Construction of VGG16 Convolution Neural Network (VGG16_CNN) Classifier with NestNet-Based Segmentation Paradigm for Brain Metastasis Classification
Abstract
1. Introduction
2. Associated Studies
3. Background of Predictive Attributes for BM Classification
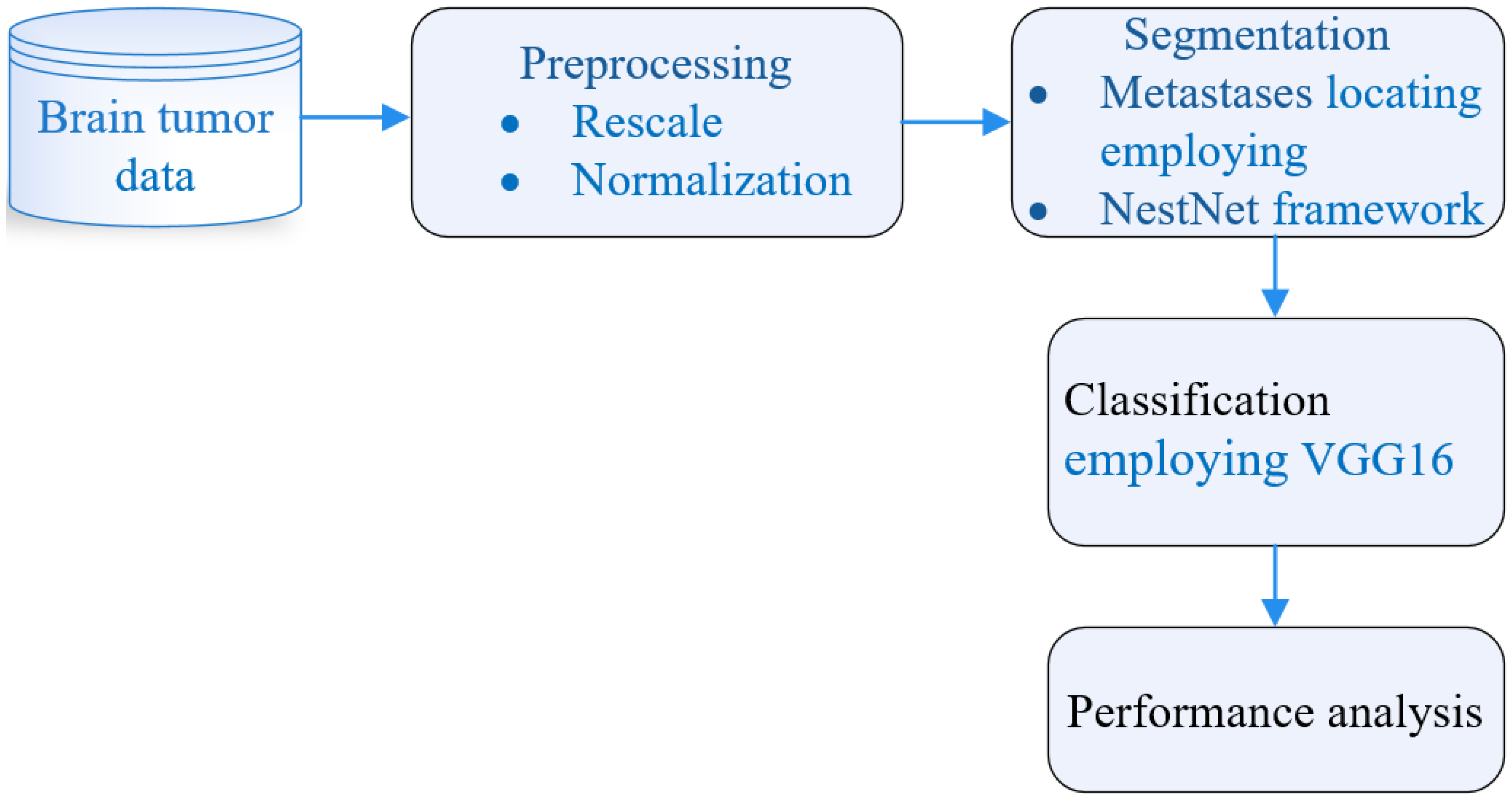
4. System Paradigm
4.1. Image Initialization
4.2. Brain Image Preprocessing
4.3. MtS Localization Employing NtNt Framework
- Left/right hemisphere or central form,
- Cerebral lobes: frontal, parietal, temporal, and occipital lobe,
- Insular cortex,
- Subcortical forms: basal ganglia, thalamus, brainstem, corpus callosum, and cerebellum,
- Eloquent brain regions: vision center (the region surrounding Sulcus calcarinus), auditory center (Gyri temporales transverse), Wernicke’s region (from Gyrus temporalis superior’s dorsal area to the parietal lobe’s Gyri angularis et supramarginal), Broca’s region (Gyrus frontalis inferior’s Pars triangularis et opercularis), the primary somatosensory cortex (Gyrus postcentralis), and primary somatomotor cortex (Gyrus praecentralis).
