Abstract
Within the literature concerning modern machine learning techniques applied to the medical field, there is a growing interest in the application of these technologies to the nephrological area, especially regarding the study of renal pathologies, because they are very common and widespread in our society, afflicting a high percentage of the population and leading to various complications, up to death in some cases. For these reasons, the authors have considered it appropriate to collect, using one of the major bibliographic databases available, and analyze the studies carried out until February 2022 on the use of machine learning techniques in the nephrological field, grouping them according to the addressed pathologies: renal masses, acute kidney injury, chronic kidney disease, kidney stone, glomerular disease, kidney transplant, and others less widespread. Of a total of 224 studies, 59 were analyzed according to inclusion and exclusion criteria in this review, considering the method used and the type of data available. Based on the study conducted, it is possible to see a growing trend and interest in the use of machine learning applications in nephrology, becoming an additional tool for physicians, which can enable them to make more accurate and faster diagnoses, although there remains a major limitation given the difficulty in creating public databases that can be used by the scientific community to corroborate and eventually make a positive contribution in this area.
1. Introduction
Kidney diseases, such as renal tumors, acute kidney injury (AKI), and chronic kidney disease (CKD), are important issues for nephrology and public health worldwide, as they are associated with high mortality and morbidity rates [1,2]. These diseases, if not identified and treated preventively, can degenerate and lead to severe renal dysfunction, comorbidities, and, in the worst case, death [3,4,5]. Currently, in order to detect and prevent the degeneration of kidney disease, continuous monitoring of specific parameters obtained through diagnostic tests is performed [6]. Given that statistical models are used to determine the actual presence or absence of disease [7], its severity [8], or its degeneration [9], it is natural to think that models based on artificial intelligence (AI) and machine learning (ML) [10] could also be used to achieve this same goal, to obtain statistically better results or more high-performing solutions.
In the last decade, machine learning (ML) techniques have been increasingly employed in a variety of research areas. The consolidation of these methodologies, as well as the benefits of their employment, have occasionally made them the primary mode of operation in many sectors, such as object detection [11], speech recognition [12], emotion recognition [13], and sentiment analysis [14,15]. ML techniques have also captured the interest of the medical community, and multiple positive results have been achieved; some examples of healthcare applications are real-time prioritization and triage [16,17,18,19], personalized medications and care [20,21,22], and patient data analytics [23,24].
In nephrology, ML techniques are used for several purposes:
- -
- segmentation and identification of the anatomy of interest within the diagnostic images (e.g., kidney masses such as tumors, cysts, etc.);
- -
- classification of a kidney mass type, or of the stage in which a specific tumor is found;
- -
- prediction of the evolution of kidney functionality, which can highlight the presence of pathologies.
Among others, ML techniques can be used in the analysis of suspicious renal masses. In such cases, it is nowadays necessary to surgically remove the tumor to identify if it is of a malignant or benign nature, but, due to its position, surgical removal is impossible without risking permanently compromising the patient’s urological function. For this reason, by working directly with diagnostic data and images, machine learning techniques can be crucial alternative solutions for segmenting and identifying masses.
Furthermore, some techniques can be used to help physicians to distinguish between particular cases of some pathologies that are very difficult to distinguish. In these cases, features obtained from diagnostic exams are used to classify the single cases; in this way, the physicians can reach a more precise diagnosis.
In addition to these applications, there are also techniques realized to prescribe specific therapies, or to detect a pathology in advance, in order to prevent it or any of the possible degenerative side effects (e.g., chronic kidney disease, acute kidney injury). In these applications are included also tasks with the aim to predict the compatibility and the outcome of a surgical operation, such as a kidney transplant.
Recently, the number of works related to this area has dramatically increased, rising from a few dozen papers before 2018, to a few hundred presently (based on papers indexed on the Scopus® database from Elsevier). For this reason, it is crucial to carry out an updated survey summarizing the most promising opportunities offered by ML in this area. Accordingly, the present work aims to propose an updated and schematic survey of the most effective existing techniques and to draft possible future research lines based on ML.
First, the most promising articles are selected from the overall literature and classified based on their different applications. Then, in Section 4, there is a description and a comparison of all the used datasets relative to the works selected. Then, in Section 5, the implemented methods and the possible future developments are analyzed. Finally, in Section 6, conclusions are drafted.
