpro was assessed by determining the extent of enzyme inhibition (% inhibition); the results are summarized in Table 1 and Table 2. In these experiments, 40 μM or 100 μM of 2-oxoamide, 0.5 μM of SARS-CoV-2 Mpro, and 10 μM of the fluorescence resonance energy transfer (FRET) substrate Dabcyl-KTSAVLQ↓SGFRKM-E(Edans)-NH2 in 20 mM tris(hydroxymethyl)aminomethane (Tris) buffer (pH 7.3), 100 mM NaCl, 1 mM ethylene diamine tetra-acetic acid (EDTA), were used.

Table 2.
In vitro inhibition of SARS-CoV-2 Mpro by GK241 analogs.
As shown in Table 1, 2-oxoamide GIVA cPLA2 inhibitors AX109 and AX074 weakly inhibited SARS-CoV-2 Mpro at concentrations of 40 μM and 100 μM (entries 1 and 2, Table 1), but none of them higher than 50% even at 100 μM. Interestingly, the 2-oxoamide GIIA sPLA2 inhibitor GK241 inhibited SARS-CoV-2 Mpro almost completely at 100 μM and by 76.9% at 40 μM (entry 3, Table 1). Pentafluoroethyl ketone GK187, which is a selective and potent GVIA iPLA2 inhibitor, showed a very weak effect even at 100 μM. Overall, the selective GIIA sPLA2 inhibitor GK241 was found to inhibit SARS-CoV-2 Mpro. The IC50 value for GK241, determined by the inhibition curve, was found to be 24 μM.
The results for the in vitro inhibition of SARS-CoV-2 Mpro by the analogs of GK241 are summarized in Table 2. Conversion of the free carboxyl to the corresponding amide 7d or to a hydroxymethyl-protected group 7c, 7b resulted in abolishment of the inhibitory potency (entries 2–4, Table 2), indicating that a free carboxyl group was necessary for the inhibition. When the long chain of GK241 was replaced by a shorter one (reduction by four carbon atoms), the inhibitory potency of 8d on SARS-CoV-2 Mpro was abolished (entry 9, Table 2).
When the valine residue of GK241 was replaced by alanine, 8a (entry 7, Table 2) was found to inhibit SARS-CoV-2 Mpro by 65.49% at 40 μM, while the conversion of the free carboxyl group to an ester (7a, entry 1, Table 2) again led to the abolishment of the inhibitory potency. The derivatives 8b and 7f, based on an Ala-Ala dipeptide, presented almost no activity (entries 8 and 6, Table 2). Finally, compound 7e, based on a glutamine surrogate, did not present any activity (entry 5, Table 2).
2.4. Molecular Mechanics Docking and Molecular Dynamics Calculations
To obtain a better insight into the interactions between SARS-CoV-2 Mpro and the most active compound GK241, we applied molecular mechanics docking and molecular dynamics (MD) calculations. More specifically, to simulate the specific interaction, compound GK241 was subjected to covalent docking calculations as implemented in the Maestro Schroedinger suite by creating a covalent bond between Cys145 and the 2-carbonyl carbon of the 2-oxoamide moiety. The procedure transformed carbonyl to an sp3 carbon atom and the adjacent oxygen to a hydroxyl group. The crystal structure of SARS-CoV-2 Mpro protein PDB 6Y2F [15] was adequately prepared for simulations by adding the two missing flexible residues E47 and D48, as this loop was near the binding cavity and initial calculations showed that it could interact with the ligand. Moreover, initial calculations showed that the formation of the covalent bond could result in both R and S configurations for the 2-carbon atom, which is not surprising, since there are examples in the literature reporting inhibitors that may lead to both R and S configurations [28]. Thus, docking calculations were finally performed to collect 100 structures for each 2-carbon atom configuration. These structures were further ranked according to the ligand binding energy calculated using the MM-GBSA approach, ranging between −58.69 to −10.66 kcal/mol. Common structural characteristic of the generated structures for both the R and S configuration groups was the tendency to orient the long aliphatic chain to S2, S3, S4 protease clefts (Figure 2A,B), while differences were mainly observed in the conformation of the covalent bond formatted between Cys145 and the 2-oxoamide carbonyl, as expressed by the dihedral angle Cβcys145-Scys145-C2-O2. The lowest energy structures of up to 3 kcal relative binding energy were grouped according to common conformational characteristics of the ligand; representative structures are shown in Figure 2C and Figure S1 (Supplementary Materials). The resulting docking structures appeared to sample the available conformational space by orienting the valine moiety carboxylate (structures 3 and 5) or isopropyl (structures 2 and 4) to the S1 cavity, while in structures 1 and 6 the –OH group or the initial part of the aliphatic chain were oriented in this cleft. Another interesting structural characteristic was that only structures 2, 5 and 7 interacted with H41 specifically through the –OH group. These seven selected structures were further subjected to 50 nsec MD calculations to assess the stability of the protein-ligand interaction. The calculated binding energies and mean RMSD for both protein and ligand are summarized in Table S1 (Supplementary Materials) along with major interactions between the ligand and specific protein residues. Visual inspection of the MD trajectories and conformational flexibility of both the protein and the ligand showed that, in most of the structures, the long aliphatic chain had the tendency to widely explore the conformational space, probably inducing protein conformational changes. The RMSF calculated for the protein Cα showed that the loop between residues 45–50, in particular, exhibited increased flexibility (Figure S2, Supplementary Materials) which appeared to be related to interactions with part of the long aliphatic chain. A relative flexibility was also observed for residues 185–192, specifically in the case of structure 1. Among the seven structures, the most stable during MD simulations was found to be structure 7, and, thus, MD simulations were extended to 150 nsec to validate the initial observation (Figure 2D). In structure 7, the 2-carbon atom exhibited an S configuration and both the –OH and amide moieties aligned, in general, very well with the ketoamide crystal structure reported by Hilgenfeld and coworkers [15] (Figure 2A,B). The –OH group interacted with H41 for 50% of the MD sampled structures, while the valine side-chain occupied the S1′ cavity with the carboxylate interacting with Q166 and the amide NH forming an H-bond with the T26 backbone carbonyl during the whole MD simulation, both directly and through a water molecule. Both protein and ligand RMSD appeared to be low during the whole MD simulation and the protein RMSF validated the stability of all parts of the protein (Figure 2C,D).
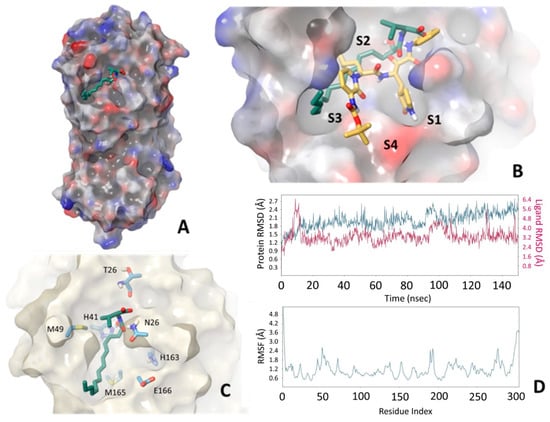
Figure 2.
Structural characteristics of SARS-CoV-2 Mpro-GK241 interaction as provided by docking and MD simulations. (A) GK241 occupying the main cavity of the enzyme active site. (B) Comparison of structure 7 with crystal structure PDB 6Y2F. The formation of the covalent bond produces exactly the same configuration, and the amide bond has similar orientation, while the long aliphatic chain occupies part of the S2 and S3 and S4 cavities. (C) Interactions of structure 7 with different protein residues. (D) RMSD for the protein (blue) and the ligand (magenta), as well as protein residue RMSF during the 150 nsec MD simulation.