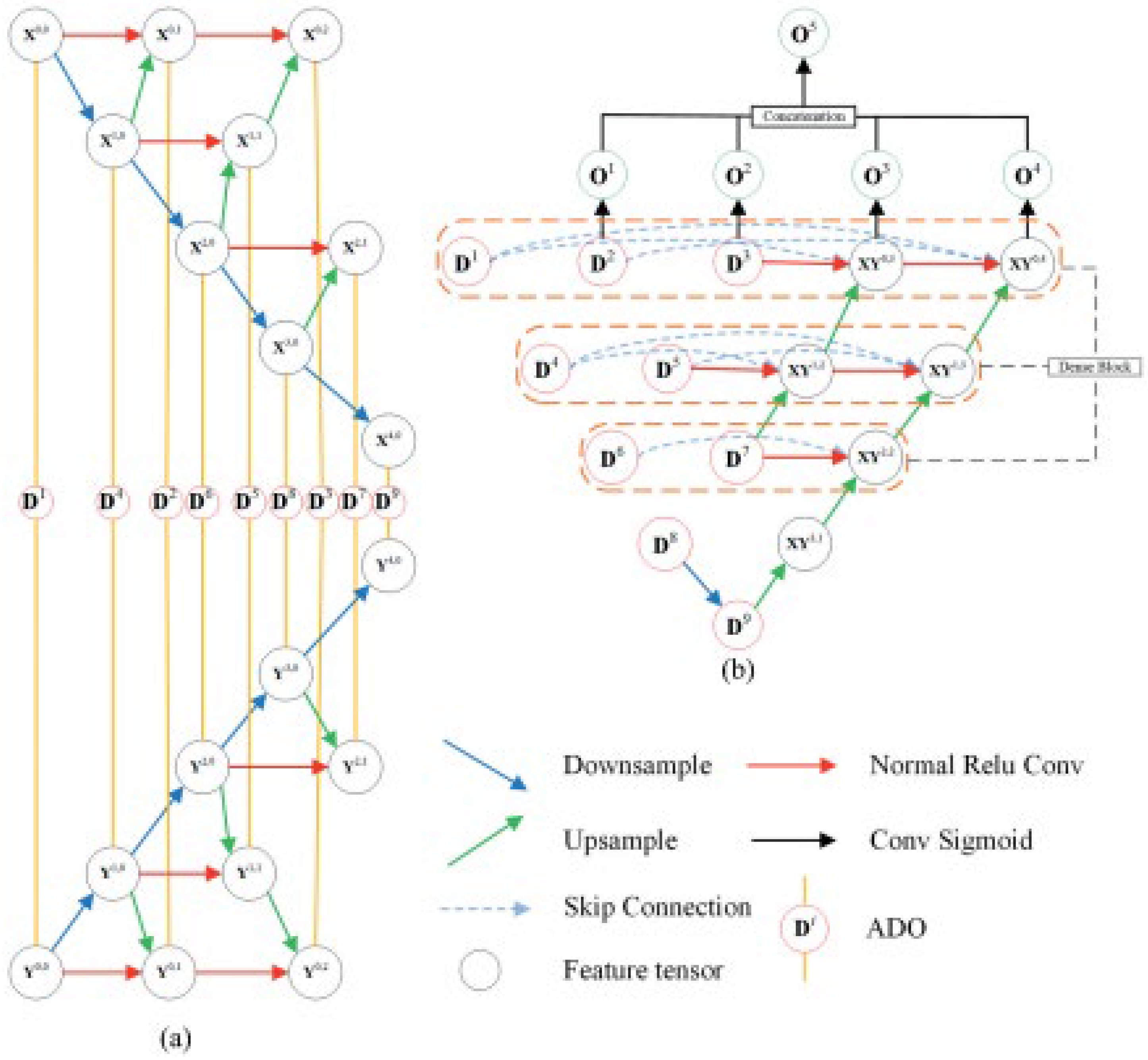
4.4. MtS Area Segmentation Employing NtNt Framework
4.5. Encoding Module
4.6. Decoding Module
4.7. Classification Employing VGG16 Conv NN
4.8. Shared Core Layer
4.9. Atrous Convolution Block
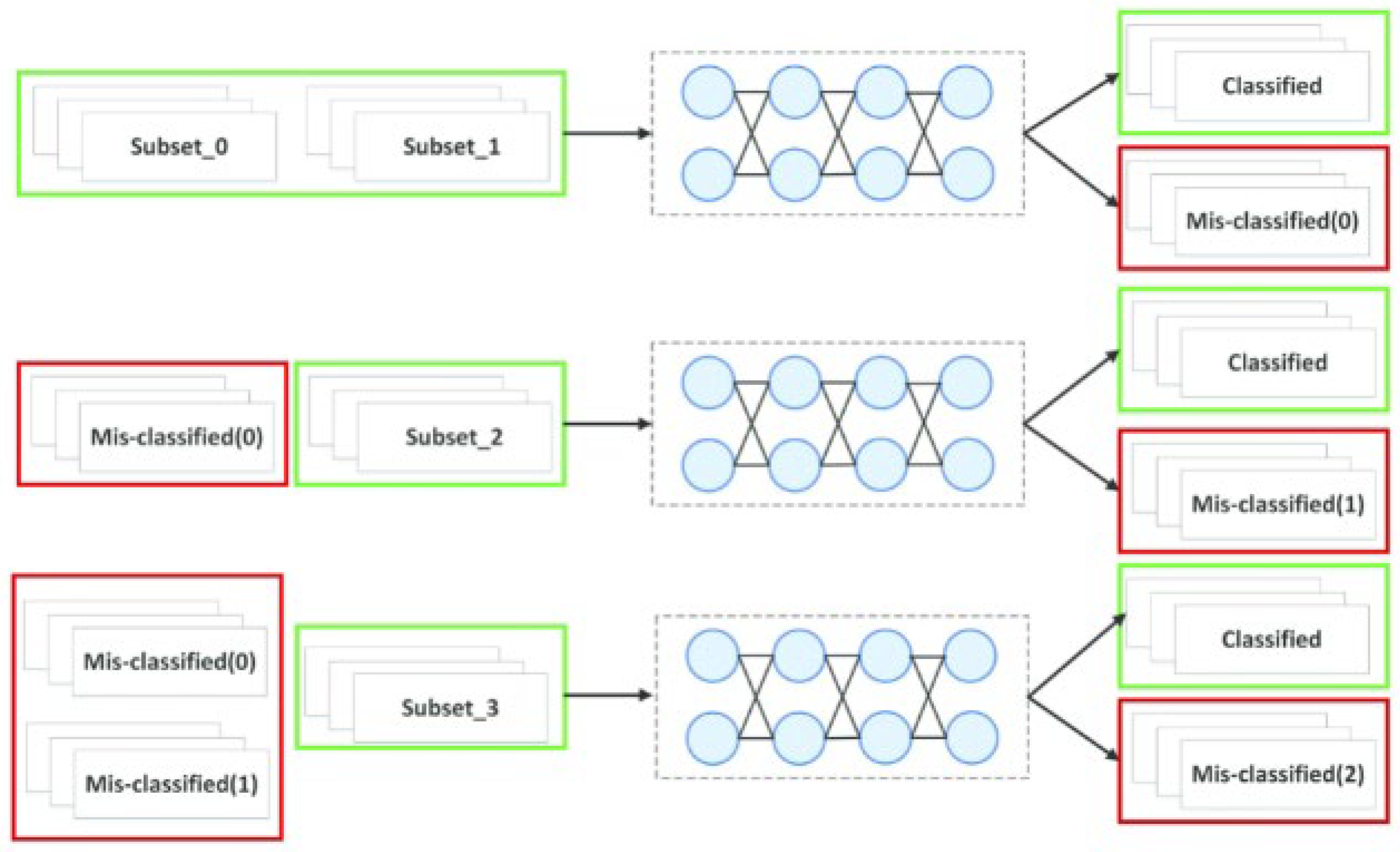
4.10. Optimized Boosting Strategy
4.11. Weighted Softmax Function
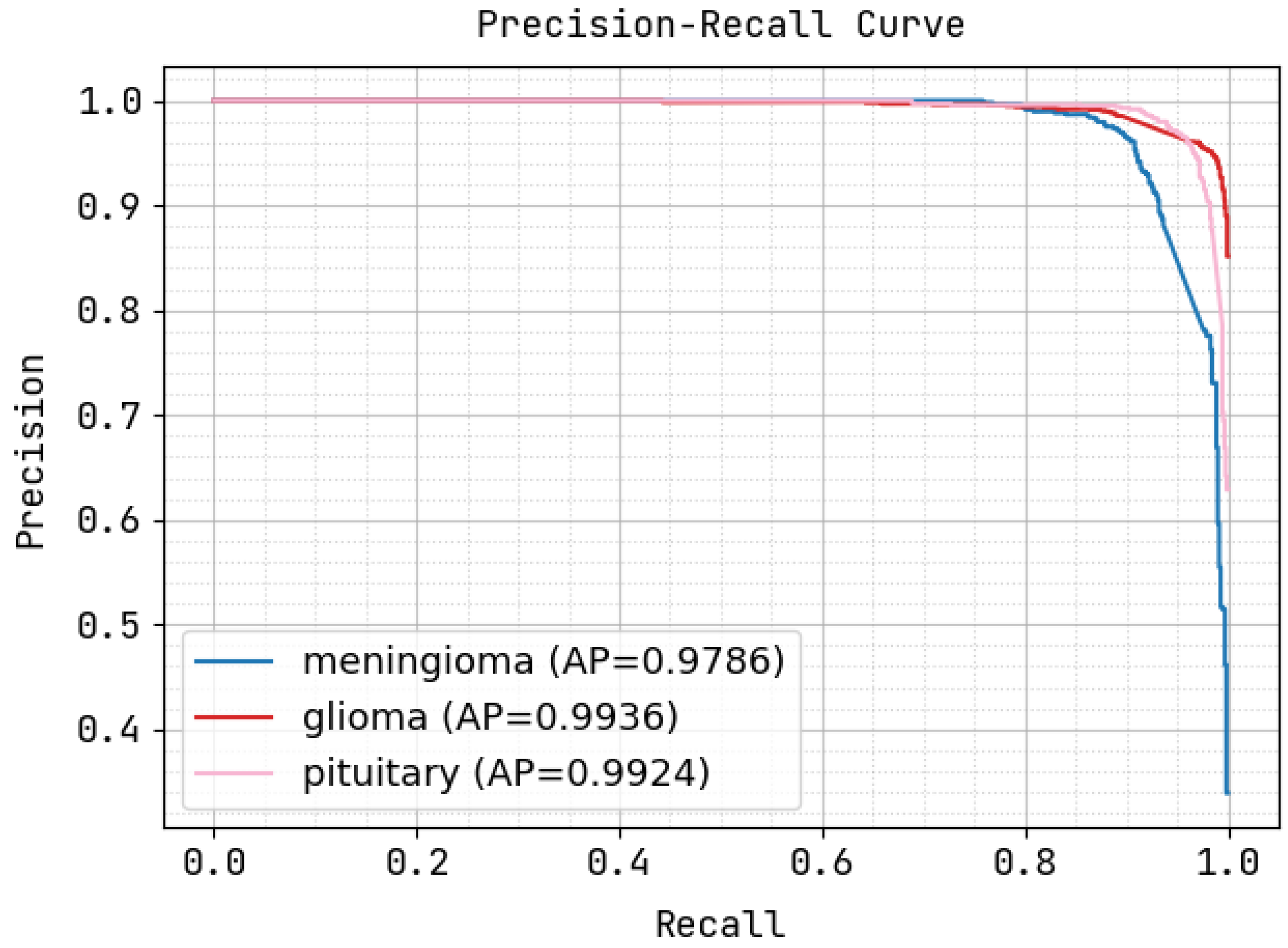
5. Experimental Analysis
5.1. Database Explanation
5.2. Execution Metrics
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Tabouret, E.; Chinot, O.; Metellus, P.; Tallet, A.; Viens, P.; Goncalves, A. Recent Trends in Epidemiology of Brain Metastases: An Overview. Anticancer Res. 2012, 32, 4655–4662. [Google Scholar] [PubMed]
- Steinmann, D.; Schäfer, C.; van Oorschot, B.; Wypior, H.J.; Bruns, F.; Bölling, T.; Sehlen, S.; Hagg, J.; Bayerl, A.; Geinitz, H.; et al. Effects of Radiotherapy for Brain Metastases on Quality of Life (QoL). Strahlenther. Onkol. 2009, 185, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Le Rhun, E.; Guckenberger, M.; Smits, M.; Dummer, R.; Bachelot, T.; Sahm, F.; Galldiks, N.; de Azambuja, E.; Berghoff, A.S.; Metellus, P.; et al. EANO–ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-up of Patients with Brain Metastasis from Solid Tumours. Ann. Oncol. 2021, 32, 1332–1347. [Google Scholar] [CrossRef] [PubMed]
- Chang, E.L.; Wefel, J.S.; Hess, K.R.; Allen, P.K.; Lang, F.F.; Kornguth, D.G.; Arbuckle, R.B.; Swint, J.M.; Shiu, A.S.; Maor, M.H.; et al. Neurocognition in Patients with Brain Metastases Treated with Radiosurgery or Radiosurgery Plus Whole-brain Irradiation: A Randomised Controlled Trial. Lancet Oncol. 2009, 10, 1037–1044. [Google Scholar] [CrossRef]