The contribution that the authors intend to make with this work is to give a macroscopic view of the existing works concerning nephrology. In particular, the aim is to understand the state of the art of the methods that employ ML techniques to deal with some of the most common kidney diseases, reporting the various resulting metrics for each method. In addition, dimensional analysis of the various types of existing datasets that have been used so far is carried out and a generic comparison is made from the point of view of the type of data.
2. Article Selection
A study of the literature related to publications spanning from 1992 to February 2022 was carried out using Elsevier’s abstract and citation database, Scopus®, by entering keywords, “Artificial Intelligence”, “Machine Learning”, “Kidney”, to identify the most common and effective artificial intelligence (AI) and ML techniques that directly involve the kidney. In particular, the entered query was as follows:
TITLE-ABS (artificial OR intelligence OR machine OR learning OR kidney)
AND
KEY (artificial AND intelligence AND machine AND learning AND kidney)
AND
KEY (artificial AND intelligence AND machine AND learning AND kidney)
The research thus performed allowed the identification of papers that use AI and ML in kidney analysis contexts. Figure 1 shows a significant increase in recent years (after 2017) in the interest and production of papers by the scientific community—in general, there was an overall number of 224 papers dealing with the selected topic.
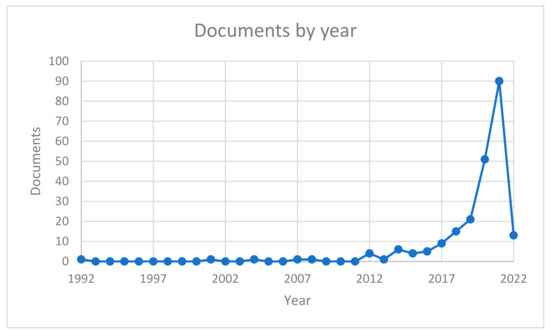
Figure 1.
Trend of documents per year.
To focus on the most relevant works, the literature analysis was carried out according to the following inclusion and exclusion criteria.
Inclusion criteria: (1) articles dealing with ML and AI techniques applied to the kidney were considered; (2) original articles concerning one or more of the following aspects were taken into consideration—segmentation, classification, and prediction of diseases directly related to the kidney; (3) reviews related to these topics were studied to perform a final check of the selected articles.
Exclusion criteria: (1) editorials, commentaries, and abstracts were not included in this study; (2) studies related to animals or carried out only at a laboratory level were excluded; (3) research studies that were not applied in clinical practice were not considered.
According to the aforementioned procedure, fifty-nine studies were found to be eligible to be part of this survey.
3. Machine Learning Approaches for Nephrology
In the following, the studies are grouped based on the nature of the kidney disease. In detail, the analyzed pathologies are “kidney masses”, “acute kidney injury”, “chronic kidney disease”, “kidney stone”, “glomerular disease”, “kidney transplant”, and “other kidney pathologies”. From the analysis of the selected articles, three main research tasks are identified across the application areas:
- (1)
- segmentation and identification, which intends to analyze diagnostic images with the purpose of highlighting or detecting one or more specific elements;
- (2)
- classification, which aims to perform a diagnosis or to determine the degree of severity of disease;
- (3)
- prediction, which aims to prevent or forecast some future event, e.g., predict either the degeneration of a disease or the outcome of a specific therapy.
In the next subsections are reported, for each disease, a brief description of the symptoms to provide the reader with a simple explanation of the clinical scenario, and the various ML techniques used in the state of the art, grouped according to the research tasks described above, highlighting the type of database used. Figure 2 shows a graph schematically outlining the several analyzed pathologies (red color). From each pathology, one or two branches may be amplified according to the type of data available in the available studies (green color), and finally from these as many branches as the ML methods used on that type of data for that specific renal pathology (blue color). The following sections are based on the schematization depicted in the graph.
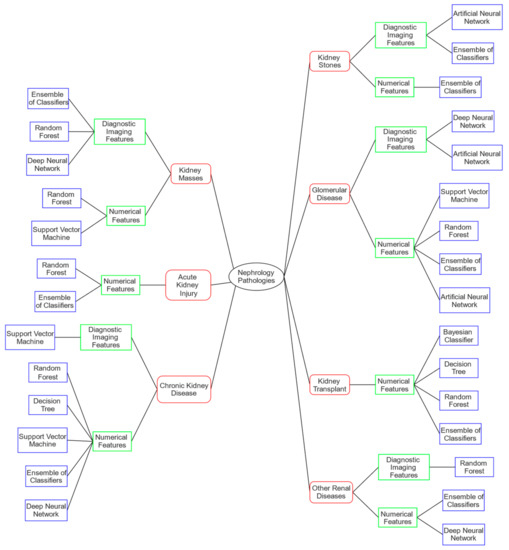
Figure 2.