In order to further rationalize the experimental results, the derivatives 8d and 8a were subjected to covalent docking following the same procedure as above, resulting in 100 structures for each of the R and S 2-carbon configurations. The calculated MM-GBSA interaction energies were higher in both cases than those observed for GK241, ranging between −50.91 and −8.35 kcal/mol for 8d and −56.99 and −25.98 kcal/mol for 8a. For comparison with GK241, we selected structures, similar to structure 7, specifically having an S configuration and forming an H bond between the –OH and residue H41, which are presented in Figure S3 (Supplementary Materials) along with the crystal structure and structure 7 for comparison. Concerning the derivative 8d, a major difference observed was that the 10-carbon-atom aliphatic chain was always oriented differently covering part of the S1 and S2 cavities, as shown in Figure S3A (Supplementary Materials). This major difference in the conformation of the aliphatic side-chain reflected a major difference in the MM-GBSA calculated binding energy of ~4 kcal compared to GK241 structure 7 and was in agreement with the experimental results showing no activity at 40 μΜ concentration. On the other hand, derivative 8a showed major similarities with GK241 as far as the aliphatic chain was concerned, mainly occupying the same part of the enzyme active site. However, the alanine moiety of 8a occupied the S1 cavity differently to GK241 as the N-Cαala bond adopted a different conformation compared to GK241, resulting in a different orientation of the methyl group of the alanine moiety compared to the corresponding valine isopropyl. These differences can explain the small differences observed in the experimental activity of these derivatives.
3. Discussion
PLA2s are a superfamily of enzymes [6,7,8] which are involved in almost any inflammatory disease [6,7,8,13,14,29]. In humans, three PLA2 types, represented by GIIA sPLA2, GIVA cPLA2 and GVIA iPLA2, are of high medicinal interest and have been targets for the development of small-molecule synthetic inhibitors [13]. Among the various classes of synthetic inhibitors, 2-oxoamides constitute a class of compounds whose members can selectively inhibit either GIVA cPLA2 or GIIA sPLA2. The results of the present study showed that the selective 2-oxamide inhibitors of GIVA cPLA2 AX109 and AX074 [22,23] did not exhibit any appreciable inhibition of SARS-CoV-2 Mpro. Similarly, the selective pentafluoroethyl ketone inhibitor of GVIA iPLA2 GK187 [25] did not show appreciable inhibition of SARS-CoV-2 Mpro. On the contrary, the potent 2-oxamide inhibitor of GIIA sPLA2 (IC50 143 nM) [24] was found to inhibit SARS-CoV-2 Mpro with an IC50 value of 24 μM. Given that GIIA sPLA2 has most recently been recognized as a factor contributing to the severity and mortality of COVID-19 [12,30], this finding is of high importance.
Peptide and peptide-mimetic 2-oxoamides have been identified as potent inhibitors of SARS-CoV-2 Mpro and their interaction with the catalytic site of the cysteine protease SARS-CoV-2 Mpro has been defined by determining the X-ray structure of the enzyme-inhibitor complex [15]. Inhibitors of SARS-CoV-2 Mpro have attracted high interest as candidate antiviral drugs [31,32], and, recently, the inhibitor PF-07321332 (nirmatrelvir) has received emergency approval by the Food and Drug Administration (FDA).
Inflammation is a critical factor in COVID-19 [5,33], and, consequently, agents able to combat virus replication, and, at the same time, regulate inflammation, could offer a new approach for the treatment of COVID-19. Since GIIA sPLA2 is associated with increased mortality by COVID-19 [12], and lipid mediators arising from the activity of PLA2 have been correlated with severe SARS-CoV-2 infection in humans [34], a therapeutic compound able to simultaneously inhibit both SARS-CoV-2 Mpro and GIIA sPLA2 would be of great value, as it would significantly reduce the risk of COVID-19 mortality. For the first time, a dual inhibitor of GIIA sPLA2 and SARS-CoV-2 Mpro is identified. GK241 shows weak inhibitory activity against SARS-CoV-2 Mpro compared to other known 2-oxoamide SARS-CoV-2 Mpro inhibitors; however, it may represent a basis for the development of a new class of potent dual-action inhibitors.
4. Materials and Methods
4.1. General Chemistry Methods
Forced-flow chromatography on Merck® (Merck, Darmstadt, Germany) Kieselgel 60 F254 230–400 mesh was used for the purification of the products, while aluminum-backed silica plates (0.2 mm, 60 F254) were used for thin-layer chromatography (TLC). The visualization of the developed chromatograms was performed by fluorescence quenching using phosphomolybdic acid, ninhydrin or potassium permanganate stains. The melting points were determined on a Buchi® 530 apparatus (Buchi, Flawil, Switzerland) and were uncorrected. Specific rotations were measured on an AA-65 series (Optical Activity Ltd., Bury, UK) polarimeter. 1H and 13C NMR spectra were recorded on a Varian® Mercury (Varian, Palo Alto, CA, USA) (200 MHz and 50 MHz, respectively) or a Bruker Avance Neo (Bruker, Faellanden, Switzerland) (400 MHz and 100 MHz, respectively) and were internally referenced to residual solvent signals. The data for 1H NMR are reported as follows: chemical shift (δ ppm), multiplicity (s = singlet, d = doublet, t = triplet, qu = quintet, m = multiplet, and br s = broad signal), coupling constant, integration and peak assignment. The data for 13C NMR are reported in terms of the chemical shift (δ ppm). High-resolution mass spectrometry (HRMS) spectra were recorded on a Bruker® Maxis Impact QTOF (Bruker Daltonics, Bremen, Germany) spectrometer.
4.2. General Procedure for the Coupling of α-Hydroxycarboxylic Acids 4a,b with Amines 5a–g
To a stirred solution of amine 5a–g (1.0 mmol), cooled to 0 °C, triethylamine (2.2 mmol, 0.31 mL), EDC∙HCl (1.1 mmol, 211 mg), HOBt (1 mmol, 135 mg) and α-hydroxycarboxylic acid 4a,b (1.0 mmol) were added consecutively. After stirring for 1 h at 0 °C and for 16 hrs at room temperature (r.t.), the solvent was removed under reduced pressure. The residue was diluted in EtOAc and washed with brine (10 mL), an aqueous solution of 1N HCl (10 mL), brine (10 mL), an aqueous solution of 5% NaHCO3 (10 mL) and brine (10 mL), consecutively. The organic layer was dried over Na2SO4 and concentrated under reduced pressure. The resulting α-hydroxyamide 6a–g was further purified (if necessary) by flash chromatography eluting with the appropriate mixture of EtOAc:petroleum ether (40–60 °C).
4.2.1. (2S)-tert-Butyl 2-(2-hydroxyhexadecanamido)propanoate (6a)
Yield 85%; White solid; mp: 56–57 °C; Diastereoisomer 1: 1H NMR (200 MHz, CDCl3): δ = 7.23–7.16 (m, 1H, NH), 4.40–4.23 (m, 2H, CH, OH), 4.02–3.97 (m, 1H, CH), 1.67–1.07 (m, 38H, 13 × CH2, 4 × CH3), 0.76 (t, J = 5.9 Hz, 3H, CH3CH2); 13C NMR (50 MHz, CDCl3): δ = 174.3, 172.4, 82.0, 71.9, 48.2, 34.7, 31.9, 29.7, 29.64, 29.58, 29.5, 29.3, 27.9, 25.0, 22.7, 18.4, 14.1; Diastereoisomer 2: 1H NMR (200 MHz, CDCl3): δ = 7.11 (d, J = 7.6 Hz, 1H, NH), 4.43 (dt, J = 14.5, 7.2 Hz, 1H, CH), 4.10–4.04 (m, 1H, CH), 3.40 (br s, 1H, OH), 1.78–1.12 (m, 38H, 13 × CH2, 4 × CH3), 0.85 (t, J = 6.4 Hz, 3H, CH3CH2); 13C NMR (50 MHz, CDCl3): δ = 173.9, 172.2, 82.2, 72.1, 48.4, 34.9, 32.0, 29.79, 29.75, 29.7, 29.6, 29.53, 29.46, 28.0, 25.1, 22.8, 18.6, 14.2; HRMS (ESI) [M + Na]+ m/z: 422.3243; (calculated for [C23H45NNaO4]+ 422.3241).