- Kocher, M.; Wittig, A.; Piroth, M.D.; Treuer, H.; Seegenschmiedt, H.; Ruge, M.; Grosu, A.-L.; Guckenberger, M. Stereotactic Radiosurgery for Treatment of Brain Metastases. Strahlenther. Onkol. 2014, 190, 521–532. [Google Scholar] [CrossRef]
- Brown, P.D.; Jaeckle, K.; Ballman, K.V.; Farace, E.; Cerhan, J.H.; Anderson, S.K.; Carrero, X.W.; Barker, F.G.; Deming, R.; Burri, S.H.; et al. Effect of Radiosurgery Alone vs. Radiosurgery with Whole Brain Radiation Therapy on Cognitive Function in Patients with 1 to 3 Brain Metastases: A Randomized Clinical Trial. JAMA 2016, 316, 401–409. [Google Scholar] [CrossRef]
- Sperduto, P.W.; Mesko, S.; Li, J.; Cagney, D.; Aizer, A.; Lin, N.U.; Nesbit, E.; Kruser, T.J.; Chan, J.; Braunstein, S.; et al. Beyond an Updated Graded Prognostic Assessment (breast GPA): A Prognostic Index and Trends in Treatment and Survival in Breast Cancer Brain Metastases from 1985 to Today. Int. J. Radiat. Oncol. Biol. Phys. 2020, 107, 334–343. [Google Scholar] [CrossRef]
- Kocher, M.; Ruge, M.I.; Galldiks, N.; Lohmann, P. Applications of Radiomics and Machine Learning for Radiotherapy of Malignant Brain Tumors. Strahlenther. Onkol. 2020, 196, 856–867. [Google Scholar] [CrossRef]
- Razzak, M.I.; Naz, S.; Zaib, A. Deep learning for medical image processing: Overview, challenges and the future. Classif. BioApps 2018, 323–350. [Google Scholar]
- Zhang, Y.; Li, H.; Du, J.; Qin, J.; Wang, T.; Chen, Y.; Liu, B.; Gao, W.; Ma, G.; Lei, B. 3D Multi-attention Guided Multi-task Learning Network for Automatic Gastric Tumor Segmentation and Lymph Node Classification. IEEE Trans. Med. Imaging 2021, 40, 1618–1631. [Google Scholar] [CrossRef]