Scheme of pathologies with ML techniques applied according to the type of dataset available. Kidney disease addressed in red; type of available data in green; ML technique used in blue.
3.1. Kidney Masses
Kidney masses are abnormal growths within the kidney. They are mainly subdivided into two main categories: solid and cystic. Generally, the presence of a kidney mass is determined by relying on imaging techniques such as computed tomography (CT), magnetic resonance imaging (MRI), or ultrasound (US).
In general, cystic kidney masses are, in most cases, benign [25], while solid kidney masses are generally malignant; therefore, the kidney is generally partially or totally removed to perform the histological exam. However, approximately 16% of surgically removed solid kidney masses are benign [26] and surgical removal would not have been necessary. Unfortunately, the distinction of the nature of the solid renal mass, using diagnostic imaging, is very complex, even for specialized physicians, given the significant similarities in the appearance of some types of malignant and benign renal masses, in terms of texture, size, volume, and position. To face this challenge, modern ML techniques have been employed to process image data, proving to help physicians in making a more precise and accurate diagnosis. To classify and distinguish between malignant and benign masses, [27] some use a Bayesian classifier [28], a learning algorithm based on the statistical relationship between radiomics features (relational functional gradient boosting), and [29] an algorithm based on CT texture analysis. Many works focus on the analysis of renal cell carcinoma (RCC), which is the cause of 80% of kidney cancer deaths [25], either to distinguish different types of RCCs or to differentiate them from benign tumors. In [30,31,32,33,34], the main goal is to diagnose the most common malignant tumor, the clear cell RCC, using radiomic features and ML-based classifiers (e.g., random forest, CatBoost). Using radiomic features extracted from multiphoton microscopy images of kidney tissue sections, [35] try to distinguish RCC chromophobes and oncocytomas, while [36] try to classify the stage of a particular type of malignant tumor, the papillary RCC, using microarray datasets [37] and clinical information of the patients. Some more recent research, such as that of [38,39,40], focuses not only on tumor classification, but also on automatic tumor identification through diagnostic images, by using three-dimensional image processing with ML techniques such as 3D U-Net, and 3D V-Net; with these solutions, they are able to automatically segment the tumor inside the CT. Table 1 shows these works, explaining the main objective of each one, the adopted ML techniques, the database exploited, the best result achieved, and finally the year of publication; the reported metrics should be read from the perspective that the higher the reported value, the better the obtained performance.

Table 1.
Renal mass research.
3.2. Acute Kidney Injury
During an episode of acute kidney injury (AKI), the kidneys show difficulty in maintaining the proper fluid balance in the body, due to an accumulation of waste products. Given the speed with which it strikes and the damage that it causes, being able to detect it early can be of great significance. In this type of critical situation, AI is demonstrated to be one of the best solutions to correctly identify a patient with AKI. In studies by [45,46,47], the goal is to predict AKI based on early symptoms to prevent a possible degeneration of the disease, analyzing electronic health records (HER) and other clinical data, such as laboratory tests, vital signs, and patient demographics. AI techniques, thanks also to the speed of response, can be decisive, as in the case of [48], in which the authors try to detect AKI in burn patients using a k-Nearest Neighbor classifier on numerical features obtained from plasma creatinine testing [49].
Some research, such as [50,51], focuses on predicting an episode of AKI in patients undergoing examinations that require contrast agents, specifically coronary angiography. It has been observed that the use of such agents can lead to AKI episodes; in these studies, the authors aim to predict the AKI episode with AI approaches by using clinical variables collected before the examination and by the results of the coronary angiography that they undergo [52].
Recent studies focus on predicting AKI episodes’ insurgence within different periods from its manifestation. The most common prediction time intervals vary from 48 h to a maximum of 90 days, as in [53]; in this work, the authors evaluate their solution based on the analysis of time-series data over these time intervals. It is possible to find another example in [54], in which the authors, through numerical features extracted from multiple blood tests per single patient, attempt to predict AKI within 30 days from its manifestation. Finally, in [55], the authors, using daily collected patients’ clinical data, propose a particular type of deep learning algorithm, based on time series, which is able to predict AKI within 48 h from its occurrence, as well as classify the stage of the AKI disease if it is already present. In Table 2, analogously to Table 1, are reported all the related objectives, methods, used databases, and results.

