Identification of Antibacterial Peptide Candidates Encrypted in Stress-Related and Metabolic Saccharomyces cerevisiae Proteins
Abstract
1. Introduction
2. Results
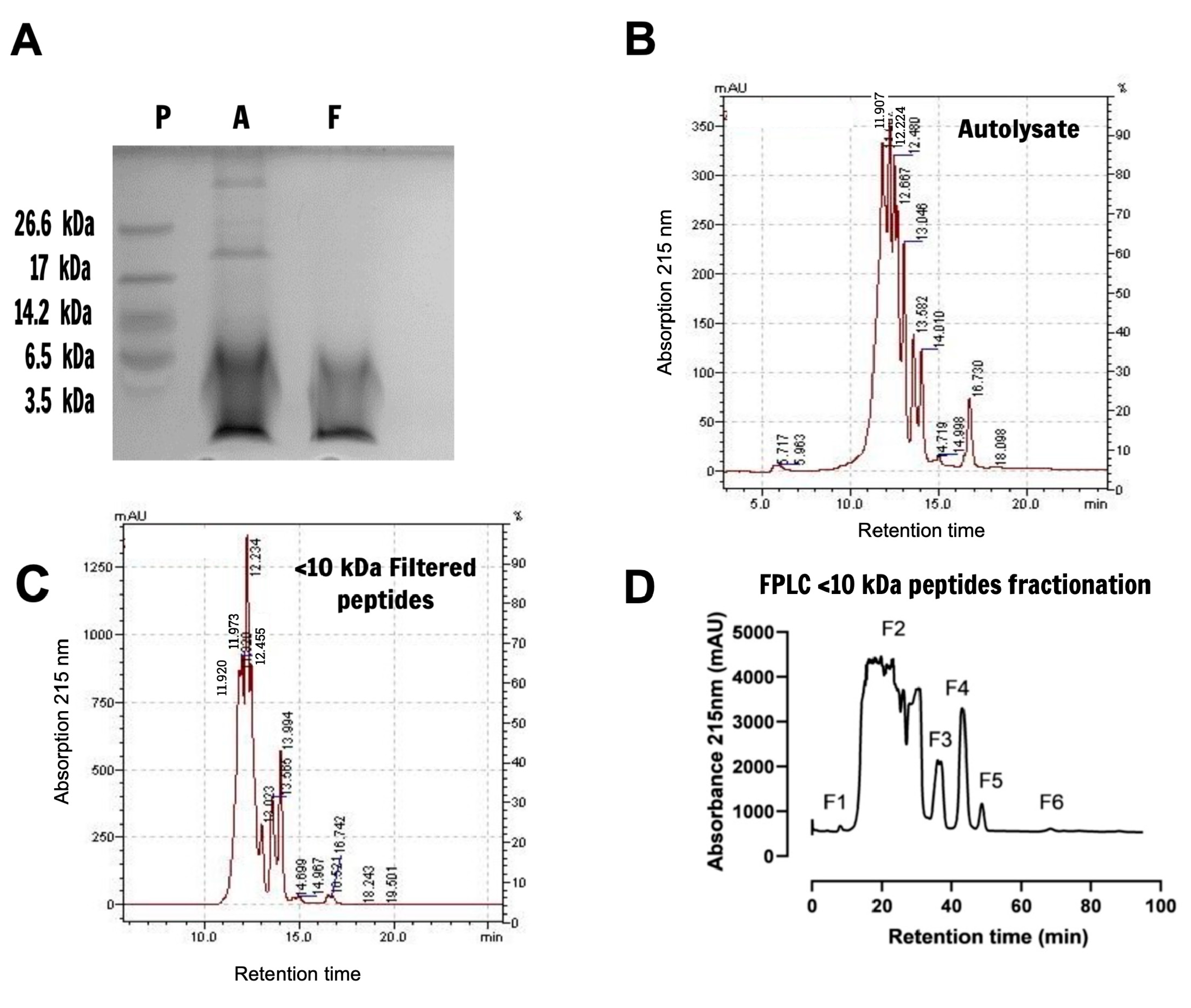
2.1. Characterization and Fractionation of Yeast Extract Peptides
2.2. Antimicrobial Activity Displayed by the Autolysate and <10 kDa Filtered Peptide Extracts and FPLC Gel Filtration Fractions
2.3. Toxicological Evaluation of the <10 kDa Filtered Peptide Extract
2.4. Peptide Identification and In Silico Screening for AMP Candidates
3. Discussion
4. Material and Methods
4.1. Organisms
4.2. Autolysate Preparation
4.3. Preparation of the Ultrafiltered Extract Containing <10 kDa Peptides
4.4. Protein/Peptide Content Determination
4.5. Protein and Peptide Size Distribution Profile in the Autolysate and <10 kDa Filtered Peptides
4.6. Fractionation of the <10 kDa Filtered Peptides
4.7. Evaluation of Antimicrobial Activity in the Autolysate, <10 kDa Filtered Peptide Extract, and FPLC Gel Filtration Fractions
4.8. Investigation of In Vitro Toxicity against Human Cell Lineages
4.9. Nano-Liquid Chromatography and Mass Spectrometry (Nano-LC-MS/MS)
4.10. Data Analysis and Protein Identification
4.11. In Silico Screening for Antimicrobial Peptide Candidates
4.12. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parapouli, M.; Vasileiadis, A.; Afendra, A.-S.; Hatziloukas, E. Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 2020, 6, 1. [Google Scholar] [CrossRef]
- Rakowska, R.; Sadowska, A.; Dybkowska, E.; Swiderski, F. Spent yeast as natural source of functional food additives. Rocz. Państwowego Zakładu Hig. 2017, 68, 115–121. [Google Scholar]
- e-CFR. Title 21: Food and Drugs. Available online: https://www.ecfr.gov/cgi-bin/ECFR?page=browse (accessed on 20 September 2020).