- Chen, J.; Jiao, J.; He, S.; Han, G.; Qin, J. Few-shot Breast Cancer Metastases Classification via Unsupervised Cell Ranking. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 18, 1914–1923. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Song, T.; Katayama, T.; Jiang, X.; Shimamoto, T.; Leu, J.S. Deep Regional Metastases Segmentation for Patient-Level Lymph Node Status Classification. IEEE Access 2021, 9, 129293–129302. [Google Scholar] [CrossRef]
- Isensee, F.; Petersen, J.; Kohl, S.A.; Jäger, P.F.; Maier-Hein, K.H. nnU-net: Breaking the Spell on Successful Medical Image Segmentation. arXiv 2019, arXiv:190408128. [Google Scholar]
- Kickingereder, P.; Isensee, F.; Tursunova, I.; Petersen, J.; Neuberger, U.; Bonekamp, D.; Brugnara, G.; Schell, M.; Kessler, T.; Foltyn, M.; et al. Automated Quantitative Tumour Response Assessment of MRI in Neuro-oncology with Artificial Neural Networks: A multicentre, retrospective study. Lancet Oncol. 2019, 20, 728–740. [Google Scholar] [CrossRef]
- Xue, J.; Wang, B.; Ming, Y.; Liu, X.; Jiang, Z.; Wang, C.; Liu, X.; Chen, L.; Qu, J.; Xu, S.; et al. Deep-Learning-Based Detection and Segmentation-Assisted Management on Brain Metastases. Neuro-Oncology 2019, 22, 505–514. [Google Scholar] [CrossRef]
- McBee, M.P.; Awan, O.A.; Colucci, A.T.; Ghobadi, C.W.; Kadom, N.; Kansagra, A.P.; Tridandapani, S.; Auffermann, W.F. Deep Learning in Radiology. Acad. Radiol. 2018, 25, 1472–1480. [Google Scholar] [CrossRef]
- Samani, Z.R.; Parker, D.; Wolf, R.; Hodges, W.; Brem, S.; Verma, R. Distinct Tumor Signatures using Deep Learning-based Characterization of the Peritumoral Microenvironment in Glioblastomas and Brain Metastases. Sci. Rep. 2021, 11, 14469. [Google Scholar] [CrossRef]