4.2.2. N-((S)-1-((tert-Butyldimethylsilyl)oxy)-3-methylbutan-2-yl)-2-hydroxyhexadecanamide (6b)
Yield 80%; Colorless solid of low melting point; 1H NMR (200 MHz, CDCl3): δ = 6.81–6.73 (m, 1H, NH), 4.11–4.04 (m, 1H, CH), 3.76–3.40 (m, 5H, CH2O, 2 × CH, OH), 1.94–1.18 (m, 26H, 13 × CH2), 0.93–0.74 (m, 18H, 6 × CH3), 0.02 (s, 6H, 2 × SiCH3); 13C NMR (50 MHz, CDCl3): δ = 173.9, 173.7, 72.2, 72.0, 62.9, 55.4, 35.3, 35.2, 32.0, 29.81, 29.77, 29.7, 29.6, 29.5, 28.9, 25.9, 25.2, 25.0, 22.8, 19.7, 19.0, 18.3, 14.2, −5.5; HRMS (ESI) [M + Na]+ m/z: 494.4000; (calculated for [C27H57NNaO3Si]+ 494.4000).
4.2.3. N-((S)-1-(Benzyloxy)-3-methylbutan-2-yl)-2-hydroxyhexadecanamide (6c)
Yield 77%; White solid; mp: 63–64 °C; 1H NMR (200 MHz, CDCl3): δ = 7.38–7.18 (m, 5H, 5 × ArH), 6.80–6.68 (m, 1H, NH), 4.54–4.41 (m, 2H, CH2Ar), 4.12–3.99 (m, 1H, CHOH), 3.91–3.79 (m, 1H, CHNH), 3.61–3.32 (m, 3H, CH, CH2O), 2.09–1.13 (m, 26H, 13 × CH2), 0.93–0.84 (m, 9H, 3 × CH3); 13C NMR (50 MHz, CDCl3): δ = 173.99, 173.96, 138.15, 138.10, 128.5, 127.9, 127.81, 127.76, 73.3, 72.3, 72.0, 70.21, 70.18, 54.0, 53.9, 35.2, 32.0, 29.81, 29.77, 29.72, 29.69, 29.6, 29.5, 29.3, 25.1, 25.0, 22.8, 19.6, 19.0, 14.2; HRMS (ESI) [M + H]+ m/z: 448.3785; (calculated for [C28H50NO3]+ 448.3785).
4.2.4. N-((S)-1-(Butylamino)-3-methyl-1-oxobutan-2-yl)-2-hydroxyhexadecanamide (6d)
Yield 76%; White solid; mp: 95–97 °C; 1H NMR (200 MHz, CDCl3): δ = 7.53 (d, J = 9.3 Hz, 1H, NHCH), 7.19 (t, J = 5.8 Hz, 0.5H, NHCH2), 7.02 (t, J = 5.7 Hz, 0.5H, NHCH2), 4.97 (d, J = 4.9 Hz, 0.5H, CH), 4.50 (d, J = 5.3 Hz, 0.5H, CH), 4.32–4.20 (m, 1H, CH), 4.17–4.04 (m, 1H, CH), 3.36–3.01 (m, 3H, NHCH2, OH), 2.17–2.00 (m, 2H, CH2), 1.87–1.12 (m, 28H, 14 × CH2), 0.95–0.82 (m, 12H, 4 × CH3); 13C NMR (50 MHz, CDCl3): δ = 175.03, 174.96, 171.8, 171.6, 72.3, 58.7, 58.4, 39.42, 39.38, 35.1, 32.0, 31.5, 31.1, 29.83, 29.78, 29.5, 25.3, 25.2, 22.8, 20.23, 20.20, 19.4, 18.7, 18.6, 14.2, 13.8; HRMS (ESI) [M + Na]+ m/z: 449.3715; (calculated for [C25H50N2NaO3]+ 449.3714).
4.2.5. (2S)-Methyl 2-(2-hydroxyhexadecanamido)-3-(2-oxopyrrolidin-3-yl)propanoate (6e)
Yield 57%; Yellow oil; 1H NMR (200 MHz, CDCl3): δ = 8.45 (d, J = 7.8 Hz, 0.17H, NH), 8.27 (d, J = 8.6 Hz, 0.17H, NH), 7.97 (d, J = 7.4 Hz, 0.33H, NH), 7.84 (d, J = 7.9 Hz, 0.33H, NH), 6.69 (s, 0.17H, NH), 6.59 (s, 0.17H, NH), 6.54 (s, 0.33H, NH), 6.46 (s, 0.33H, NH), 4.64–4.49 (m, 1H, CH), 4.14–4.04 (m, 1H, CH), 3.71 (s, 3H, OCH3), 3.51–3.40 (m, 1H, CHH), 3.36–3.28 (m, 1H, CHH), 2.54–2.04 (m, 4H, CH2, CH, OH), 1.94–1.53 (m, 4H, 2 × CH2), 1.49–0.94 (m, 24H, 12 × CH2), 0.85 (t, J = 5.9 Hz, 3H, CH3); HRMS (ESI) [M + Na]+ m/z: 463.3139; (calculated for [C24H44N2NaO5]+ 463.3142).
4.2.6. (2S)-tert-Butyl 2-((2S)-2-(2-hydroxyhexadecanamido)propanamido)propanoate (6f)
Yield 75%; White solid; mp: 65–66 °C; 1H NMR (200 MHz, CDCl3): δ = 7.41 (d, J = 7.9 Hz, 1H, NH), 7.28–7.23 (m, 1H, NH), 4.65–4.51 (m, 1H, CH), 4.42–4.28 (m, 1H, CH), 4.18–4.03 (m, 2H, CH, OH), 2.01–1.10 (m, 41H, 13 × CH2, 5 × CH3), 0.84 (t, J = 6.5 Hz, 3H, CH2CH3); 13C NMR (50 MHz, CDCl3): δ = 174.7, 174.6, 172.3, 172.1, 171.9, 171.8, 81.98, 81.95, 72.1, 72.0, 48.9, 48.4, 34.9, 32.0, 29.8, 29.74, 29.66, 29.6, 29.4, 28.0, 25.1, 22.8, 18.8, 18.6, 18.2, 18.1, 14.2; HRMS (ESI) [M + Na]+ m/z: 493.3612; (calculated for [C26H50N2NaO5]+ 493.3612).
4.2.7. (2S)-tert-Butyl 2-(2-hydroxydodecanamido)-3-methylbutanoate (6g)
Yield 87%; White solid of low melting point; 1H NMR (400 MHz, CDCl3): δ = 6.94 (d, J = 9.3 Hz, 0.5H, NH), 6.76 (d, J = 9.4 Hz, 0.5H, NH), 4.45 (dd, J = 9.1, 4.5 Hz, 1H, CH), 4.14 (dd, J = 7.8, 3.8 Hz, 1H, CH), 2.24–2.14 [m, 1H, CH(CH3)2], 1.87–1.79 (m, 1H, CHHCHOH), 1.69–1.59 (m, 1H, CHHCHOH), 1.47–1.25 (m, 25H, 8 × CH2, 3 × CH3), 0.98–0.86 (m, 9H, 3 × CH3); 13C NMR (100 MHz, CDCl3): δ =173.94, 173.89, 171.3, 171.1, 82.2, 72.5, 72.1, 57.3, 57.1, 35.3, 35.1, 32.0, 31.6, 31.5, 29.72, 29.70, 29.65, 29.6, 29.52, 29.46, 28.2, 25.1, 25.0, 22.8, 19.12, 19.07, 17.8, 17.7, 14.2; HRMS (ESI) [M + Na]+ m/z: 394.2928; (calculated for [C21H41NNaO4]+ 394.2928).
