Development and Validation of [3H]OF-NB1 for Preclinical Assessment of GluN1/2B Candidate Drugs
Abstract
:1. Introduction
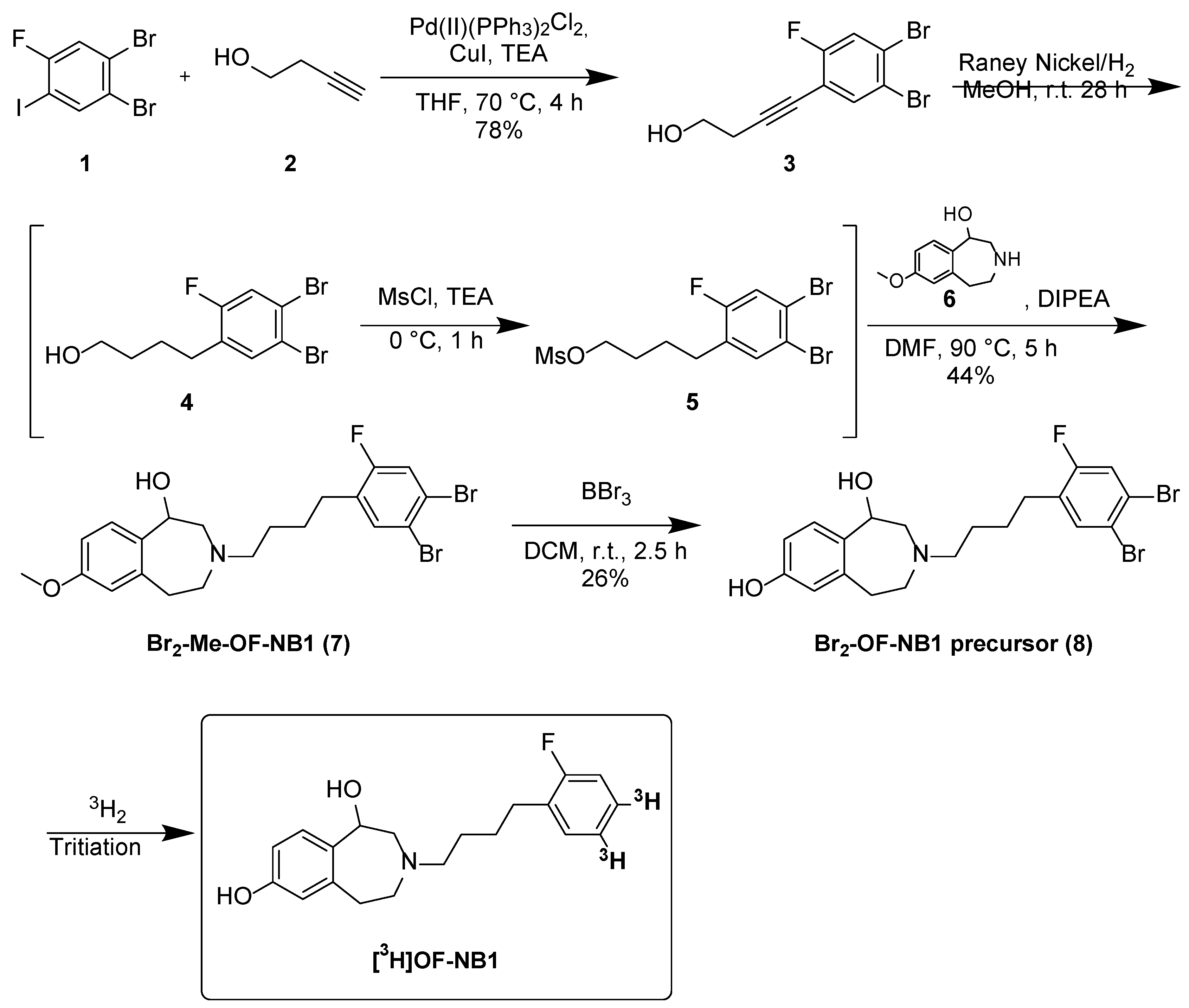
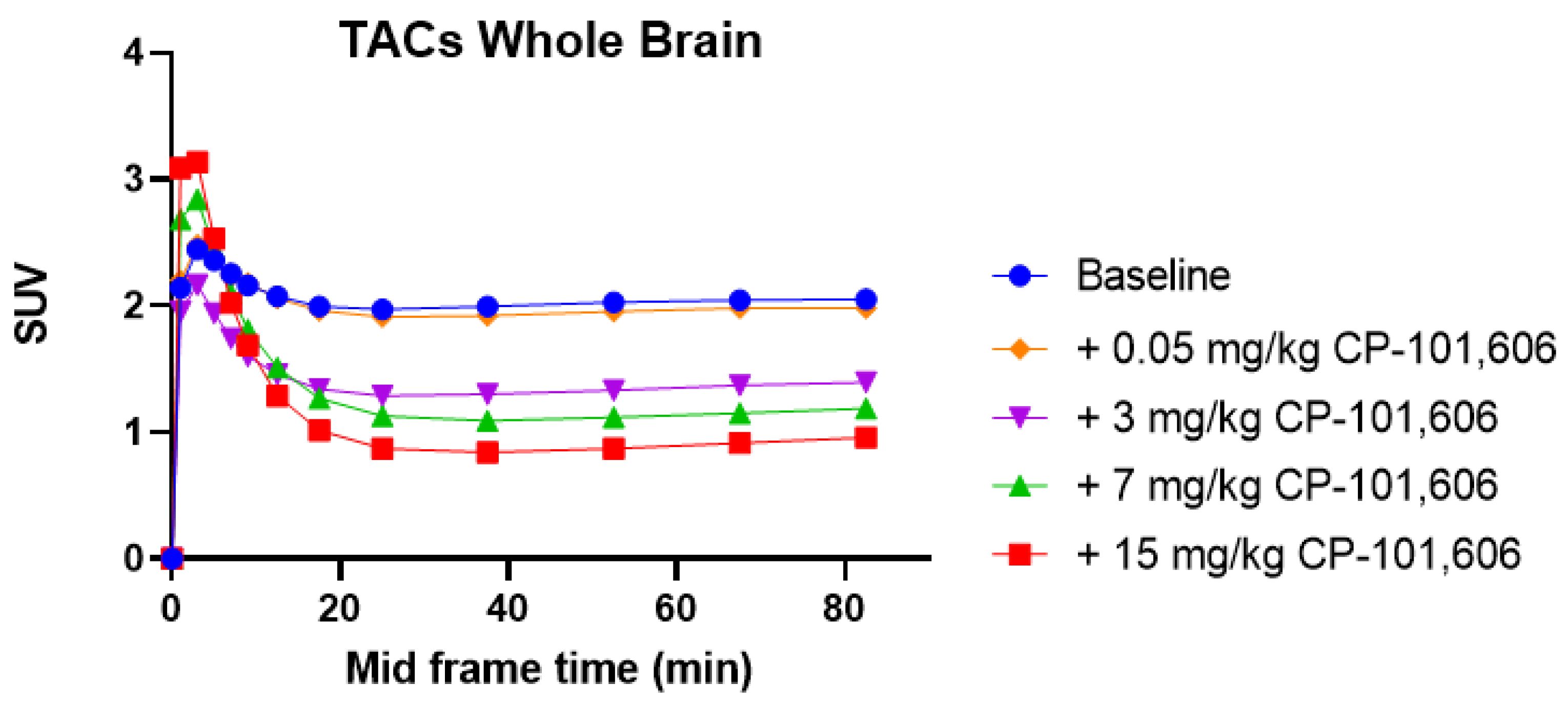
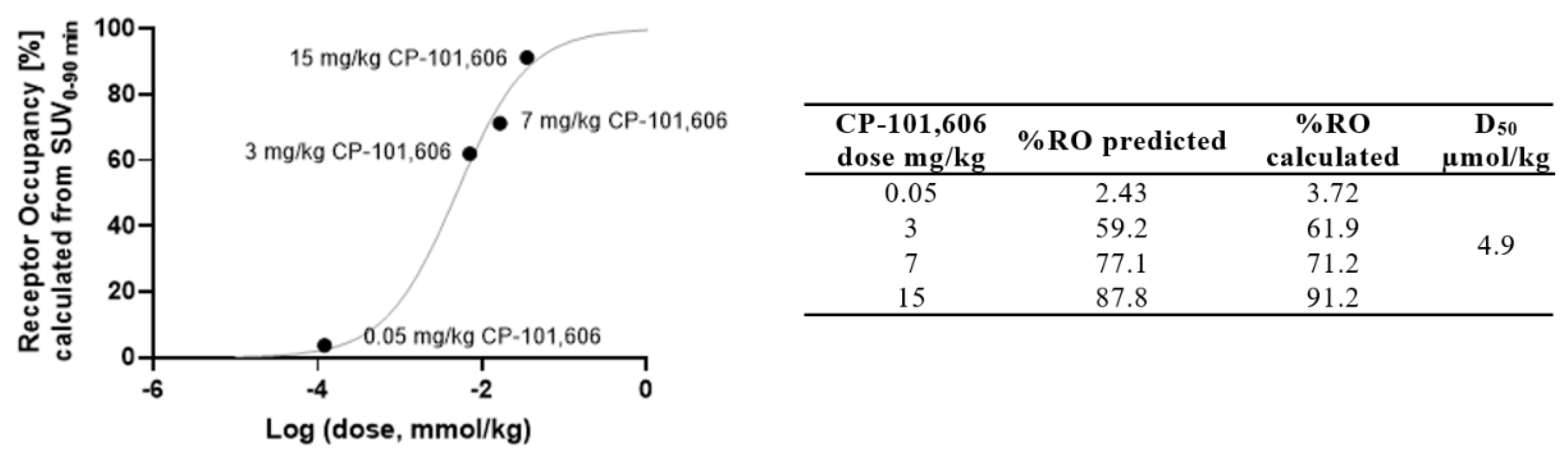
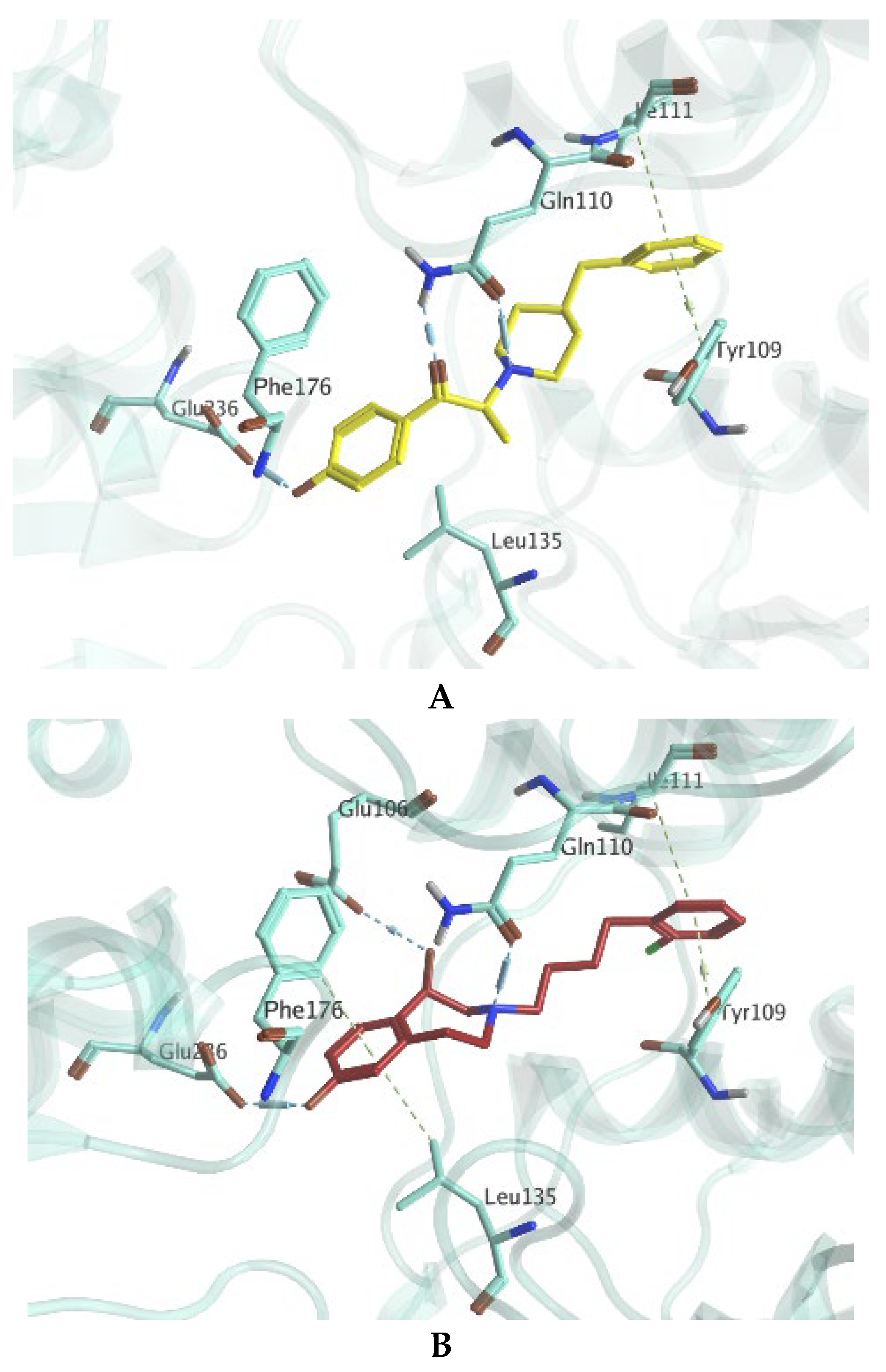
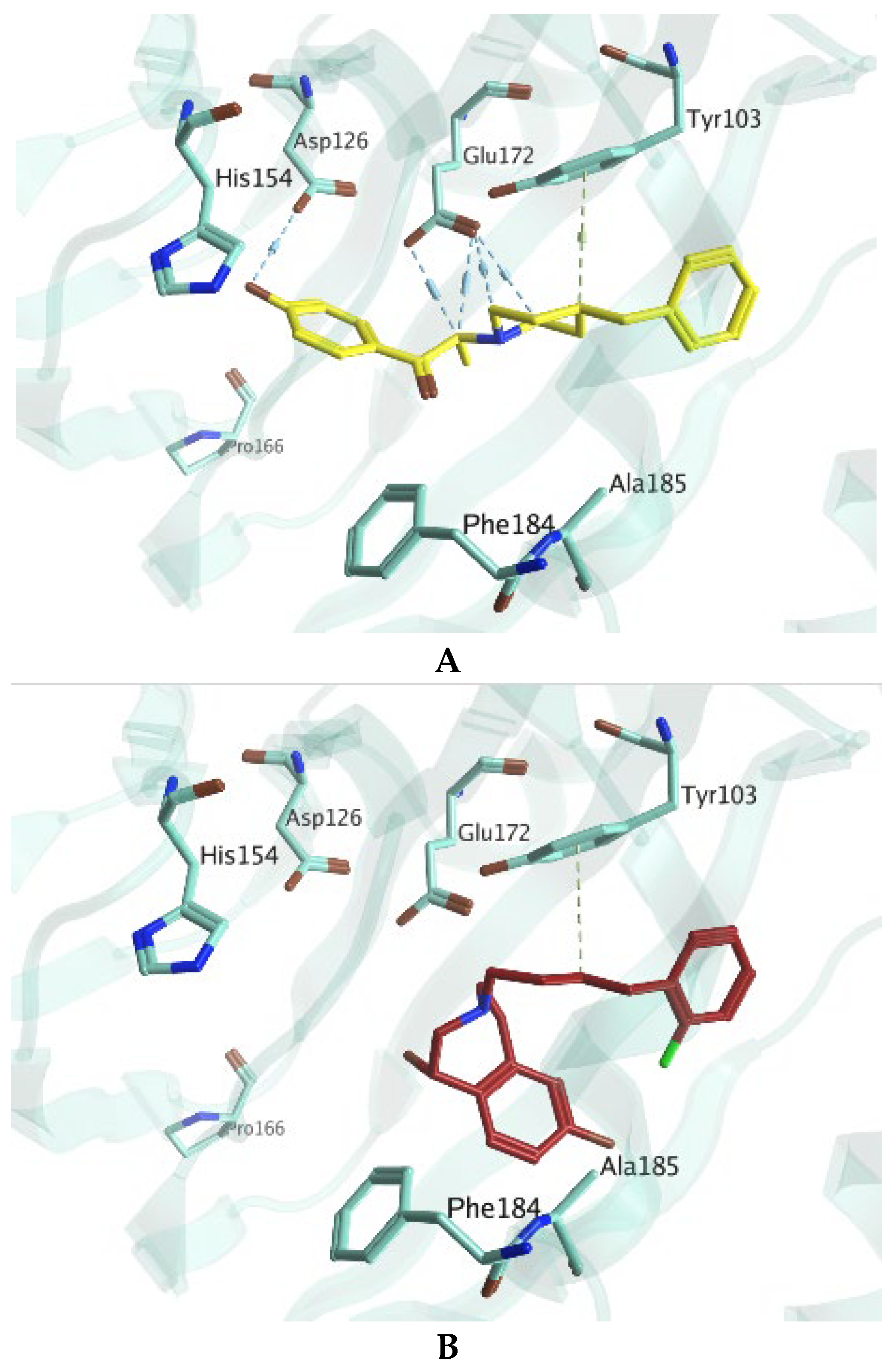
2. Results and Discussion
3. Materials and Methods
3.1. General Methods
3.2. Mass Spectrometry
3.3. NMR Spectroscopy
3.4. Synthetic Procedures
3.4.1. 4-(4,5-Dibromo-2-fluorophenyl)but-3yn-1-ol (3)
3.4.2. 4-(4,5-Dibromo-2-fluorophenyl)butan-1-ol (4)
3.4.3. 4-(4,5-Dibromo-2-fluorophenyl)butan-1-ol (5)
3.4.4. 3-(4-(4,5-Dibromo-2-fluorophenyl)butyl)-7-methoxy-2,3,4,5-tetrahydro-1h-benzo[d]azepin-1-ol (7)
3.4.5. 3-(4-(4,5-Dibromo-2-fluorophenyl)butyl)-2,3,4,5-tetrahydro-1h-benzo[d]azepine-1,7-diol (Br2-OF-NB1, 8)
3.5. In Vitro GluN1/2B Competitive Binding Assay
3.6. In Vitro σ1R Competitive Binding Assay
3.7. In Silico Simulation
3.7.1. Preparation of Co-Crystallized Protein Structure of 3QEL and 6DK1
3.7.2. Validation of the Docking Protocol
3.7.3. Ligands Dataset Curation for Docking Simulations
3.7.4. Docking Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Paoletti, P.; Bellone, C.; Zhou, Q. NMDA receptor subunit diversity: Impact on receptor properties, synaptic plasticity and disease. Nat. Rev. Neurosci. 2013, 14, 383–400. [Google Scholar] [CrossRef] [PubMed]
- Lau, C.G.; Zukin, R.S. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat. Rev. Neurosci. 2007, 8, 413–426. [Google Scholar] [CrossRef]