- Dong, F.; Li, Q.; Jiang, B.; Zhu, X.; Zeng, Q.; Huang, P.; Chen, S.; Zhang, M. Differentiation of supratentorial single brain metastasis and glioblastoma by using peri-enhancing oedema region-derived radiomic features and multiple classifiers. Eur. Radiol. 2020, 30, 3015–3022. [Google Scholar] [CrossRef]
- Shin, I.; Kim, H.; Ahn, S.; Sohn, B.; Bae, S.; Park, J.; Kim, H.; Lee, S.-K. Development and Validation of a Deep Learning–Based Model to Distinguish Glioblastoma from Solitary Brain Metastasis Using Conventional MR Images. Am. J. Neuroradiol. 2021, 42, 838–844. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition in Computer Vision and Pattern Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2016, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; der Maaten, L.V.; Weinberger, K.Q. Densely Connected Convolutional Networks in Computer Vision and Pattern Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2017, Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Li, J.; Ng, W.W.Y.; Tian, X.; Kwong, S.; Wang, H. Weighted Multi-deep Ranking Supervised Hashing for Efficient Image Retrieval. Int. J. Mach. Learn. Cybern. 2020, 11, 883–897. [Google Scholar] [CrossRef]
- Yu, X.; Fan, J.; Chen, J.; Zhang, P.; Zhou, Y.; Han, L. NestNet: A multiscale convolutional neural network for remote sensing image change detection. Int. J. Remote Sens. 2021, 42, 4898–4921. [Google Scholar] [CrossRef]
- Khaleghian, S.; Ullah, H.; Kræmer, T.; Hughes, N.; Eltoft, T.; Marinoni, A. Sea Ice Classification of SAR Imagery Based on Convolution Neural Networks. Remote Sens. 2021, 13, 1734. [Google Scholar] [CrossRef]
- Losch, M. Detection and Segmentation of Brain Metastases with Deep Convolutional Networks. Master’s Thesis, KTH, Computer Vision and Active Perception, CVAP. 2015. Available online: http://urn.kb.se/resolve?urn=urn:nbn:se:kth:diva-173519 (accessed on 15 June 2022).