4.3. General Procedure for the Oxidation of Hydroxyamides 6a–g to Oxoamides 7a–g
To a stirred solution of α-hydroxy-amides (6a–g) (1 mmol) in dry CH2Cl2 (0.2 M), under an inert argon atmosphere, Dess–Martin periodinane (1.3 mmol, 551 mg) was added. After stirring for 1 h, the solvent was removed under reduced pressure and Et2O (30 mL) was added. The organic layer was washed with a saturated solution of aqueous NaHCO3 (20 mL) containing Na2S2O3 (1.5 g, 9.5 mmol), H2O (20 mL), and dried over Na2SO4. After removal of the organic solvent under reduced pressure, the residue was purified by flash chromatography eluting with the appropriate mixture of EtOAc:petroleum ether (40–60 °C).
4.3.1. (S)-tert-Butyl 2-(2-oxohexadecanamido)propanoate (7a)
Yield 87%; White solid; mp: 44–45 °C; [α]D = +5 (c = 1 in CHCl3); 1H NMR (400 MHz, CDCl3): δ = 7.42 (d, J = 7.5 Hz, 1H, NH), 4.35 (qu, J = 7.2 Hz, 1H, CHNH), 2.83 (t, J = 7.3 Hz, 2H, CH2CO), 1.57–1.50 (m, 2H, CH2CH2CO), 1.41 (s, 9H, 3 × CCH3), 1.35 (d, J = 7.1 Hz, 3H, CHCH3), 1.27–1.09 (m, 22H, 11 × CH2), 0.81 (t, J = 6.5 Hz, 3H, CH2CH3); 13C NMR (100 MHz, CDCl3): δ = 198.5, 171.1, 159.6, 82.2, 48.6, 36.7, 31.9, 29.70, 29.69, 29.67, 29.6, 29.5, 29.38, 29.35, 29.1, 27.9, 23.2, 22.7, 18.2, 14.1; HRMS (ESI) [M + Na]+ m/z: 420.3084; (calculated for [C23H43NNaO4]+ 420.3084).
4.3.2. (S)-N-(1-((tert-Butyldimethylsilyl)oxy)-3-methylbutan-2-yl)-2-oxohexadecanamide (7b)
Yield 89%; Colorless oil; [α]D = −19 (c = 1 in CHCl3); 1H NMR (400 MHz, CDCl3): δ = 7.12 (d, J = 9.6 Hz, 1H, NH), 3.73 (dd, J = 10.2, 3.3 Hz, 1H, CHHO), 3.64 (ddd, J = 13.3, 7.4, 3.6 Hz, 1H, CHNH), 3.56 (dd, J = 10.2, 3.9 Hz, 1H, CHHO), 2.89 (t, J = 7.4 Hz, 2H, CH2CO), 1.99–1.87 (m, 1H, CHCHNH), 1.62–1.55 (m, 2H, CH2CH2CO), 1.34–1.18 (m, 22H, 11 × CH2), 0.93 (d, J = 6.8 Hz, 3H, CH3CH2), 0.89–0.84 (m, 15H, 2 × CH3CH, 3 × CH3C), 0.01 (d, J = 1.9 Hz, 6H, 2 × CH3Si); 13C NMR (100 MHz, CDCl3): δ = 199.6, 160.0, 62.7, 56.1, 36.9, 32.0, 29.80, 29.79, 29.77, 29.7, 29.6, 29.5, 29.2, 29.0, 25.9, 23.4, 22.8, 19.6, 19.0, 18.3, 14.2, −5.45, −5.47; HRMS (ESI) [M + Na]+ m/z: 492.3843; (calculated for [C27H55NNaO3Si]+ 492.3843).
4.3.3. (S)-N-(1-(Benzyloxy)-3-methylbutan-2-yl)-2-oxohexadecanamide (7c)
Yield 93%; White solid; mp: 47–48 °C; [α]D = −34 (c = 1 in CHCl3); 1H NMR (400 MHz, CDCl3): δ = 7.37–7.28 (m, 5H, 5 × ArH), 7.17 (d, J = 9.7 Hz, 1H, NH), 4.51 (d and d, J = 12.1 Hz, 2H, CH2Ph), 3.87–3.80 (m, 1H, CHNH), 3.61 (dd, J = 9.7, 4.2 Hz, 1H, CHHO), 3.46 (dd, J = 9.7, 4.0 Hz, 1H, CHHO), 3.00–2.86 (m, 2H, CH2CO), 2.05–1.97 (m, 1H, CHCHNH), 1.66–1.59 (m, 2H, CH2CH2CO), 1.39–1.20 (m, 22H, 11 × CH2), 0.96–0.89 (m, 9H, 2 × CH3CH, CH3CH2); 13C NMR (100 MHz, CDCl3): δ = 199.5, 160.1, 138.0, 128.5, 127.8, 127.7, 73.3, 69.7, 54.6, 36.9, 32.0, 29.8, 29.75, 29.73, 29.68, 29.5, 29.43, 29.36, 29.2, 23.3, 22.8, 19.5, 18.9, 14.2; HRMS (ESI) [M + Na]+ m/z: 468.3448; (calculated for [C28H47NNaO3]+ 468.3448).
4.3.4. (S)-N-(1-(Butylamino)-3-methyl-1-oxobutan-2-yl)-2-oxohexadecanamide (7d)
Yield 89%; White solid; mp: 78–79 °C; [α]D = −20 (c = 1 in CHCl3); 1H NMR (400 MHz, CDCl3): δ = 7.56 (d, J = 9.2 Hz, 1H, NHCO), 6.52 (br s, 1H, NHCH2), 4.18–4.14 (m, 1H, CHNH), 3.33–3.25 (m, 1H, CHHNH), 3.18–3.10 (m, 1H, CHHNH), 2.91–2.76 (m, 2H, CH2CO), 2.17–2.07 (m, 1H, CHCHNH), 1.61–1.53 (m, 2H, CH2), 1.49–1.42 (m, 2H, CH2), 1.34–1.13 (m, 24H, 12 × CH2), 0.94–0.82 (m, 12H, 4 × CH3); 13C NMR (100 MHz, CDCl3): δ = 198.3, 170.1, 160.3, 59.0, 39.4, 36.9, 32.0, 31.7, 31.3, 29.74, 29.73, 29.71, 29.66, 29.5, 29.4, 29.2, 23.2, 22.7, 20.2, 19.3, 18.4, 14.2, 13.8; HRMS (ESI) [M + Na]+ m/z: 447.3557; (calculated for [C25H48N2NaO3]+ 447.3557).
4.3.5. (2S)-Methyl 2-(2-oxohexadecanamido)-3-(2-oxopyrrolidin-3-yl)propanoate (7e)
Yield 57%; White solid; mp: 59–61 °C; [α]D = −3 (c = 1 in CHCl3); 1H NMR (400 MHz, CDCl3): δ = 8.91 (d, J = 7.8 Hz, 0.25H, NH), 8.19 (d, J = 7.9 Hz, 0.75H, NH), 7.07 (s, 0.25H, NH), 6.88 (s, 0.75H, NH), 4.66–4.61 (m, 0.25H, NHCH), 4.54–4.48 (m, 0.75H, NHCH), 3.72 (s, 3H, CH3O), 3.37–3.27 (m, 2H, NHCH2), 2.85 (t, J = 7.4 Hz, 2H, CH2CO), 2.50–2.14 (m, 3H, CH2, CH), 2.02–1.78 (m, 2H, CH2), 1.60–1.52 (m, 2H, CH2), 1.32–1.18 (m, 22H, 11 × CH2), 0.84 (t, J = 6.9 Hz, 3H, CH3CH2); 13C NMR (100 MHz, CDCl3): δ = 198.4, 198.2, 179.9, 179.6, 171.5, 171.4, 160.7, 160.6, 52.7, 52.6, 51.5, 51.3, 40.8, 40.6, 38.5, 38.4, 37.0, 36.9, 33.4, 32.6, 32.0, 29.74, 29.72, 29.70, 29.66, 29.52, 29.51, 29.4, 29.1, 28.5, 28.4, 23.21, 23.18, 22.7, 14.2; HRMS (ESI) [M + Na]+ m/z: 461.2990; (calculated for [C24H42N2NaO5]+ 461.2986).