- Heitmann, M.; Zannini, E.; Arendt, E. Impact of Saccharomyces cerevisiae metabolites produced during fermentation on bread quality parameters: A review. Crit. Rev. Food Sci. Nutr. 2018, 58, 1152–1164. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Pastor, R.; Pérez-Torrado, R.; Garre, E.; Matallana, E. Recent advances in yeast biomass production. In Biomass—Detection, Production and Usage; InTech: Rijeka, Croatia, 2011. [Google Scholar]
- Pérez-Torrado, R.; Gamero, E.; Gómez-Pastor, R.; Garre, E.; Aranda, A.; Matallana, E. Yeast biomass, an optimised product with myriad applications in the food industry. Trends Food Sci. Technol. 2015, 46, 167–175. [Google Scholar] [CrossRef]
- Pereira, P.R.; Freitas, C.S.; Paschoalin, V.M.F. Saccharomyces cerevisiae biomass as a source of next-generation food preservatives: Evaluating potential proteins as a source of antimicrobial peptides. Compr. Rev. Food Sci. Food Saf. 2021, 20, 4450–4479. [Google Scholar] [CrossRef]
- Spohn, R.; Daruka, L.; Lázár, V.; Martins, A.; Vidovics, F.; Grézal, G.; Méhi, O.; Kintses, B.; Számel, M.; Jangir, P.K. Integrated evolutionary analysis reveals antimicrobial peptides with limited resistance. Nat. Commun. 2019, 10, 4538. [Google Scholar] [CrossRef]
- Peschel, A.; Sahl, H.G. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 2006, 4, 529–536. [Google Scholar] [CrossRef]
- Hong, J.; Hu, J.; Ke, F. Experimental Induction of Bacterial Resistance to the Antimicrobial Peptide Tachyplesin I and Investigation of the Resistance Mechanisms. Antimicrob. Agents Chemother. 2016, 60, 6067–6075. [Google Scholar] [CrossRef]
- Lewies, A.; Du Plessis, L.H.; Wentzel, J.F. Antimicrobial Peptides: The Achilles’ Heel of Antibiotic Resistance? Probiotics Antimicrob. Proteins 2019, 11, 370–381. [Google Scholar] [CrossRef]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial peptides: An emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed]
- Bahar, A.A.; Ren, D. Antimicrobial Peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef] [PubMed]
- Pinhati, F.R.; Del Aguila, E.M.; Torres, A.P.R.; Sousa, M.P.d.; Santiago, V.M.J.; Silva, J.T.; Paschoalin, V.M.F. Evaluation of the efficiency of deterioration of aromatic hydrocarbons by bacteria from wastewater treatment plant of oil refinery. Química Nova 2014, 37, 1269–1274. [Google Scholar] [CrossRef]
- Indrayanto, G.; Putra, G.S.; Suhud, F. Validation of in-vitro bioassay methods: Application in herbal drug research. Profiles Drug Subst. Excip. Relat. Methodol. 2021, 46, 273–307. [Google Scholar] [CrossRef] [PubMed]
- Waghu, F.H.; Gopi, L.; Barai, R.S.; Ramteke, P.; Nizami, B.; Idicula-Thomas, S. CAMP: Collection of sequences and structures of antimicrobial peptides. Nucleic Acids Res. 2014, 42, D1154–D1158. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Karnik, S.; Barai, R.S.; Jayaraman, V.K.; Idicula-Thomas, S. CAMP: A useful resource for research on antimicrobial peptides. Nucleic Acids Res. 2010, 38, D774–D780. [Google Scholar] [CrossRef]
- Rajarajaran, A.; Dakshanamoorthy, A. Beta-Glucans: A Biomimetic Approach for Reducing Chronicity in Delayed Wound Healing. J. Dermatol. Ski. Sci. 2020, 2, 16–21. [Google Scholar]
- Vetvicka, V.; Vetvickova, J. β (1-3)-D-glucan affects adipogenesis, wound healing and inflammation. Orient. Pharm. Exp. Med. 2011, 11, 169–175. [Google Scholar] [CrossRef]