- Zhong, L.; Meng, Q.; Chen, Y.; Du, L.; Wu, P. A laminar augmented cascading flexible neural forest model for classification of cancer subtypes based on gene expression data. BMC Bioinform. 2021, 22, 1–17. [Google Scholar]
- Pang, S.; Fan, M.; Wang, X.; Wang, J.; Song, T.; Wang, X.; Cheng, X. VGG16-T: A novel deep convolutional neural network with boosting to identify pathological type of lung cancer in early stage by CT images. Int. J. Comput. Intell. Syst. 2020, 13, 771. [Google Scholar] [CrossRef]
- Renjith, V.S.; Jose, P.S.H. Efficacy of Deep Learning Approach for Automated Melanoma Detection. In Proceedings of the 2021 International Conference on Decision Aid Sciences and Application (DASA), Virtual, 7–8 December 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 471–478. [Google Scholar]
- Cheng, X.; Kadry, S.; Meqdad, M.N.; Crespo, R.G. CNN supported framework for automatic extraction and evaluation of dermoscopy images. J. Supercomput. 2022, 78, 17114–17131. [Google Scholar] [CrossRef]
- Arcos-García, Á.; Alvarez-Garcia, J.A.; Soria-Morillo, L.M. Deep neural network for traffic sign recognition systems: An analysis of spatial transformers and stochastic optimisation methods. Neural Netw. 2018, 99, 158–165. [Google Scholar] [CrossRef] [PubMed]







| Name | Filter | Feature Map | Weights | Biases |
|---|---|---|---|---|
| Conv3-64 | 3 × 3 × 64 | 50 × 50 × 64 | 1728 | 65 |
| Conv3-64 | 3 × 3 × 64 | 50 × 50 × 64 | 3456 | 56 |