4.3.6. (S)-tert-Butyl 2-((S)-2-(2-oxohexadecanamido)propanamido)propanoate (7f)
Yield 82%; White solid; mp: 92–94 °C; [α]D = −12 (c = 1 in CHCl3); 1H NMR (400 MHz, CDCl3): δ = 7.58 (d, J = 7.3 Hz, 1H, NHCOCO), 6.82–6.71 (m, 1H, NHCHCOO), 4.49 (qu, J = 7.1 Hz, 1H, CHNHCOCO), 4.41 (qu, J = 7.2 Hz, 1H, CHCOO), 2.86 (t, J = 7.2 Hz, 2H, CH2CO), 1.60–1.53 (m, 2H, CH2CH2CO), 1.43 (s, 9H, 3 × CCH3), 1.41 (d, J = 7.0 Hz, 3H, CH3CHNHCOCO), 1.33 (d, J = 7.2 Hz, 3H, CH3CHCOO), 1.29–1.16 (m, 22H, 11 × CH2), 0.84 (t, J = 6.7 Hz, 3H, CH3CH2); 13C NMR (100 MHz, CDCl3): δ = 198.4, 171.9, 170.7, 160.0, 82.2, 48.9, 36.9, 32.0, 29.8, 29.74, 29.71, 29.66, 29.5, 29.4, 29.1, 28.0, 23.2, 22.8, 18.54, 18.46, 14.2; HRMS (ESI) [M + Na]+ m/z: 491.3455; (calculated for [C26H48N2NaO5]+ 491.3455).
4.3.7. (S)-tert-Butyl 3-methyl-2-(2-oxododecanamido)butanoate (7g)
Yield 67%; Colorless oil; [α]D = +14 (c = 1 in CHCl3); 1H NMR (400 MHz, CDCl3): δ = 7.34 (d, J = 9.2 Hz, 1H, NH), 4.33 (dd, J = 9.1, 4.6 Hz, 1H, CHNH), 2.86 (t, J = 7.4 Hz, 2H, CH2CO), 2.23–2.12 (m, 1H, CHCHNH), 1.61–1.53 (m, 2H, CH2CH2CO), 1.44 (s, 9H, 3 × CCH3), 1.32–1.15 (m, 14H, 7 × CH2), 0.90 (dd, J = 7.0, 5.8 Hz, 6H, 2 × CHCH3), 0.84 (t, J = 6.9 Hz, 3H, CH2CH3); 13C NMR (100 MHz, CDCl3): δ = 198.7, 170.1, 160.1, 82.4, 57.6, 36.8, 32.0, 31.6, 29.6, 29.5, 29.39, 29.36, 29.1, 28.1, 23.3, 22.7, 19.0, 17.6, 14.2; HRMS (ESI) [M + Na]+ m/z: 392.2769; (calculated for [C21H39NNaO4]+ 392.2771).
4.4. General Procedure of Deprotection of tert-Butyl Esters to Carboxylic Acids 8a–c
To a stirred solution of tert-butyl ester 7a,f,g (1 mmol) in dry CH2Cl2 (1 mL), TFA (1 mL) was added and the reaction mixture was left stirring for 3 hrs. After removal of the solvent, the residue was diluted in diethyl ether and precipitation by petroleum ether (40–60 °C) and filtration afforded the desired product.
4.4.1. (S)-2-(2-Oxohexadecanamido)propanoic Acid (8a)
Yield 96%; White solid; mp: 101–103 °C; [α]D = +13 (c = 1 in CHCl3); 1H NMR (400 MHz, CDCl3): δ = 8.88 (br s, 1H, COOH), 7.42 (d, J = 7.6 Hz, 1H, NH), 4.58 (qu, J = 7.3 Hz, 1H, CHCH3), 2.90 (t, J = 7.3 Hz, 2H, CH2CO), 1.65–1.56 (m, 2H, CH2CH2CO), 1.52 (d, J = 7.2 Hz, 3H, CHCH3), 1.36–1.20 (m, 22H, 11 × CH2), 0.87 (t, J = 6.6 Hz, 3H, CH2CH3); 13C NMR (100 MHz, CDCl3): δ = 198.4, 176.8, 160.0, 48.1, 36.8, 32.0, 29.77, 29.75, 29.7, 29.54, 29.45, 29.4, 29.1, 23.2, 22.8, 17.8, 14.2; HRMS (ESI) [M-H]- m/z: 340.2492; (calculated for [C19H34NO4]− 340.2493).
4.4.2. (S)-2-((S)-2-(2-Oxohexadecanamido)propanamido)propanoic Acid (8b)
Yield 84%; White solid; mp: 157–158 °C; [α]D = +8 (c = 1 in DMF); 1H NMR (400 MHz, DMSO-d6): δ = 12.52 (br s, 1H, COOH), 8.35 (d, J = 7.9 Hz, 1H, NHCOCO), 8.25 (d, J = 7.3 Hz, 1H, NHCHCOOH), 4.32 (qu, J = 7.1 Hz, 1H, CHNHCOCO), 4.20 (qu, J = 7.3 Hz, 1H, CHCOOH), 2.78 (t, J = 7.3 Hz, 2H, CH2CO), 1.55–1.43 (m, 2H, CH2CH2CO), 1.30–1.20 (m, 28H, 11 × CH2, 2 × CH3CH), 0.86 (t, J = 6.8 Hz, 3H, CH3CH2); 13C NMR (100 MHz, DMSO-d6) δ = 198.8, 173.8, 171.1, 160.6, 47.9, 47.5, 36.5, 31.3, 29.03, 29.02, 29.00, 28.99, 28.96, 28.84, 28.78, 28.7, 28.4, 22.7, 22.1, 18.0, 17.1, 13.9; HRMS (ESI) [M-H]− m/z: 411.2864; (calculated for [C22H39N2O5]− 411.2864).
4.4.3. (S)-3-Methyl-2-(2-oxododecanamido)butanoic Acid (8c)
Yield 59%; White solid; mp: 53–54 °C; [α]D = +6 (c = 1 in CHCl3); 1H NMR (400 MHz, CDCl3): δ = 10.59 (s, 1H, COOH), 7.39 (d, J = 9.1 Hz, 1H, NH), 4.57–4.44 (m, 1H, CHNH), 2.89 (t, J = 7.5 Hz, 2H, CH2CO), 2.36–2.23 (m, 1H, CH(CH3)2), 1.65–1.52 (m, 2H, CH2CH2CO), 1.36–1.16 (m, 14H, 7 × CH2), 1.03–0.79 (m, 9H, 3 × CH3); 13C NMR (100 MHz, CDCl3): δ = 198.5, 176.1, 160.3, 57.2, 36.9, 32.0, 31.2, 29.6, 29.5, 29.43, 29.40, 29.1, 23.3, 22.8, 19.1, 17.6, 14.2; HRMS (ESI) [M-H]− m/z: 312.2176; (calculated for [C17H30NO4]− 312.2180).