- Paoletti, P.; Neyton, J. NMDA receptor subunits: Function and pharmacology. Curr. Opin. Pharmacol. 2007, 7, 39–47. [Google Scholar] [CrossRef]
- Zhang, B.; Fang, W.; Ma, W.; Xue, F.; Ai, H.; Lu, W. Differential Roles of GluN2B in Two Types of Chemical-induced Long Term Potentiation-mediated Phosphorylation Regulation of GluA1 at Serine 845 in Hippocampal Slices. Neuroscience 2020, 433, 144–155. [Google Scholar] [CrossRef]
- Yashiro, K.; Philpot, B.D. Regulation of NMDA receptor subunit expression and its implications for LTD, LTP, and metaplasticity. Neuropharmacology 2008, 55, 1081–1094. [Google Scholar] [CrossRef] [PubMed]
- Goebel, D.J.; Poosch, M.S. NMDA receptor subunit gene expression in the rat brain: A quantitative analysis of endogenous mRNA levels of NR1Com, NR2A, NR2B, NR2C, NR2D and NR3A. Mol. Brain Res. 1999, 69, 164–170. [Google Scholar] [CrossRef]
- Zhang, X.M.; Luo, J.H. GluN2A versus GluN2B: Twins, but quite different. Neurosci. Bull. 2013, 29, 761–772. [Google Scholar] [CrossRef]
- Mony, L.; Kew, J.N.; Gunthorpe, M.J.; Paoletti, P. Allosteric modulators of NR2B-containing NMDA receptors: Molecular mechanisms and therapeutic potential. Br. J. Pharmacol. 2009, 157, 1301–1317. [Google Scholar] [CrossRef] [PubMed]
- Hardingham, G.E.; Bading, H. Synaptic versus extrasynaptic NMDA receptor signalling: Implications for neurodegenerative disorders. Nat. Rev. Neurosci. 2010, 11, 682–696. [Google Scholar] [CrossRef]