- Vlassopoulou, M.; Yannakoulia, M.; Pletsa, V.; Zervakis, G.I.; Kyriacou, A. Effects of fungal beta-glucans on health—A systematic review of randomized controlled trials. Food Funct. 2021, 12, 3366–3380. [Google Scholar] [CrossRef]
- Zykova, S.N.; Balandina, K.A.; Vorokhobina, N.V.; Kuznetsova, A.V.; Engstad, R.; Zykova, T.A. Macrophage stimulating agent soluble yeast β-1,3/1,6-glucan as a topical treatment of diabetic foot and leg ulcers: A randomized, double blind, placebo-controlled phase II study. J. Diabetes Investig. 2014, 5, 392–399. [Google Scholar] [CrossRef]
- Avramia, I.; Amariei, S. Spent Brewer’s Yeast as a Source of Insoluble β-Glucans. Int. J. Mol. Sci. 2021, 22, 825. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, S.D.V.; Cordeiro, S.L.; Cavalcanti, J.E.C.; Melchuna, K.M.; Lima, A.M.d.S.; Filho, I.A.; Medeiros, A.C.; Rocha, K.B.F.; Oliveira, E.M.; Faria, E.D.B.; et al. Effects of Purified Saccharomyces cerevisiae (1 → 3)-β-Glucan on Venous Ulcer Healing. Int. J. Mol. Sci. 2012, 13, 8142–8158. [Google Scholar] [CrossRef] [PubMed]
- Takalloo, Z.; Nikkhah, M.; Nemati, R.; Jalilian, N.; Sajedi, R.H. Autolysis, plasmolysis and enzymatic hydrolysis of baker’s yeast (Saccharomyces cerevisiae): A comparative study. World J. Microbiol. Biotechnol. 2020, 36, 68. [Google Scholar] [CrossRef] [PubMed]
- Fakruddin, M.D.; Hossain, M.N.; Ahmed, M.M. Antimicrobial and antioxidant activities of Saccharomyces cerevisiae IFST062013, a potential probiotic. BMC Complement. Altern. Med. 2017, 17, 64. [Google Scholar] [CrossRef] [PubMed]
- Al-Sahlany, S.T.G.; Altemimi, A.B.; Al-Manhel, A.J.A.; Niamah, A.K.; Lakhssassi, N.; Ibrahim, S.A. Purification of bioactive peptide with antimicrobial properties produced by Saccharomyces cerevisiae. Foods 2020, 9, 324. [Google Scholar] [CrossRef]
- Dasgupta, S.; Yang, C.; Castro, L.M.; Tashima, A.K.; Ferro, E.S.; Moir, R.D.; Willis, I.M.; Fricker, L.D. Analysis of the Yeast Peptidome and Comparison with the Human Peptidome. PLoS ONE 2016, 11, e0163312. [Google Scholar] [CrossRef] [PubMed]
- Branco, P.; Francisco, D.; Chambon, C.; Hébraud, M.; Arneborg, N.; Almeida, M.G.; Caldeira, J.; Albergaria, H. Identification of novel GAPDH-derived antimicrobial peptides secreted by Saccharomyces cerevisiae and involved in wine microbial interactions. Appl. Microbiol. Biotechnol. 2014, 98, 843–853. [Google Scholar] [CrossRef]
- Branco, P.; Francisco, D.; Monteiro, M.; Almeida, M.G.; Caldeira, J.; Arneborg, N.; Prista, C.; Albergaria, H. Antimicrobial properties and death-inducing mechanisms of saccharomycin, a biocide secreted by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2017, 101, 159–171. [Google Scholar] [CrossRef]
- Branco, P.; Kemsawasd, V.; Santos, L.; Diniz, M.; Caldeira, J.; Almeida, M.G.; Arneborg, N.; Albergaria, H. Saccharomyces cerevisiae accumulates GAPDH-derived peptides on its cell surface that induce death of non-Saccharomyces yeasts by cell-to-cell contact. FEMS Microbiol. Ecol. 2017, 93, fix055. [Google Scholar] [CrossRef]
- Branco, P.; Viana, T.; Albergaria, H.; Arneborg, N. Antimicrobial peptides (AMPs) produced by Saccharomyces cerevisiae induce alterations in the intracellular pH, membrane permeability and culturability of Hanseniaspora guilliermondii cells. Int. J. Food Microbiol. 2015, 205, 112–118. [Google Scholar] [CrossRef]