Table 2.
AKI research.
3.3. Chronic Kidney Disease
Chronic kidney disease (CKD) is a condition characterized by the gradual loss of kidney function over time. CDK damages the kidneys by decreasing their ability to filter waste from the blood. In severe conditions, waste can reach high levels and lead to the development of other complications, which, in the most extreme cases, will require periodic medical treatment, such as dialysis, or even a kidney transplant [62]. CDK is a disease that can be diagnosed by physicians through the study and analysis of a variety of indices (e.g., eGFR [63]); thus, it is suitable for the application of ML methods. An example of using AI for this purpose can be seen in the study by [26], where the stage of pathology is classified using radiomic features obtained from ultrasound images of the kidney.
The general interest and applications to diagnose CKD underwent an abrupt increase with the creation and public release in 2015 of a database containing characteristic features (i.e., age, blood pressure, specific gravity, albumin, sugar, red blood cells, pus cell, pus cell clumps, bacteria, blood glucose random, blood urea, serum creatinine, sodium, potassium, hemoglobin, packed cell volume, white blood cell count, red blood cell count, hypertension, diabetes mellitus, coronary artery disease, appetite, pedal edema, and anemia) related to 400 patients during the early symptoms of the disease [64]. Different methods based on the analysis and classification of patient features are adopted by [26,65,66,67,68,69,70,71,72,73].
In addition to the diagnosis of CKD, there are some related studies in the literature, such as [74], in which the authors try to predict a possible plan for the patients’ diet, given the fact that following a proper and suitable diet plan can help to slow down the progress of CKD [75]. In [76], since maintaining appropriate hemoglobin levels during treatment for CKD is critical, the authors try to predict the hemoglobin level in the blood during anemia treatment in predialysis CKD patients, to intervene more quickly.
This information, the used databases, and the obtained accuracy results are shown in Table 3, analogously to the others.

Table 3.
CKD research.
3.4. Kidney Stone
Nephrolithiasis, or kidney stones, is a condition characterized by the presence of deposits in the kidney, caused by an alteration in the balance between the solubility and precipitation of salts in the urinary tract and kidneys [78]. One crucial point is given by the fact that surgery is required in 20% of patients with this condition [79]. In this context, AI is applied to identify the correct type of treatment to be followed based on parameters such as sediment composition, location, and size [80]. Some research focuses on the detection of kidney stones, such as [81,82], which use radiomic features extracted from manually segmented CT, with the goal of the early detection of stone deposits before they reach a size greater than 2 cm, allowing the use of non-invasive treatments. Other research, such as [83,84,85], focuses on predicting the outcome of shock wave treatment without the use of diagnostic imaging techniques, by analyzing the preoperative parameters of patients (such as age, sex, presence of related diseases, and stone characteristics including stone laterality, location, and maximum length). Similar to the other tables, Table 4 reports this information, the databases used, and the accuracy of the obtained results.

Table 4.
Kidney stone research.
3.5. Glomerular Diseases
Glomerular diseases are diseases that affect the glomeruli, whose function is to filter blood and, at the same time, to retain proteins and blood that the body needs. Many diseases, such as diabetes, affect kidney function by attacking the glomeruli [86]. In this regard [87,88,89], use methods based on the analysis of patients’ clinical data to predict type II diabetes. Some studies focus on specific conditions and causes of glomerular diseases, such as Immunoglobulin A Nephropathy (IgAN), which is the most common biopsy-proven primary glomerulonephritis in the world [90]; it damages not only the kidneys, but also the immune system response [91]. In [92,93,94], the authors implement applications able to predict IgAN using a renal immunofluorescent image obtained by fluorescence microscopes relative to a renal biopsy. Other works, such as [95,96,97], focus on detecting type II diabetes directly from diagnostic images, using radiomic features. Finally, [98] try to predict the weight of children with glomerular disease to avoid possibly dangerous weight loss, using diagnostic numerical features obtained from blood monitoring and analysis.
All the useful information is reported in Table 5, analogously to the previous tables.

Table 5.
Glomerular disease research.