4.5. (S)-1-((tert-Butyldimethylsilyl)oxy)-3-methylbutan-2-amine (5b)
To a solution of benzyl (S)-(1-((tert-butyldimethylsilyl)oxy)-3-methylbutan-2-yl)carbamate (1.0 mmol, 353 mg) in MeOH (10 mL), 10% Pd/C (0.05 mmol, 53 mg) was added, and the reaction was left stirring under H2 for 16 hrs. Upon completion, the reaction mixture was filtered through celite, and the solvent was evaporated under reduced pressure to afford the desired product. Yield 89%; Pale yellow solid of low melting point; [α]D = +4 (c = 1 in CHCl3); 1H NMR (200 MHz, CDCl3): δ = 3.65 (dd, J = 9.9, 4.2 Hz, 1H, CHH), 3.43 (dd, J = 9.9, 7.2 Hz, 1H, CHH), 3.01 (br s, 2H, NH2), 2.66–2.57 (m, 1H, CHNH2), 1.77–1.61 (m, 1H, CH(CH3)2), 1.04–0.71 (m, 15H, 5 × CH3), 0.03 (s, 6H, 2 × CH3Si); HRMS (ESI) [M + H]+ m/z: 218.1934; (calculated for [C11H28NOSi]+ 218.1935).
4.6. (S)-tert-Butyl (1-(benzyloxy)-3-methylbutan-2-yl)carbamate (10)
To a flame-dried flask, under argon, NaH 60% (1.3 mmol, 52 mg) and dry N,N-dimethylformamide (DMF) (1.3 mL) were added. A solution of alcohol 9 (1.0 mmol, 203 mg) in dry DMF (0.7 mL) was added dropwise at 0 °C. The reaction mixture was stirred for 30 min at 0 °C and then benzyl bromide (1.1 mmol, 0.13 mL) was added dropwise. The reaction mixture was left stirring for 16 hrs at room temperature. Upon completion, the reaction mixture was quenched with a saturated aqueous solution of NH4Cl (1 mL), H2O (3 mL) was added, the aqueous layer was extracted with ethyl acetate (2 × 10 mL) and the combined organic layers were washed with H2O (15 mL). The organic layer was collected, and after drying over Na2SO4, the solvent was removed under reduced pressure. The product was purified by flash chromatography eluting with a mixture of EtOAc:petroleum ether (40–60 °C)–5:95. Yield 47%; Colorless oil; [α]D = −20 (c = 1 in CHCl3); 1H NMR (200 MHz, CDCl3): δ = 7.44–7.13 (m, 5H, 5 × ArH), 4.78 (d, J = 7.9 Hz, 1H, NH), 4.56–4.42 (m, 2H, CH2Ph), 3.61–3.37 (m, 3H, CH2CH, CHNH), 1.99–1.82 (m, 1H, CH(CH3)2), 1.45 (s, 9H, C(CH3)3), 0.92 (d, J = 6.8 Hz, 3H, CH3), 0.91 (d, J = 6.8 Hz, 3H, CH3); 13C NMR (50 MHz, CDCl3): δ = 155.9, 138.3, 128.4, 127.62, 127.58, 78.9, 73.1, 70.5, 55.5, 29.6, 28.4, 19.6, 18.7; HRMS (ESI) [M + H]+ m/z: 294.2064; (calculated for [C17H28NO3]+ 294.2064).
4.7. (S)-1-(Benzyloxy)-3-methylbutan-2-aminium chloride (5c)
N-Boc-Protected amine 10 (1.0 mmol, 294 mg) was stirred for 2 hrs with a 4N solution of HCl in MeOH (50.0 mmol, 12.5 mL). Upon completion of the reaction, Et2O was added, and the solvents were removed under reduced pressure. The latter was repeated until complete removal of HCl, to afford the desired product. Yield 100%; White solid; mp: 115–118 °C; [α]D = +20 (c = 1 in MeOH); 1H NMR (200 MHz, CD3OD): δ = 7.41–7.25 (m, 5H, 5 × ArH), 4.65–4.52 (m, 2H, CH2Ph), 3.73–3.56 (m, 2H, CH2CH), 3.16–3.08 (m, 1H, CHNH2), 2.11–1.93 (m, 1H, CH(CH3)2), 1.03 (d, J = 6.8 Hz, 3H, CH3), 0.97 (d, J = 6.8 Hz, 3H, CH3); HRMS (ESI) [M + H]+ m/z: 194.1535; (calculated for [C12H20NO]+ 194.1539).
4.8. Enzyme Assay
The enzyme inhibition assay was performed as previously described [15]. A buffer composed of 20 mM Tris, 100 mM NaCl, 1 mM EDTA, pH 7.3 was used for the enzyme inhibition assay. For the determination of the inhibition rate, 0.5 μM of SARS-CoV-2 Mpro was incubated with 40 μM or 100 μM of 2-oxoamide in the buffer at 37 °C for 10 min. The FRET substrate was then added to each well at a final concentration of 10 μM and a final total volume of 50 μL, to initiate the reaction. The GraphPad Prism 6.0 software (GraphPad) was used for the calculation of % inhibition rate. Measurements of the inhibition rate for the compounds were performed in triplicate and are presented as mean ± SD.
4.9. Covalent Docking Calculations
4.9.1. Protein Preparation
Docking calculations were performed using the SARS-CoV-2 Mpro crystallographic structure in complex with the covalent α-ketoamide inhibitor 13b (PDB ID: 6Y2F) [15]. Preparation and minimization of Mpro, using the Protein Preparation Wizard tool within the Maestro Schrodinger suite, were performed to ensure structural correctness. Hydrogen addition, bond orders and steric clashes correction, water molecules and HetAtoms deletion, charge optimization and restrained minimization supported by the OPLS3 force field were achieved [35]. Moreover, addition of the missing residues E47 and D48 was performed using the Crosslink Proteins tool, in the Maestro Schrodinger suite. The inhibitor GK241 and analogs 8a and 8d were prepared for docking using the LigPrep tool, in the Maestro Schrodinger suite [36].
4.9.2. Covalent Docking
Covalent docking is a multiple step process, that is designed upon Schroedinger’s Glide and Prime, capable to determine ligands activity against a protein target taking into account both non-covalent interactions and covalent bond formation. Covalent docking calculations were carried out using the Covalent Docking application, implemented in Maestro Schrodinger suite. Initially, pose selection was carried out using non-covalent docking simulations (Glide) and positional constraints. Specifically, ligand docking was performed in a mutated binding site. The reactive residue was transformed to alanine and the ligand warhead (the ligand moiety able to form covalent bond) was docked closely to the catalytic residue avoiding unfavorable clashes. Subsequently, the mutation was reversed, and receptor sampling was performed. Covalent bond formation was achieved based on geometric criteria and structural optimization. The following step involved both minimization of protein–ligand complexes in vacuum, and clustering of the optimized poses. This early selection was used as a basis for the further minimization, scoring and ranking of covalent docking poses using the Prime VSGB2.0 energy model. Finally, an additional scoring function generated the affinity score that represents an average value of both the pre-reaction and post-reaction Glide Scores assessing the overall covalent docking procedure. Simultaneously, MM-GBSA energy property calculations for the structure of the receptor, the ligand and the protein-ligand complex were carried out for every docking pose [37]. In this case, Cys145 was identified as the reactive residue in Mpro and nucleophilic addition to a double bond as the reaction type. MM-GBSA scoring was selected and an output of 100 poses per ligand reaction site was achieved [37]. A covalent bond was formed between the reactive residue, Cys145 and the C2 carbonyl of the inhibitor. Subsequently, Mpro–ligand complexes with thiohemiketal formation between ligand the 2-oxo group and the catalytic Cys145 were selected.
4.10. Molecular Dynamics Simulations
MD simulations were performed using Desmond software, which offers a simple setup. Each calculation comprised an eight-step, automated workflow, divided into two categories. A seven-step protocol enabling the system to be minimized and equilibrated was followed by simulation during the last step of the process. This workflow process allowed for the study of protein–ligand interactions and their conformational variations over time and evaluation of the effects of water molecules in the complex [38]. Initially, the System Builder tool in Desmond was used for the preparation of the complexes. TIP3P was selected as a solvent model, OPLS_2005 was assigned as the force field and the system was embedded in a triclinic shaped box. The volume of the box was minimized, and the negative charges were neutralized by the addition of Na+ ions and 0.15 M of salt was added. Subsequently, MD simulations of 50 ns were performed in Desmond2020-1 using an NPT ensemble. A Nose–Hoover chain thermostat and Martyna–Tobias–Klein barostat were applied to maintain the temperature and pressure constant at 300 Kelvin and 1.01325 bar, respectively.