| Conv3-128 | 3 × 3 × 128 | 50 × 50 × 128 | 34,554 | 78 |
| Conv3-256 | 3 × 3 × 256 | 50 × 50 × 256 | 34,579 | 88 |
| Conv3-256 | 3 × 3 × 256 | 50 × 50 × 256 | 75,235 | 34 |
| Conv3-512 | 3 × 3 × 512 | 12 × 12 × 512 | 57,561 | 36 |
| Conv3-512 | 3 × 3 × 512 | 12 × 12 × 512 | 43,575 | 56 |
| Atrous | 3 × 3 × 512 | 3 × 3 × 512 | 34,591 | 78 |
| Atrous | 3 × 3 × 512 | 3 × 3 × 512 | 45,647 | 79 |
| Conv 1 × 1 | 1 × 1 × 4 | 1 × 1 × 512 | 45,890 | 89 |
| Conv 1 × 1 | 1 × 1 × 4 | 1 × 1 × 512 | 87,902 | 65 |
| Epochs | 10 | 20 | 30 | 45 | 65 |
|---|---|---|---|---|---|
| moU-Net | 88.4 | 89.3 | 90.2 | 91.1 | 92.02 |
| DSNet | 87.2 | 88.2 | 91.4 | 93.3 | 94.3 |
| U-Net | 88.1 | 89.3 | 90.1 | 92.2 | 93.04 |
| VGG16_CNN | 90.2 | 91.3 | 93.2 | 94.3 | 95.53 |
| Epochs | 10 | 20 | 30 | 45 | 65 |
|---|---|---|---|---|---|
| moU-Net | 73.4 | 74.3 | 75.2 | 77.1 | 79.4 |
| DSNet | 77.2 | 78.2 | 81.4 | 83.3 | 84.72 |
| U-Net | 80.1 | 82.3 | 82.1 | 84.2 | 86.42 |
| VGG16_CNN | 89.2 | 90.3 | 93.2 | 94.3 | 95.94 |
| Epochs | 10 | 20 | 30 | 45 | 65 |
|---|---|---|---|---|---|
| moU-Net | 86.4 | 87.3 | 88.2 | 89.1 | 90.7 |
| DSNet | 85.2 | 86.2 | 87.4 | 88.3 | 91.44 |
| U-Net | 84.1 | 86.3 | 88.1 | 89.2 | 91.58 |
| VGG16_CNN | 89.2 | 90.3 | 92.2 | 93.3 | 94.44 |
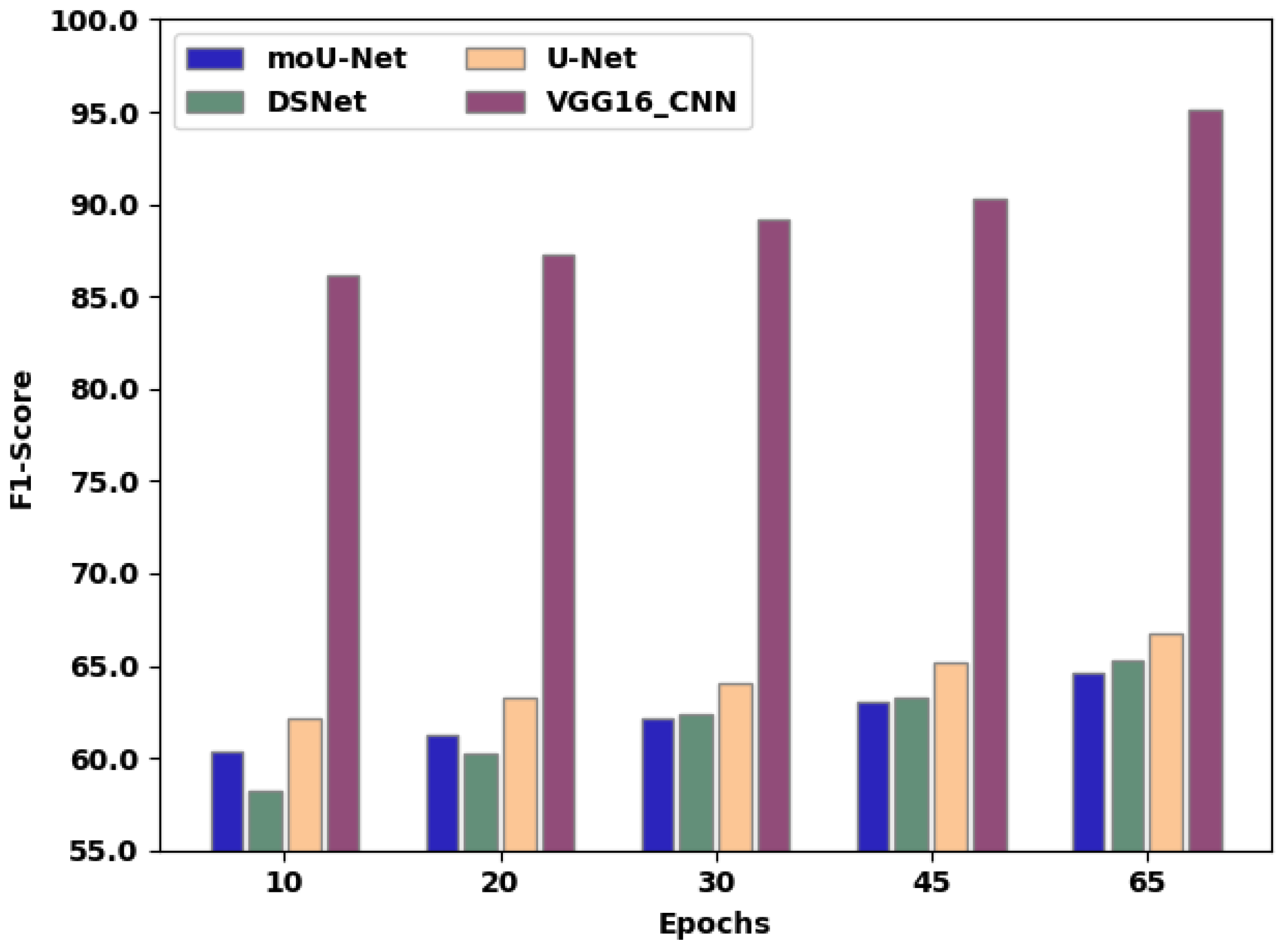
| Epochs | 10 | 20 | 30 | 45 | 65 |
|---|---|---|---|---|---|
| moU-Net | 60.4 | 61.3 | 62.2 | 63.1 | 64.62 |