3.6. Kidney Transplant
Even if kidney transplantation is not a pathology but rather a specific surgical treatment, some authors considered creating a dedicated section since there are several studies regarding this topic, and it is one of the most common treatments for patients with severe kidney pathologies.
In detail, kidney transplantation is a surgical procedure that involves taking a healthy kidney from a living or cadaveric donor and implanting it into the recipient patient. For the transplant to be successful, many factors must be considered, including the compatibility of the donor with the human leukocyte antigen (HLA) proteins of the recipient. Although, nowadays, there is a method that reduces the risk of rejection, in the case of mismatched HLA [99,100], approximately 40% of donated kidneys are rejected [101]. The ML techniques applied by [102,103,104,105,106] focus on predicting the probability of success and survival in these types of interventions using numerical features (e.g., age, sex, time in dialysis, donor type, donor age, HLA mismatches, delayed graft function, acute rejection episode, and chronic allograft nephropathy). Table 6 reports all the necessary information, analogously to the others.

Table 6.
Kidney transplant research.
3.7. Other Renal Diseases
In this group are reported other renal diseases that do not fit within the classification provided so far. These studies focus on uncommon objectives, such as [108], which aims to predict the level of hemoglobin in patients with renal dysfunction, using numerical characteristics obtained from clinical data related to dialysis [109]; in [110], an application is developed that intends to define the need to perform or not a renal biopsy by analyzing physicians’ annotations through a natural language processing ML algorithm; [111] try to predict the survival of hemodialysis patients using numerical characteristics (age, sex, diabetes mellitus, chronic glomerulonephritis or nephrosclerosis, body mass index, albumin, sodium, potassium, calcium, phosphorus, creatinine, total cholesterol, etc.). In [112], the authors extract radiomics features from three-dimensional ultrasound images to identify renal and liver tissue in patients with hydronephrosis. Finally, [113] use numerical features extracted from patients’ EHRs with the corresponding acquisition time, to predict the risk of stratification of renal function deterioration.
Table 7 is presented analogously to the previous ones.

Table 7.
Other renal diseases research.
4. Databases Used in Reviewed Research
In this section, two tables contain information about the databases used in the research considered. This information includes the name of the database, when available, or otherwise a distinctive name related to the type of data and the organization in which they were collected; the number of elements that make up the dataset; a brief description of the type of data present; the year in which the database was made public, when available, otherwise the year in which it was used for the first time in a paper; and, finally, whether the database is open access.
Specifically, in Table 8 are reported all databases that have as the data type diagnostic images; this can be CT, MRI, US, or images obtained through analysis in the laboratory with instruments such as a digital microscope. This second type of technique is mainly used for the detection of masses or malformations within the kidneys. It is possible to note that these types of databases have very different volumes; in the case of 3D US images, there are, for example, databases of nine patients; for CT and MRI, there are databases with a minimum of 50 cases up to a few hundred, and finally, with regard to other imaging techniques, there are databases from a minimum of 24 up to a maximum of 1321 cases. This discordance at the numerical level is given mainly by the effectiveness and invasiveness of the different examinations and therefore by the frequency of their use in clinical practice. US is a less effective imaging technique in this field, compared to CT and MRI, and, therefore, the studies concerning the application of this technique are very small and dated. As for the examinations performed on biopsies, the number of samples is much larger because it is an examination that is compulsorily performed in every case to define with absolute certainty the type of mass removed. Among the reported databases, only two are publicly accessible: the CPTAC Clear Cell Renal Cell Carcinoma Discovery Study [41,114], released in 2018 by the U.S. National Cancer Institute, and kits2019 [44], released in 2019 by grand-challenge.org, hosted by MICCAI.

Table 8.
Diagnostic image databases.
In Table 9 are reported all the databases exclusive of numerical type, relating to information obtained from diagnostic tests, such as blood tests, genetic tests of kidney tissues, or data from patient history. For these databases, the volume varies; for more complex tests, such as genetic tests, there is a variation ranging from a few tens up to a few hundred cases; for medical histories, this ranges from a few hundred up to 269,999 cases; for simpler diagnostic tests, from a few tens up to several thousand cases. Of these databases, only three are publicly available; for some, access is limited to a specific country (in the table, these are reported as “only in the USA”). Among the public databases, two contain RNA sequences of renal tumors, which are used to identify the pathological stage of the tumor. Finally, the third public database contains data on blood tests, patient history, and information about CDK-related diseases.