5. Conclusions
In this paper, we present our results on the identification of a synthetic 2-oxoamide inhibitor, which is a known potent inhibitor of GIIA sPLA2 (IC50 143 nM) [24], and, at the same time, can weakly inhibit SARS-CoV-2 Mpro (IC50 24 μM). The known 2-oxoamides AX109 and AX074, which selectively inhibit GIVA cPLA2, as well as pentafluoroethyl ketone GK187, which is a selective and potent inhibitor of GVIA iPLA2, did not exhibit any appreciable inhibition of SARS-CoV-2 Mpro. The free carboxyl group and the long chain of the inhibitor GK241 were necessary for the inhibition of SARS-CoV-2 Mpro. Since the role of GIIA sPLA2 in mortality from COVID-19 has recently been recognized [12], the development of dual inhibitors of GIIA sPLA2 and SARS-CoV-2 Mpro appears to represent an attractive strategy for the development of novel agents to treat COVID-19. 2-Oxoamide GK241 may provide a lead structure for the development of such dual inhibitors.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ph15080961/s1: Table S1. Structural details of structures 1–7 concerning the configuration of 2-carbon and the dihedral angle CβC145-SC145-C2-O2 related to the covalent bond formatted between Cys145 and 2-oxoamide moiety. Molecular dynamics RMSDs for the protein and the ligand along with MM-GBSA binding energy; Figure S1. Structural details for the Mpro-GK241 interactions in structures 1–6 resulting from covalent docking calculations; Figure S2. Protein residues root mean square fluctuation during the 50 nsec MD simulation showing local changes in the protein chain. Figure S3. Structures of 8d (A) in magenta and 8a (B) in blue bound to Mpro in comparison with GK241 (C) in green and crystal structure PDB 6Y2F (D) in khaki. The side-chain in derivative 8d adopts a curved conformation exploring part of S1 and S2 protease cavities. 1H NMR and 13C NMR spectra of the compounds synthesized.
Author Contributions
Conceptualization, G.K.; investigation, M.A.T., G.S.K., L.Z. and I.A.; data curation, M.A.T. and L.Z.; writing—original draft preparation, M.A.T., E.M. and G.K.; writing—review and editing, G.K. and R.H.; supervision, G.K. and R.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a Stavros Niarchos Foundation (SNF) grant to the National and Kapodistrian University of Athens. This project has received funding from the European Union’s Horizon 2020 research and Innovation program under grant No 10100362 (the SCORE project). The research was supported by the Hellenic Foundation for Research and Innovation (HFRI) under the 3rd Call for HFRI PhD Fellowships (Fellowship Number: 18603). Ι.A. was funded by the European Regional Development Fund (ERDF) and Greek national funds through the Operational Program “Competitiveness, Entrepreneurship and Innovation”, under the heading “RESEARCH–CREATE-INNOVATE” (project code: 5134646).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are available in the main text or the Supplementary Materials.
Acknowledgments
E.M. would like to thank V. Myrianthopoulos for helpful discussions.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| AA | arachidonic acid |
| DMF | N,N-dimethylformamide |
| EDC∙HCl | 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride |
| EDTA | ethylene diamine tetra-acetic acid |
| FDA | Food and Drug Administration |
| FFAs | free fatty acids |
| FRET | fluorescence resonance energy transfer |
| GIIA sPLA2 | secreted PLA2 |
| GIVA cPLA2 | cytosolic PLA2 |
| GVIA iPLA2 | calcium-independent PLA2 |
| HOBt | 1-hydroxybenzotriazole |
| HRMS | high-resolution mass spectrometry |
| Mpro | main protease |
| MD | molecular dynamics |
| OA | oleic acid |
| PCs | phosphatidylcholines |
| PLA2 | phospholipase A2 |
| RdRp | RNA-dependent RNA polymerase |
| r.t. | room temperature |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus-2 |
| TFA | trifluoroacetic acid |
| THF | tetrahydrofuran |
| TLC | thin-layer chromatography |
| Tris | tris(hydroxymethyl)aminomethane |
References
- Coronavirus Disease (COVID-19) Pandemic. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 26 March 2022).
- Cannalire, R.; Cerchia, C.; Beccari, A.R.; Saverio Di Leva, F.; Summa, V. Targeting SARS-CoV-2 proteases and polymerase for COVID-19 treatment: State of the art and future opportunities. J. Med. Chem. 2022, 65, 2716–2746. [Google Scholar] [CrossRef] [PubMed]
- Citarella, A.; Scala, A.; Piperno, A.; Micale, N. SARS-CoV-2 Mpro: A potential target for peptidomimetics and small-molecule inhibitors. Biomolecules 2021, 11, 607. [Google Scholar] [CrossRef]
- Casari, I.; Manfredi, M.; Metharom, P.; Falasca, M. Dissecting lipid metabolism alterations in SARS-CoV-2. Prog. Lipid Res. 2021, 82, 101092. [Google Scholar] [CrossRef]
- Pimentel, L.L.; Rodríguez-Alcalá, L.M. Cholesterol, inflammation, and phospholipids: COVID-19 share traits with cardiovascular disease. BBA—Mol. Cell Biol. Lipids 2021, 1866, 158839. [Google Scholar] [CrossRef]
- Murakami, M.; Taketomi, Y.; Miki, Y.; Sato, H.; Yamamoto, K.; Lambeau, G. Emerging roles of secreted phospholipase A2 enzymes: The 3rd edition. Biochimie 2014, 107, 105–113. [Google Scholar] [CrossRef]
- Mouchlis, V.D.; Dennis, E.A. Phospholipase A2 catalysis and lipid mediator lipidomics. BBA—Mol. Cell Biol. Lipids 2019, 1864, 766–771. [Google Scholar] [CrossRef]
- Vasquez, A.M.; Mouchlis, V.D.; Dennis, E.A. Review of four major distinct types of human phospholipase A2. Adv. Biol. Regul. 2018, 67, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Bock, J.-O.; Ortea, I. Re-analysis of SARS-CoV-2-infected host cell proteomics time-course data by impact pathway analysis and network analysis: A potential link with inflammatory response. Aging 2020, 12, 11277–11286. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Shu, T.; Yang, X.; Song, J.-X.; Zhang, M.; Yao, C.; Liu, W.; Huang, M.; Yu, Y.; Yang, Q.; et al. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl. Sci. Rev. 2020, 7, 1157–1168. [Google Scholar] [CrossRef]
- Barberis, E.; Timo, S.; Amede, E.; Vanella, V.V.; Puricelli, C.; Cappellano, G.; Raineri, D.; Cittone, M.G.; Rizzi, E.; Pedrinelli, A.R.; et al. Large-scale plasma analysis revealed new mechanisms and molecules associated with the host response to SARS-CoV-2. Int. J. Mol. Sci. 2020, 21, 8623. [Google Scholar] [CrossRef]
- Snider, J.M.; You, J.K.; Wang, X.; Snider, A.J.; Hallmark, B.; Zec, M.M.; Seeds, M.C.; Sergeant, S.; Johnstone, L.; Wang, Q.; et al. Group IIA secreted phospholipase A2 is associated with the pathobiology leading to COVID-19 mortality. J. Clin. Investig. 2021, 131, 149236. [Google Scholar] [CrossRef] [PubMed]