- Leite, A.M.O.; Mayo, B.; Rachid, C.T.C.C.; Peixoto, R.S.; Silva, J.T.; Paschoalin, V.M.F.; Delgado, S. Assessment of the microbial diversity of Brazilian kefir grains by PCR-DGGE and pyrosequencing analysis. Food Microbiol. 2012, 31, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Bastos, V.; Santos, M.; Gomes, L.; Leite, A.; Paschoalin, V.; Mere Del Aguila, E. Analysis of the cocobiota and metabolites of Moniliophthora perniciosa-resistant Theobroma cacao beans during spontaneous fermentation in Southern Brazil: Dynamic cocobiota in the spontaneous fermentation of bean-pulp mass in Brazil. J. Sci. Food Agric. 2018, 98, 4963–4970. [Google Scholar] [CrossRef] [PubMed]
- Vrancken, G.; De Vuyst, L.; Van der Meulen, R.; Huys, G.; Vandamme, P.; Daniel, H.-M. Yeast species composition differs between artisan bakery and spontaneous laboratory sourdoughs. FEMS Yeast Res. 2010, 10, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Rozpędowska, E.; Hellborg, L.; Ishchuk, O.P.; Orhan, F.; Galafassi, S.; Merico, A.; Woolfit, M.; Compagno, C.; Piškur, J. Parallel evolution of the make–accumulate–consume strategy in Saccharomyces and Dekkera yeasts. Nat. Commun. 2011, 2, 302. [Google Scholar] [CrossRef]
- Lin, Z.; Wang, T.-Y.; Tsai, B.-S.; Wu, F.-T.; Yu, F.-J.; Tseng, Y.-J.; Sung, H.-M.; Li, W.-H. Identifying Cis-Regulatory Changes Involved in the Evolution of Aerobic Fermentation in Yeasts. Genome Biol. Evol. 2013, 5, 1065–1078. [Google Scholar] [CrossRef][Green Version]
- Pfeiffer, T.; Morley, A. An evolutionary perspective on the Crabtree effect. Front. Mol. Biosci. 2014, 1, 17. [Google Scholar] [CrossRef] [PubMed]
- Escalera-Fanjul, X.; Quezada, H.; Riego-Ruiz, L.; González, A. Whole-Genome Duplication and Yeast’s Fruitful Way of Life. Trends Genet. 2019, 35, 42–54. [Google Scholar] [CrossRef]
- Aslankoohi, E.; Rezaei, M.N.; Vervoort, Y.; Courtin, C.M.; Verstrepen, K.J. Glycerol production by fermenting yeast cells is essential for optimal bread dough fermentation. PLoS ONE 2015, 10, e0119364. [Google Scholar] [CrossRef]
- Gasch, A.P. The environmental stress response: A common yeast response to diverse environmental stresses. In Yeast Stress Responses. Topics in Current Genetics; Hohmann, S., Mager, W.H., Eds.; Springer: Berlin/Heidelberg, Germany, 2003; Volume 1, pp. 11–70. [Google Scholar]
- Verghese, J.; Abrams, J.; Wang, Y.; Morano, K.A. Biology of the heat shock response and protein chaperones: Budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol. Mol. Biol. Rev. 2012, 76, 115–158. [Google Scholar] [CrossRef]
- Gasch, A.P.; Spellman, P.T.; Kao, C.M.; Carmel-Harel, O.; Eisen, M.B.; Storz, G.; Botstein, D.; Brown, P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 2000, 11, 4241–4257. [Google Scholar] [CrossRef]
- SGD. Saccharomyces Genome Database (SGD). Available online: www.yeastgenome.org (accessed on 20 September 2020).
- Heinisch, J.J.; Rodicio, R. Stress responses in wine yeast. In Biology of Microorganisms on Grapes, in Must and in Wine; König, H., Unden, G., Fröhlich, J., Eds.; Springer: Cham, Switzerland, 2017; pp. 377–395. [Google Scholar]
- Kong, I.I.; Turner, T.L.; Kim, H.; Kim, S.R.; Jin, Y.-S. Phenotypic evaluation and characterization of 21 industrial Saccharomyces cerevisiae yeast strains. FEMS Yeast Res. 2018, 18, foy001. [Google Scholar] [CrossRef] [PubMed]