- Liu, Y.; Wong, T.P.; Aarts, M.; Rooyakkers, A.; Liu, L.; Lai, T.W.; Wu, D.C.; Lu, J.; Tymianski, M.; Craig, A.M.; et al. NMDA receptor subunits have differential roles in mediating excitotoxic neuronal death both in vitro and in vivo. J. Neurosci. 2007, 27, 2846–2857. [Google Scholar] [CrossRef]
- Wang, R.; Reddy, P.H. Role of Glutamate and NMDA Receptors in Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 57, 1041–1048. [Google Scholar] [CrossRef] [PubMed]
- Adell, A. Brain NMDA Receptors in Schizophrenia and Depression. Biomolecules 2020, 10, 947. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.; Haider, A.; Ametamey, S.M. N-Methyl-D-Aspartate (NMDA) receptor modulators: A patent review (2015-present). Expert Opin. Ther. Pat. 2020, 30, 743–767. [Google Scholar] [CrossRef]
- Liu, W.; Jiang, X.; Zu, Y.; Yang, Y.; Liu, Y.; Sun, X.; Xu, Z.; Ding, H.; Zhao, Q. A comprehensive description of GluN2B-selective N-methyl-D-aspartate (NMDA) receptor antagonists. Eur. J. Med. Chem. 2020, 200, 112447. [Google Scholar] [CrossRef]
- Garner, R.; Gopalakrishnan, S.; McCauley, J.A.; Bednar, R.A.; Gaul, S.L.; Mosser, S.D.; Kiss, L.; Lynch, J.J.; Patel, S.; Fandozzi, C.; et al. Preclinical pharmacology and pharmacokinetics of CERC-301, a GluN2B-selective N-methyl-D-aspartate receptor antagonist. Pharmacol. Res. Perspect. 2015, 3, e00198. [Google Scholar] [CrossRef] [PubMed]