- Nikolaou, A.; Kokotou, M.G.; Vasilakaki, S.; Kokotos, G. Small-molecule inhibitors as potential therapeutics and as tools to understand the role of phospholipases A2. BBA—Mol. Cell Biol. Lipids 2019, 1864, 941–956. [Google Scholar] [CrossRef]
- Scott, K.F.; Mann, T.J.; Fatima, S.; Sajinovic, M.; Razdan, A.; Kim, R.R.; Cooper, A.; Roohullah, A.; Bryant, K.J.; Gamage, K.K.; et al. Human group IIA phospholipase A2—Three decades on from its discovery. Molecules 2021, 26, 7267. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef]
- Ma, C.; Sacco, M.D.; Hurst, B.; Townsend, J.A.; Hu, Y.; Szeto, T.; Zhang, X.; Tarbet, B.; Marty, M.T.; Chen, Y.; et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020, 30, 678–692. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Zhang, B.; Jiang, X.-M.; Su, H.; Li, J.; Zhao, Y.; Xie, X.; Jin, Z.; Peng, J.; Liu, F.; et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 2020, 368, 1331–1339. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.T.H.; Moesser, M.A.; Walters, R.K.; Malla, T.R.; Twidale, R.M.; John, T.; Deeks, H.M.; Johnston-Wood, T.; Mikhailov, V.; Sessions, R.B.; et al. Discovery of SARS-CoV-2 Mpro peptide inhibitors from modelling substrate and ligand binding. Chem. Sci. 2021, 12, 13686–13703. [Google Scholar] [CrossRef]
- Breidenbach, J.; Lemke, C.; Pillaiyar, T.; Schäkel, L.; Al Hamwi, G.; Diett, M.; Gedschold, R.; Geiger, N.; Lopez, V.; Mirza, S.; et al. Targeting the main protease of SARS-CoV-2: From the establishment of high throughput screening to the design of tailored inhibitors. Angew. Chem. Int. Ed. 2021, 60, 10423–10429. [Google Scholar] [CrossRef]
- Dampalla, C.S.; Rathnayake, A.D.; Perera, K.D.; Jesri, A.-R.M.; Nguyen, H.N.; Miller, M.J.; Thurman, H.A.; Zheng, J.; Kashipathy, M.M.; Battaile, K.P.; et al. Structure-guided design of potent inhibitors of SARS-CoV-2 3CL protease: Structural, biochemical, and cell-based studies. J. Med. Chem. 2021, 64, 17846–17865. [Google Scholar] [CrossRef] [PubMed]
- Dampalla, C.S.; Kim, Y.; Bickmeier, N.; Rathnayake, A.D.; Nguyen, H.N.; Zheng, J.; Kashipathy, M.M.; Baird, M.A.; Battaile, K.P.; Lovell, S.; et al. Structure-guided design of conformationally constrained cyclohexane inhibitors of severe acute respiratory syndrome coronavirus-2 3CL protease. J. Med. Chem. 2021, 64, 10047–10058. [Google Scholar] [CrossRef]
- Stephens, D.; Barbayianni, E.; Constantinou-Kokotou, V.; Peristeraki, A.; Six, D.A.; Cooper, J.; Harkewicz, R.; Deems, R.A.; Dennis, E.A.; Kokotos, G. Differential inhibition of group IVA and group VIA phospholipases A2 by 2-oxoamides. J. Med. Chem. 2006, 49, 2821–2828. [Google Scholar] [CrossRef][Green Version]
- Six, D.A.; Barbayianni, E.; Loukas, V.; Constantinou-Kokotou, V.; Hadjipavlou-Litina, D.; Stephens, D.; Wong, A.C.; Magrioti, V.; Moutevelis-Minakakis, P.; Baker, S.F.; et al. Structure-activity relationship of 2-oxoamide inhibition of group IVA cytosolic phospholipase A2 and group V secreted phopholipase A2. J. Med. Chem. 2007, 50, 4222–4235. [Google Scholar] [CrossRef] [PubMed]
- Vasilakaki, S.; Barbayianni, E.; Leonis, G.; Papadopoulos, M.G.; Mavromoustakos, T.; Gelb, M.H.; Kokotos, G. Development of a potent 2-oxoamide inhibitor of secreted phospholipase A2 guided by molecular docking calculations and molecular dynamics simulations. Bioorg. Med. Chem. 2016, 24, 1683–1695. [Google Scholar] [CrossRef] [PubMed]
- Magrioti, V.; Nikolaou, A.; Smyrniotou, A.; Shah, I.; Constantinou-Kokotou, V.; Dennis, E.A.; Kokotos, G. New potent and selective polyfluoroalkyl ketone inhibitors of GVIA calcium-independent phospholipase A2. Bioorg. Med. Chem. 2013, 21, 5823–5829. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Burkhart, J.P.; Peet, N.P.; Bey, P. Oxidation of α-hydroxy esters to α-keto esters using the Dess-Martin periodinane reagent. Tetrahedron Lett. 1988, 29, 3433–3436. [Google Scholar] [CrossRef]
- So, R.C.; Ndonye, R.; Izmirian, D.P.; Richardson, S.K.; Guerrera, R.L.; Howell, A.R. Straightforward synthesis of sphinganines via a serine-derived Weinreb amide. J. Org. Chem. 2004, 69, 3233–3235. [Google Scholar] [CrossRef]
- Sacco, M.D.; Ma, C.; Lagarias, P.; Gao, A.; Townsend, J.A.; Meng, X.; Dube, P.; Zhang, X.; Hu, Y.; Kitamura, N.; et al. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mpro and cathepsin L. Sci. Adv. 2020, 6, eabe0751. [Google Scholar] [CrossRef]
- Dennis, E.A.; Norris, P.C. Eicosanoid storm in infection and inflammation. Nat. Rev. Immunol. 2015, 15, 511–523. [Google Scholar] [CrossRef]
- Pungerčar, J.; Bihl, F.; Lambeau, G.; Križaj, I. What do secreted phospholipases A2 have to offer in combat against different viruses up to SARS-CoV-2? Biochimie 2021, 189, 40–50. [Google Scholar] [CrossRef]
- Hoffman, R.; Kania, R.S.; Brothers, M.A.; Davies, J.F.; Ferre, R.A.; Gajiwala, K.S.; He, M.; Hogan, R.J.; Kozminski, K.; Li, L.Y.; et al. Discovery of ketone-based covalent inhibitors of coronavirus 3CL proteases for the potential therapeutic treatment of COVID-19. J. Med. Chem. 2020, 63, 12725–12747. [Google Scholar] [CrossRef]
- Owen, D.R.; Allerton, C.M.N.; Anderson, A.S.; Aschenbrenner, L.; Avery, M.; Berritt, S.; Boras, B.; Cardin, R.D.; Carlo, A.; Coffman, K.J.; et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science 2021, 374, 1586–1593. [Google Scholar] [CrossRef]
- Merad, M.; Martin, J.C. Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat. Rev. Immunol. 2020, 20, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, B.; Sharma, L.; Roberts, L.; Peng, X.; Bermejo, S.; Leighton, I.; Casanovas-Massana, A.; Minasyan, M.; Farhadian, S.; Ko, A.I.; et al. Cutting edge: Severe SARS-CoV-2 infection in humans is defined by a shift in the serum lipidome, resulting in dysregulation of eicosanoid immune mediators. J. Immunol. 2021, 206, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Schrödinger, LLC. Schrödinger Release 2021-4. LigPrep; Schrödinger, LLC: New York, NY, USA, 2021. [Google Scholar]
- Zhu, K.; Borrelli, K.W.; Greenwood, J.R.; Day, T.; Abel, R.; Farid, R.S.; Harder, E. Docking covalent inhibitors: A parameter free approach to pose prediction and scoring. J. Chem. Inf. Model. 2014, 54, 1932–1940. [Google Scholar] [CrossRef]
- D. E. Shaw Research. Schrödinger Release 2021-4. Desmond Molecular Dynamics System; Maestro-Desmond Interoperability, Tools; D. E. Shaw Research: New York, NY, USA; Schrödinger: New York, NY, USA, 2021. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).