- Del Aguila, E.M.; Gomes, L.P.; Freitas, C.S.; Pereira, P.R.; Paschoalin, V.M.F. Natural antimicrobials in food processing: Bacteriocins, peptides and chitooligosaccharides. Front. Anti-Infect. Drug Discov. 2017, 5, 55–108. [Google Scholar]
- Smith, P.E.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Wiechelman, K.J.; Braun, R.D.; Fitzpatrick, J.D. Investigation of the bicinchoninic acid protein assay: Identification of the groups responsible for color formation. Anal. Biochem. 1988, 175, 231–237. [Google Scholar] [CrossRef]
- Kessler, R.J.; Fanestil, D.D. Interference by lipids in the determination of protein using bicinchoninic acid. Anal. Biochem. 1986, 159, 138–142. [Google Scholar] [CrossRef]
- Brown, R.E.; Jarvis, K.L.; Hyland, K.J. Protein measurement using bicinchoninic acid: Elimination of interfering substances. Anal. Biochem. 1989, 180, 136–139. [Google Scholar] [CrossRef]
- Schägger, H. Tricine–sds-page. Nat. Protoc. 2006, 1, 16. [Google Scholar] [CrossRef]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 10th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015; Volume 32, p. 88. [Google Scholar]
- McMillian, M.; Li, L.; Parker, J.; Patel, L.; Zhong, Z.; Gunnett, J.; Powers, W.; Johnson, M. An improved resazurin-based cytotoxicity assay for hepatic cells. Cell Biol. Toxicol. 2002, 18, 157–173. [Google Scholar] [CrossRef]
- Corrêa, A.C.; Vericimo, M.A.; Dashevskiy, A.; Pereira, P.R.; Paschoalin, V.M. Liposomal Taro Lectin Nanocapsules Control Human Glioblastoma and Mammary Adenocarcinoma Cell Proliferation. Molecules 2019, 24, 471. [Google Scholar] [CrossRef]
- Freitas, C.S.; Vericimo, M.A.; da Silva, M.L.; da Costa, G.C.V.; Pereira, P.R.; Paschoalin, V.M.F.; Del Aguila, E.M. Encrypted antimicrobial and antitumoral peptides recovered from a protein-rich soybean (Glycine max) by-product. J. Funct. Foods 2019, 54, 187–198. [Google Scholar] [CrossRef]
- Hunt, D.F.; Yates, J.R.; Shabanowitz, J.; Winston, S.; Hauer, C.R. Protein sequencing by tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 1986, 83, 6233. [Google Scholar] [CrossRef] [PubMed]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2016, 44, D1094–D1097. [Google Scholar] [CrossRef] [PubMed]
- Zar, J. Biostatistical Analysis, 2nd ed.; Prentice-Hall: Englewood-Cliffs, NJ, USA, 1984. [Google Scholar]
| Microorganism | Gram Staining | Yeast Extracts (μg/mL) | FPLC Gel Filtration Fractions (μg/mL) | Antibiotic Reference (μg/mL) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Autolysate | <10 kDa Filtered Peptides | F1 | F2 | F3 | F4 | F5 | F6 | CPL | ||
| Acinetobacter genomospecies 3 * | negative | 9088 | 8484 | 88.34 **** | - | - | - | - | - | 29.84 |
| Aeromonas hydrophila ATCC 7966 | negative | 994.2 ** | 1939 | 10.04 **** | 436.3 **** | 109.1 **** | 37.25 **** | - | - | - |
| Bacillus cereus ATCC 11778 | positive | 428.5 | 547.9 | 66.96 ** | - | 665.3 | 78.76 * | 508.8 | - | 0.57 |
| Escherichia coli CDC EDL-933 | negative | 7468 | 6439 | 103.4 **** | 2424 | - | 48.56 **** | 6816 | - | - |
| Escherichia coli CDC O55 | negative | 6717 | 9939 | 124.5 **** | 7680 | - | 435.3 *** | 578.9 *** | - | 0.64 |
| Escherichia coli DH5α | negative | 3549 | 5527 | 453.4 **** | 2840 | - | 96.32 **** | 206.5 **** | - | - |