Table 9.
Numerical databases.
5. Discussion
After having reported in the previous sections the existing methods in the literature to address renal pathologies with machine learning methods and analyzed the available databases, we summarize in this section what has been found for each pathology; in particular, the limitations of the studies carried out so far and possible future developments will be indicated.
Regarding renal masses, the goal of the analyzed works is to find a method to non-invasively discriminate benign and malignant masses [29], and artificial intelligence has the potential to become a very important tool for assisted diagnosis. This is motivated by the results of identified research, in which are obtained accuracies ranging from 79% [32] to a peak of approximately 90% [29] (these results are from private single-center databases). Currently, the gold standard for the detection of a renal mass is based on the analysis, by an experienced physician, of CT images before and after dosing with a contrast medium [115]. AI can perform the discrimination function because it can analyze diagnostic images, such as CT, at a very high or equal level of detail as an expert [116]. This is because it can also take into account multidimensional characteristic features, such as texture. However, using CT, the various parameters used for the acquisition and the timing with which it is done assume an important role [29]. In fact, from the articles analyzed, it emerges that, according to the CT acquisition phase taken into consideration, the results obtained change; specifically, the most used phase is the corticomedullary phase [30]. Furthermore, as regards the use of CT for the extraction of characteristic features, the literature considers the three-dimensional use of CT to be better and more representative [117], but in the research identified [27,28,29,30,31,32,33,34,35,36], to reduce the workload of manual segmentation and facilitate the repeatability of this operation, a limited number of slices or only the two-dimensional slice containing the largest portion of the mass considered is used. In addition to how CT is used, it is also important to control the method by which features are extracted; in some research [28,29,31,32,33,34,35,36], radiomic features are used, after manual segmentation by at least one experienced physician, to classify tumors. One of the major limitations introduced, in doing so, is the bias of the operator who performs the segmentation [118]. For this reason, more recent studies [38,39,40] have focused on overcoming manual segmentation by creating deep learning algorithms capable of automatically segmenting kidneys and tumors present in CT; the results obtained from these studies are positive, as they achieve a mean kidney tumor size–mean per CT of the testing set of (Kidney Sørensen-Dice + Tumor Sørensen-Dice)/2 [44], with a maximum of 0.9168. In particular, one solution proposed in the literature to deal with operator-introduced bias is for a team of clinicians to collaborate on the kits2019 database in a way that reduces the risk of bias as much as possible.
Regarding AKI, this pathology is very widespread, with consequences that, if not treated in time, can even lead to death. Currently, there is no specific intervention that can prevent AKI; there are only general measures that can be taken to delay more critical procedures such as surgery [55]. For this reason, most of the recently developed research focuses on predicting the prognosis of this disease [45,46,47,48,50,51,53,54,55], being able to predict AKI with good accuracy even 30 days in advance [54]. The solutions implemented depend not only on the task but also on the actual number of data available for each patient [119]. Maintaining a large number of data for each patient has an economic cost and features used in one center may not be available in other centers [45]. ML techniques can outperform clinical tools used to estimate AKI risk, as we see in [46], with an AUC of 0.85. The performance of solutions exploiting ML for the prediction of AKI is positive: AUC 0.76 [47], in liver transplant patients; 97% accuracy [48] and AUC 0.76 [55], for burn patients; AUC [0.79–0.843] [50,51], for patients undergoing coronary angiography. However, despite the various existing applications, there is a lack of a ML-based prediction systems that can be recognized as state of the art for AKI prediction [47].
Regarding CKD, this is a very common type of disease, which, if detected in time, can be managed through periodic therapies. Thanks to the University of California, Irvine (UCI), which made public the database known as UCI CKD [64] (containing 24 characteristics, derived from patient history and diagnostic tests, plus information regarding the presence or absence of CKD), many studies have been developed to diagnose CKD. Since this database was made public, various studies have used it to test multiple different types of solutions, obtaining increasingly impressive results for accuracy (63–100%) [65,66,67,68,69,71,72,74,76], AUC (0.995) [70], and F1 score (100%) [73]. Being the only public database available for this pathology, the research has been mainly focused on the analysis of numerical features; this is also due to the fact that patients suffering from CKD, or otherwise at risk, cannot undergo all the existing diagnostic imaging techniques. In this case, techniques that require the use of radiation, such as CT, are strongly discouraged, because they can easily worsen the patients’ condition. Therefore, imaging techniques such as US, used in [26] with 82% accuracy in predicting the stage of CKD, and MRI are preferred. The latter has been shown to have the ability to allow assessment of both renal function and structure [120]. Major future developments may shift in this direction and focus on the development of methods that take advantage of MRI to be able to determine CKD.