- Addy, C.; Assaid, C.; Hreniuk, D.; Stroh, M.; Xu, Y.; Herring, W.J.; Ellenbogen, A.; Jinnah, H.A.; Kirby, L.; Leibowitz, M.T.; et al. Single-dose administration of MK-0657, an NR2B-selective NMDA antagonist, does not result in clinically meaningful improvement in motor function in patients with moderate Parkinson’s disease. J. Clin. Pharmacol. 2009, 49, 856–864. [Google Scholar] [CrossRef]
- Alarcon, K.; Martz, A.; Mony, L.; Neyton, J.; Paoletti, P.; Goeldner, M.; Foucaud, B. Reactive derivatives for affinity labeling in the ifenprodil site of NMDA receptors. Bioorg. Med. Chem. Lett. 2008, 18, 2765–2770. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K. Activation of sigma-1 receptor chaperone in the treatment of neuropsychiatric diseases and its clinical implication. J. Pharmacol. Sci. 2015, 127, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Mantione, C.R.; Spada, M.R.; Neumeyer, J.L.; London, E.D. Further characterization of [3H]ifenprodil binding in rat brain. Eur. J. Pharmacol. 1994, 266, 67–77. [Google Scholar] [CrossRef]
- Ametamey, S.M.; Honer, M.; Schubiger, P.A. Molecular imaging with PET. Chem. Rev. 2008, 108, 1501–1516. [Google Scholar] [CrossRef] [PubMed]
- Mu, L.; Krämer, S.D.; Ahmed, H.; Gruber, S.; Geistlich, S.; Schibli, R.; Ametamey, S.M. Neuroimaging with Radiopharmaceuticals Targeting the Glutamatergic System. Chimia 2020, 74, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.; Wallimann, R.; Haider, A.; Hosseini, V.; Gruber, S.; Robledo, M.; Nguyen, T.A.N.; Herde, A.M.; Iten, I.; Keller, C.; et al. Preclinical Development of (18)F-OF-NB1 for Imaging GluN2B-Containing N-Methyl-d-Aspartate Receptors and Its Utility as a Biomarker for Amyotrophic Lateral Sclerosis. J. Nucl. Med. 2021, 62, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.; Haider, A.; Varisco, J.; Stanković, M.; Wallimann, R.; Gruber, S.; Iten, I.; Häne, S.; Müller Herde, A.; Keller, C.; et al. Structure-Affinity Relationships of 2,3,4,5-Tetrahydro-1H-3-benzazepine and 6,7,8,9-Tetrahydro-5H-benzo [7]annulen-7-amine Analogues and the Discovery of a Radiofluorinated 2,3,4,5-Tetrahydro-1H-3-benzazepine Congener for Imaging GluN2B Subunit-Containing N-Methyl-d-aspartate Receptors. J. Med. Chem. 2019, 62, 9450–9470. [Google Scholar] [CrossRef] [PubMed]
- Haider, A.; Iten, I.; Ahmed, H.; Herde, A.M.; Gruber, S.; Krämer, S.D.; Keller, C.; Schibli, R.; Wünsch, B.; Mu, L.; et al. Identification and Preclinical Evaluation of a Radiofluorinated Benzazepine Derivative for Imaging the GluN2B Subunit of the Ionotropic NMDA Receptor. J. Nucl. Med. 2019, 60, 259. [Google Scholar] [CrossRef]
- Rischka, L.; Vraka, C.; Pichler, V.; Rasul, S.; Nics, L.; Gryglewski, G.; Handschuh, P.; Murgaš, M.; Godbersen, G.M.; Silberbauer, L.R.; et al. First-in-Humans Brain PET Imaging of the GluN2B-Containing N-methyl-d-aspartate Receptor with (R)-11C-Me-NB1. J. Nucl. Med. 2022, 63, 936. [Google Scholar] [CrossRef]