| Salmonella enterica ATCC 12325 | negative | 5833 | 5590 | 164.0 **** | 13676 | - | 1117 ** | 5087 | - | 0.68 |
| Shigella flexneri ATCC 12022 | negative | 2884 | 4014 | - | 35,516 ** | - | - | - | - | 0.01 |
| Shigella sonnei ATCC 25931 | negative | 2091 | 2970 | 49.47 **** | 4641 | - | - | - | - | 0.99 |
| Staphylococcus aureus ATCC 14458 | positive | 1696 | 2177 | - | 1190 | - | 452.8 * | 331.4 ** | 333.4 ** | 36.73 |
| Estimated Indices | Cell Lineage | Yeast Extracts | FPLC Gel Filtration Fractions | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Autolysate | <10 kDa Filtered Peptides | F1 | F2 | F3 | F4 | F5 | F6 | ||
| CC50 (μg/mL) | Human HFF-1 lineage ATCC SCRC-1041 | 1464 * | 3256 | 4545 **** | 4642 **** | 3911 **** | 1622 **** | 2427 **** | 1549 **** |
| SI | Acinetobacter genomospecies 3 * | 0.16 | 0.38 | 51.45 | - | - | - | - | - |
| Aeromonas hydrophila ATCC 7966 | 1.47 | 1.68 | 452.69 | 10.64 | 35.85 | 43.54 | - | - | |
| Bacillus cereus ATCC 11778 | 3.42 | 5.94 | 67.88 | - | 5.88 | 20.59 | 4.77 | - | |
| Escherichia coli CDC EDL-933 | 0.20 | 0.51 | 43.96 | 1.92 | - | 33.40 | 0.36 | - | |
| Escherichia coli CDC O55 | 0.22 | 0.33 | 36.51 | 0.60 | - | 3.73 | 4.19 | - | |
| Escherichia coli DH5α | 0.41 | 0.59 | 10.02 | 1.63 | - | 16.84 | 11.75 | - | |
| Salmonella enterica ATCC 12325 | 0.25 | 0.58 | 27.71 | 0.34 | - | 1.45 | 0.48 | - | |
| Shigella flexneri ATCC 12022 | 0.51 | 0.81 | - | 0.13 | - | - | - | - | |
| Shigella sonnei ATCC 25931 | 0.70 | 1.10 | 91.87 | 1.00 | - | - | - | - | |
| Staphylococcus aureus ATCC 14458 | 0.86 | 1.50 | - | 3.90 | - | 3.58 | 7.32 | 4.65 | |
| Peptide Sequence | Molecular Mass | M/Z | Protein | Gene | Entry Name | F1 | F2 | F3 | F4 | F5 | F6 | AMPs Prediction |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GENVKGWKIGDYAGIK | 1733.91 | 867.9674 | Alcohol dehydrogenase ½ | ADH1/2 | ADH1_YEAST/ADH2_YEAST | x | ++++ | |||||
| IDNLLDKVDSIIIGGG | 1640.8984 | 821.4605 | Phosphoglycerate kinase | PGK1 | PGK_YEAST | x | ++++ | |||||
| IDNLLDKVDSIIIGGGM | 1771.939 | 886.9802 | Phosphoglycerate kinase | PGK1 | PGK_YEAST | x | ++++ | |||||
| IPAPRGSGIVASPA | 1291.7247 | 646.8714 | 40S ribosomal protein S2 | RPS2 | RS2_YEAST | x | ++++ | |||||
| GLIKSPIKV | 953.6273 | 477.8238 | Alcohol dehydrogenase | SCRG_01319 | B3LIX8_YEAS1 | x | ++++ | |||||
| GAPGGFPGGAPP | 980.4715 | 491.2457 | Heat shock protein SSA1 | SSA1 | HSP71_YEAST | x | ++++ | |||||
| KIGGIGTVPVGRVETGVIKPG | 2033.1997 | 1017.6146 | Elongation ator 1-alpha | TEF1/2 | EF1A_YEAST | x | ++++ | |||||
| VDIGKNEGATLITGGERLGSK | 2114.1331 | 705.7211 | Potassium-activated aldehyde dehydrogenase. Mitochondrial. EC 1.2.1.5 | ALD4 | ALDH4_YEAST | x | +++− | |||||
| GGTLNPGLAPAPVHKF | 1574.8568 | 788.4407 | Ammonia transport outward protein 2/Accumulation of dyads protein 2 | ATO2/ADY2 | ATO2_YEAST/ADY2_YEAST | x | +−++ | |||||
| LISLDGTANKSKLGAN | 1600.8784 | 534.6364 | Enolase 1 | ENO1 | ENO1_YEAST | x | +−++ | |||||
| NLLDKVDSIIIGGG | 1412.7875 | 707.4042 | Phosphoglycerate kinase | PGK1 | PGK_YEAST | x | +−++ | |||||
| NLLDKVDSIIIGGGM | 1543.828 | 772.9217 | Phosphoglycerate kinase | PGK1 | PGK_YEAST | x | +−++ | |||||
| VGKVLPELQGKL | 1279.7864 | 640.904 | Glyceraldehyde-3-Phosphate dehydrogenase 1/2/3 | TDH1/2/3 | G3P1_YEAST/G3P2_YEAST/G3P3_YEAST | x | +−++ | |||||