| DSNet | 58.2 | 60.2 | 62.4 | 63.3 | 65.24 |
| U-Net | 62.1 | 63.3 | 64.1 | 65.2 | 66.76 |
| VGG16_CNN | 86.2 | 87.3 | 89.2 | 90.3 | 95.12 |
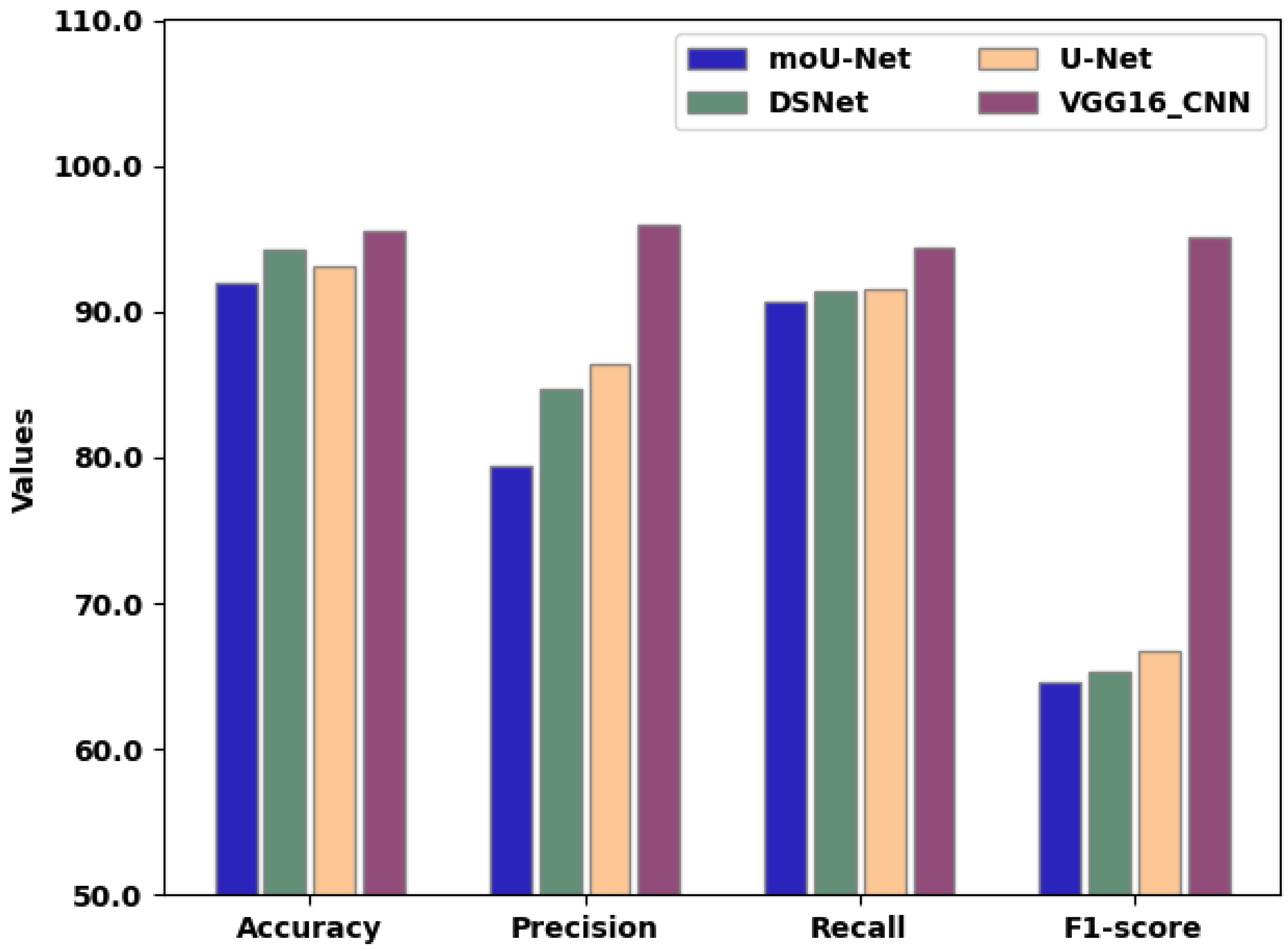
| Criteria | moU-Net | DSNet | U-Net | VGG16_CNN |
|---|---|---|---|---|
| Accuracy (%) | 92.02 | 94.3 | 93.04 | 95.53 |
| Precision (%) | 79.4 | 84.72 | 86.42 | 95.94 |
| Recall (%) | 90.7 | 91.44 | 91.58 | 94.44 |
| F1-score (%) | 64.62 | 65.24 | 66.76 | 95.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alshammari, A. Construction of VGG16 Convolution Neural Network (VGG16_CNN) Classifier with NestNet-Based Segmentation Paradigm for Brain Metastasis Classification. Sensors 2022, 22, 8076. https://doi.org/10.3390/s22208076
Alshammari A. Construction of VGG16 Convolution Neural Network (VGG16_CNN) Classifier with NestNet-Based Segmentation Paradigm for Brain Metastasis Classification. Sensors. 2022; 22(20):8076. https://doi.org/10.3390/s22208076
Chicago/Turabian StyleAlshammari, Abdulaziz. 2022. "Construction of VGG16 Convolution Neural Network (VGG16_CNN) Classifier with NestNet-Based Segmentation Paradigm for Brain Metastasis Classification" Sensors 22, no. 20: 8076. https://doi.org/10.3390/s22208076
APA StyleAlshammari, A. (2022). Construction of VGG16 Convolution Neural Network (VGG16_CNN) Classifier with NestNet-Based Segmentation Paradigm for Brain Metastasis Classification. Sensors, 22(20), 8076. https://doi.org/10.3390/s22208076