- Haider, A.; Herde, A.M.; Krämer, S.D.; Varisco, J.; Keller, C.; Frauenknecht, K.; Auberson, Y.P.; Temme, L.; Robaa, D.; Sippl, W.; et al. Preclinical Evaluation of Benzazepine-Based PET Radioligands (R)- and (S)-(11)C-Me-NB1 Reveals Distinct Enantiomeric Binding Patterns and a Tightrope Walk Between GluN2B- and σ1-Receptor-Targeted PET Imaging. J. Nucl. Med. 2019, 60, 1167–1173. [Google Scholar] [CrossRef]
- Smart, K.; Zheng, M.-Q.; Ahmed, H.; Fang, H.; Xu, Y.; Cai, L.; Holden, D.; Kapinos, M.; Haider, A.; Felchner, Z.; et al. Comparison of three novel radiotracers for GluN2B-containing NMDA receptors in non-human primates: (R)-[11C]NR2B-Me, (R)-[18F]of-Me-NB1, and (S)-[18F]of-NB1. J. Cereb. Blood Flow Metab. 2022, 42, 1398–1409. [Google Scholar] [CrossRef]
- Zheng, M.; Ahmed, H.; Smart, K.; Xu, Y.; Holden, D.; Kapinos, M.; Felchner, Z.; Haider, A.; Tamagnan, G.; Carson, R.E.; et al. Characterization in nonhuman primates of (R)-[18F]OF-Me-NB1 and (S)-[18F]OF-Me-NB1 for imaging the GluN2B subunits of the NMDA receptor. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 2153–2162. [Google Scholar] [CrossRef]
- Ahmed, H.; Zheng, M.-Q.; Smart, K.; Fang, H.; Zhang, L.; Emery, P.R.; Gao, H.; Ropchan, J.; Haider, A.; Tamagnan, G.; et al. Evaluation of (rac)-, (R)- and (S)-18F-OF-NB1 for imaging GluN2B subunit-containing N-methyl-D-aspartate receptors in non-human primates. J. Nucl. Med. 2022; published ahead of print. [Google Scholar] [CrossRef]
- Weber, F.; Brust, P.; Laurini, E.; Pricl, S.; Wünsch, B. Fluorinated PET Tracers for Molecular Imaging of σ(1) Receptors in the Central Nervous System. Adv. Exp. Med. Biol. 2017, 964, 31–48. [Google Scholar] [CrossRef]