| GNIVDVPVGPGLLGRV | 1560.8987 | 781.4649 | ATP synthase subunit alpha, mitochondrial | ATP1 | ATPA_YEAST | x | −+++ | |||||
| GAPGGAAGGAAGGAPGGFPGGAPPAPE | 2071.9709 | 1037.0002 | XXYS1_4_G0051300.mRNA.1.CDS.1 | PACBIOSEQ_LOCUS72 | A0A7I9C8D2_YEASX | x | −+++ | |||||
| PGGAAGGAAGGAPGGFPGGAPPAPE | 1943.9125 | 972.9683 | XXYS1_4_G0051300.mRNA.1.CDS.1 | PACBIOSEQ_LOCUS72 | A0A7I9C8D2_YEASX | x | −+++ | |||||
| GAPGGAAGGAPGGFPGGAPPAPE | 1815.8539 | 908.9403 | Heat shock protein SSA1 | SSA1 | HSP71_YEAST | x | x | x | −+++ | |||
| GGAPGGAAGGAPGGFPGGAPP | 1575.7429 | 788.8842 | Heat shock protein SSA1 | SSA1 | HSP71_YEAST | x | x | −+++ | ||||
| GGAPGGAAGGAPGGFPGGAPPAPE | 1872.8754 | 937.4507 | Heat shock protein SSA1 | SSA1 | HSP71_YEAST | x | −+++ | |||||
| DWRGGRTASGNIIPSSTGAAK | 2101.0664 | 701.3662 | Glyceraldehyde-3-Phosphate dehydrogenase 1/2/3 | TDH1/2/3 | G3P1_YEAST/G3P2_YEAST/G3P3_YEAST | x | −+++ | |||||
| EHTPRHHQYGSDEGEQDYHDDEQGEEQAGKQ | 3635.4805 | 728.1075 | Protein HBT1 | HBT1 | HBT1_YEAST | x | ++−− | |||||
| IVDVPVGPGLLGRV | 1389.8344 | 695.9295 | ATP synthase subunit alpha, mitochondrial | ATP1 | ATPA_YEAST | x | −++− | |||||
| IDEIDSIAPK | 1099.576 | 550.7982 | Cell division control protein 48 | CDC48 | CDC48_YEAST | x | −++− | |||||
| SPGDGATFPK | 975.4661 | 488.7423 | FK506-binding protein 1 | FPR1 | FKBP_YEAST | x | x | −++− | ||||
| IDDVDSIIKN | 1130.5819 | 566.3014 | Homocitrate synthase, cytosolic isozyme | LYS20 | HOSC_YEAST | x | −++− | |||||
| IDDVDSIIK | 1016.5389 | 509.2791 | Homocitrate synthase, cytosolic isozyme/Homocitrate synthase, mitochondrial | LYS20/21 | HOSC_YEAST/HOSM_YEAST | x | −++− | |||||
| LPANLVDLNVPAKL | 1475.8711 | 738.9482 | Pyruvate decarboxylase isozyme ½ | PDC1/5 | PDC1_YEAST/PDC5_YEAST | x | −++− | |||||
| VDLNVPAKL | 967.5702 | 484.7956 | Pyruvate decarboxylase isozyme ½ | PDC1/5 | PDC1_YEAST/PDC5_YEAST | x | −++− | |||||
| GIGTVPVGRVETGVIKPG | 1734.9991 | 868.51 | Elongation factor 1-alpha | TEF1 /2 | EF1A_YEAST | x | −++− | |||||
| IGTVPVGRVETGVIKPG | 1677.9777 | 840.0009 | Elongation factor 1-alpha | TEF1 /2 | EF1A_YEAST | x | −++− | |||||
| IIAGGVGEFEAGISKDGQTREHA | 2341.1663 | 586.3022 | Elongation factor 1-alpha | TEF1 /2 | EF1A_YEAST | x | −++− | |||||
| GIGTVPVGRV | 953.5658 | 477.7927 | Elongation factor 1-alpha, EF-1-alpha | TEF1/2 | EF1A_YEAST | x | −++− | |||||
| IGGIGTVPVGRVE | 1252.7139 | 627.3672 | Elongation factor 1-alpha | TEF1/2 | EF1A_YEAST | x | x | x | −++− | |||
| VPIGRGQRELIIGDR | 1677.9637 | 560.3348 | ATP synthase subunit alpha, mitochondrial | ATP1 | ATPA_YEAST | x | −−++ | |||||
| GGAPGGAAGGAAGGAPGGFPGGAPPAPE | 2128.9924 | 1065.5073 | XXYS1_4_G0051300.mRNA.1.CDS.1 | PACBIOSEQ_LOCUS72 | A0A7I9C8D2_YEASX | x | −−++ | |||||
| PGGAAGGAAGGAPGGFPGG | 1381.6375 | 691.8289 | XXYS1_4_G0051300.mRNA.1.CDS.1 | PACBIOSEQ_LOCUS72 | A0A7I9C8D2 (A0A7I9C8D2_YEASX) | x | −−++ | |||||
| PGGAAGGAAGGAPGGFPGGAPP | 1646.78 | 824.4052 | XXYS1_4_G0051300.mRNA.1.CDS.1 | PACBIOSEQ_LOCUS72 | A0A7I9C8D2 (A0A7I9C8D2_YEASX) | x | x | −−++ | ||||
| PGGPGGAGGAGGFPGGAGG | 1353.6061 | 677.811 | Protein SIS1 | SIS1 | SIS1_YEAST | x | −−++ | |||||
| GAPGGAAGGAPGGFPGG | 1253.5789 | 627.8004 | Heat shock protein SSA1 | SSA1 | HSP71_YEAST | x | −−++ | |||||
| GAPGGAAGGAPGGFPGGAPP | 1575.7429 | 760.3733 | Heat shock protein SSA1 | SSA1 | HSP71_YEAST | x | x | x | −−++ | |||