- Grimwood, S.; Richards, P.; Murray, F.; Harrison, N.; Wingrove, P.B.; Hutson, P.H. Characterisation of N-methyl-D-aspartate receptor-specific [3H]Ifenprodil binding to recombinant human NR1a/NR2B receptors compared with native receptors in rodent brain membranes. J. Neurochem. 2000, 75, 2455–2463. [Google Scholar] [CrossRef]
- Lever, J.R.; Gustafson, J.L.; Xu, R.; Allmon, R.L.; Lever, S.Z. σ1 and σ2 receptor binding affinity and selectivity of SA4503 and fluoroethyl SA4503. Synapse 2006, 59, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Menniti, F.; Chenard, B.; Collins, M.; Ducat, M.; Shalaby, I.; White, F. CP-101,606, a potent neuroprotectant selective for forebrain neurons. Eur. J. Pharmacol. 1997, 331, 117–126. [Google Scholar] [CrossRef]
- Zampieri, D.; Fortuna, S.; Calabretti, A.; Romano, M.; Menegazzi, R.; Schepmann, D.; Wünsch, B.; Collina, S.; Zanon, D.; Mamolo, M.G. Discovery of new potent dual sigma receptor/GluN2b ligands with antioxidant property as neuroprotective agents. Eur. J. Med. Chem. 2019, 180, 268–282. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.R.; Betz, R.M.; Dror, R.O.; Kruse, A.C. Structural basis for σ1-receptor ligand recognition. Nat. Struct. Mol. Biol. 2018, 25, 981–987. [Google Scholar] [CrossRef] [PubMed]
- Karakas, E.; Simorowski, N.; Furukawa, H. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature 2011, 475, 249–253. [Google Scholar] [CrossRef] [PubMed]

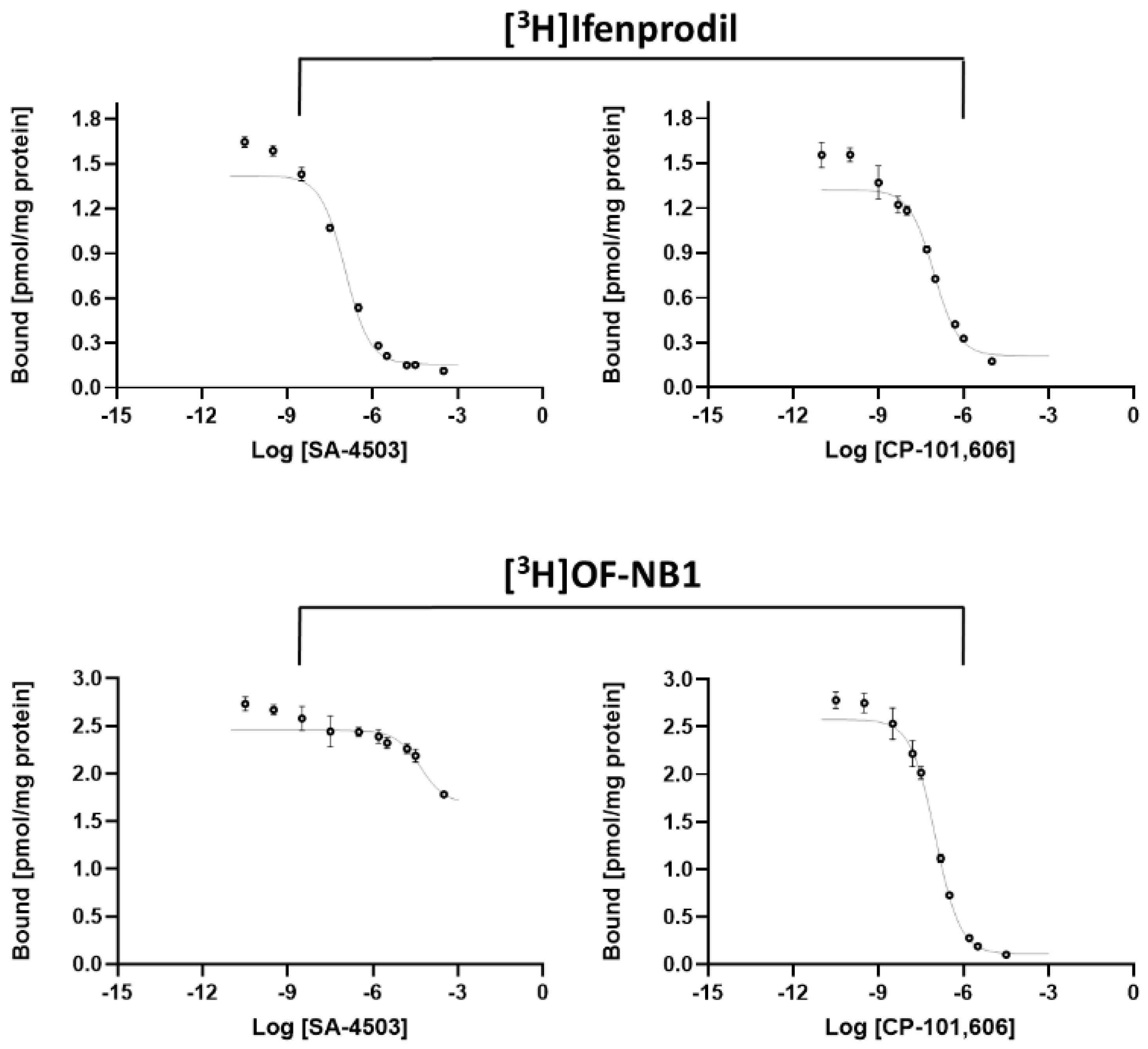
| Radioligand | Test Compound | Ki (GluN1/2B) | Ki (σ1R) |
|---|---|---|---|
| [3H]OF-NB1 | SA-4503 | >100,000 | |
| CP-101,606 | 53 ± 4.3 | ||
| [3H]Ifenprodil | SA-4503 | 51 ± 13 | |
| CP-101,606 | 37 (16 *) | ||
| [3H](+)-Pentazocine | SA-4503 | 3.8 (4.6 *) | |
| CP-101,606 | 94 ± 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, H.; Gisler, L.; Elghazawy, N.H.; Keller, C.; Sippl, W.; Liang, S.H.; Haider, A.; Ametamey, S.M. Development and Validation of [3H]OF-NB1 for Preclinical Assessment of GluN1/2B Candidate Drugs. Pharmaceuticals 2022, 15, 960. https://doi.org/10.3390/ph15080960
Ahmed H, Gisler L, Elghazawy NH, Keller C, Sippl W, Liang SH, Haider A, Ametamey SM. Development and Validation of [3H]OF-NB1 for Preclinical Assessment of GluN1/2B Candidate Drugs. Pharmaceuticals. 2022; 15(8):960. https://doi.org/10.3390/ph15080960
Chicago/Turabian StyleAhmed, Hazem, Livio Gisler, Nehal H. Elghazawy, Claudia Keller, Wolfgang Sippl, Steven H. Liang, Achi Haider, and Simon M. Ametamey. 2022. "Development and Validation of [3H]OF-NB1 for Preclinical Assessment of GluN1/2B Candidate Drugs" Pharmaceuticals 15, no. 8: 960. https://doi.org/10.3390/ph15080960
APA StyleAhmed, H., Gisler, L., Elghazawy, N. H., Keller, C., Sippl, W., Liang, S. H., Haider, A., & Ametamey, S. M. (2022). Development and Validation of [3H]OF-NB1 for Preclinical Assessment of GluN1/2B Candidate Drugs. Pharmaceuticals, 15(8), 960. https://doi.org/10.3390/ph15080960