| GGAPGGAAGGAPGGFPGG | 1310.6003 | 656.3116 | Heat shock protein SSA1 | SSA1 | HSP71_YEAST | x | −−++ | |||||
| STGAAKAVGKVLPELQGK | 1753.0098 | 585.3472 | Glyceraldehyde-3-phosphate dehydrogenase 1/2/3 | TDH1/2/3 | G3P1_YEAST/G3P2_YEAST/G3P3_YEAST | x | −−++ | |||||
| IGGIGTVPVGRV | 1123.6713 | 562.8463 | Elongation factor 1-alpha | TEF1/2 | EF1A_YEAST | x | x | −−++ | ||||
| GAPAPPPPPPPPALGGSAPKP | 1869.0148 | 935.5192 | Verprolin | VRP1 | VRP1_YEAST | x | −−++ | |||||
| GGFGGPGGPGGQGFGRQGPQG | 1827.84 | 914.9275 | Uncharacterized protein YNL208W | YNL208W, N1338 | YNU8_YEAST | x | −−++ | |||||
| EVEKEVPIPEEEKKDEEKKDEEKKDEDDKKPKLE | 4138.0688 | 690.6852 | ATP-dependent molecular chaperone HSP82 | HSP82 | HSP82_YEAST | x | −+−+ | |||||
| APGGAAGGAPGGFPGGAPPAPE | 1758.8324 | 880.4313 | Heat shock protein SSA1 | SSA1 | HSP71_YEAST | x | −+−+ | |||||
| WKIGDYAGIK | 1149.6182 | 575.8202 | Alcohol dehydrogenase ½ | ADH1/2 | ADH1_YEAST/ADH2_YEAST | x | +−+− | |||||
| APPLPRAPPVPP | 1207.7076 | 604.8652 | Myosin tail region-interacting protein MTI1 | BBC1 | BBC1_YEAST | x | +−+− | |||||
| LLSLDGTANKSKLGAN | 1600.8784 | 534.6364 | Enolase 2 | ENO2 | ENO2_YEAST | x | +−+− | |||||
| LDQEPDAGLGNGGLGRL | 1680.843 | 841.4357 | Glycogen phosphorylase | GPH1 | PHSG_YEAST | x | +−+− | |||||
| VLDQEPDAGLGNGGLGRL | 1773.8685 | 890.9691 | Glycogen phosphorylase | GPH1 | PHSG_YEAST | x | +−+− | |||||
| IGDSIFDKA | 964.4865 | 483.253 | Phosphoglycerate kinase | PGK1 | PGK_YEAST | x | x | x | +−+− | |||
| FKNPNSDKSKWLTGPQ | 1845.9373 | 923.9781 | Enolase 1 | ENO1 | ENO1_YEAST | x | +−−+ | |||||
| GSKADPYGEENQGNFPQRQQPQ | 2474.1211 | 1238.0784 | Protein GRE1 | GRE1 | GRE1_YEAST | x | +−−+ | |||||
| NNYNAIKEEHGENSEEMKKF | 2410.0859 | 804.3777 | Oligo-1,6-Glucosidase IMA1 | IMA1 | MALX3_YEAST | x | +−−+ | |||||
| PPPVFNKPPTGPPP | 1440.7765 | 721.3985 | Protein transport protein SEC31 | SEC31 | SEC31_YEAST | x | +−−+ | |||||
| SPPPVFNKPPTGPPP | 1527.8085 | 764.9156 | Protein transport protein SEC31 | SEC31 | SEC31_YEAST | x | +−−+ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, M.F.d.S.; Freitas, C.S.; Verissimo da Costa, G.C.; Pereira, P.R.; Paschoalin, V.M.F. Identification of Antibacterial Peptide Candidates Encrypted in Stress-Related and Metabolic Saccharomyces cerevisiae Proteins. Pharmaceuticals 2022, 15, 163. https://doi.org/10.3390/ph15020163
Santos MFdS, Freitas CS, Verissimo da Costa GC, Pereira PR, Paschoalin VMF. Identification of Antibacterial Peptide Candidates Encrypted in Stress-Related and Metabolic Saccharomyces cerevisiae Proteins. Pharmaceuticals. 2022; 15(2):163. https://doi.org/10.3390/ph15020163
Chicago/Turabian StyleSantos, Maria Fernanda da Silva, Cyntia Silva Freitas, Giovani Carlo Verissimo da Costa, Patricia Ribeiro Pereira, and Vania Margaret Flosi Paschoalin. 2022. "Identification of Antibacterial Peptide Candidates Encrypted in Stress-Related and Metabolic Saccharomyces cerevisiae Proteins" Pharmaceuticals 15, no. 2: 163. https://doi.org/10.3390/ph15020163
APA StyleSantos, M. F. d. S., Freitas, C. S., Verissimo da Costa, G. C., Pereira, P. R., & Paschoalin, V. M. F. (2022). Identification of Antibacterial Peptide Candidates Encrypted in Stress-Related and Metabolic Saccharomyces cerevisiae Proteins. Pharmaceuticals, 15(2), 163. https://doi.org/10.3390/ph15020163