If radiation imaging techniques cannot be used to determine CKD, the same is not true for detecting and analyzing kidney stones. In particular, for kidney stones, it is possible to use not only CT but also low-dose CT (LDCT), which exposes the patient to approximately five times less radiation than regular CT [121]. The independence of the dosage used to acquire CT is demonstrated in several studies: in [81], ML techniques are applied to process LDCT and CT and identify the composition of a kidney stone, achieving 86% accuracy for both assays used; in [82], LDCT is analyzed to differentiate between kidney stones and phleboliths in patients with acute flank pain, with 85.1% accuracy. The applicability of these methods ensures that low-dose radiation CT acquisitions can be used for the detection of a kidney stone, reducing any risks associated with the radiation exposure of normal CT. In addition to the detection and analysis of kidney stones, researchers are also studying the prediction of success in removing a kidney stone. Successful selection of the most appropriate method can lead to a higher rate of kidney stone clearance, lower risk of associated morbidities, higher probability of survival, faster recovery, and lower overall cost of care [122]. Depending on the procedure chosen [84,85], and for the prediction of stone removal, there is 60% accuracy [84] for predicting success after the first treatment, and 87.9% for predicting success when a shock wave is used for kidney stone clearance [85]. Accuracies ranging from 81% to 98.2% have been obtained for predicting a patient’s condition and possible complications following renal stone removal [83].
Since glomerular disease is a condition that worsens over time, the machine learning techniques implemented are primarily focused on predicting the prognosis of the condition and identifying the consequences caused by the presence of the disease [87,88,89,92,93,94,95,96,97,98]. The most common glomerular disease prevalent in the world is Immunoglobulin A Nephropathy (IgAN) [123]. IgAN is caused by renal dysfunction and can be diagnosed by diagnostic imaging of the kidney, particularly immunofluorescence imaging. Some researchers have focused on diagnosing IgAN from diagnostic images with different resolutions, with an accuracy of at least 80% [92] and an accuracy of 80.27% [95], using only clinical and laboratory analysis data. Around 30–40% of IgAN patients carry the risk of the disease degenerating into ESRD (end-stage renal disease) [93]; for this reason, some research tries to predict this degeneration to allow the efforts of physicians to focus mainly on patients who are more at risk, as, for example, in [93], where it predicts the degeneration of the disease in the next 5 years, with AUC of 0.82, and after 10 years with AUC of 0.89, and as in [94], with 79.8% accuracy. Another particular type of glomerular disease is caused by diabetes. Since diabetes is very common, it is very important to prevent its degeneration into diabetes kidney disease, and in [87,88,89], the authors focus precisely on this aspect by creating algorithms that can predict the prognosis, with an accuracy of 83.5–94%.
Regarding the literature inherent to renal transplantation, it is possible to identify three possible applications of AI [123]: (i) diagnosis, using AI to diagnose the level of transplant risk by detecting parameters associated with renal transplant rejections, and identifying abnormal patterns within them, as in [104], with 68.4% accuracy, and in [106]; (ii) prescription, using AI to prescribe postoperative therapies [124] to prevent complications or rejection, or to prescribe diets that may improve quality of life after renal transplantation [125]; (iii) prediction, using AI to predict mortality, and possible rejection, as in [102], with 73.8% specificity and 88.2% sensitivity; in [103], with 56% accuracy over a 3-year timeframe from possible rejection, and in [105], with 85% accuracy. It is important to note that for this specific task, the main limitation for the application of AI is given by the fact that the type of database is very patient-specific [103,104,105,106], as the values are highly dependent on both the recipient and the donor(s) available, resulting in a limitation that makes it difficult to generalize the solutions devised [126].
Before concluding, we believe that it is also necessary to analyze the ML algorithms used in nephrology, to address a possible reader interested in a specific type of algorithm rather than another, depending on the type of application that they would like to achieve. First of all, it is possible to notice that all the ML algorithms used are based on the use of supervised learning techniques. This is mainly due to the fact that the realized tasks are formulated and viewed in the form of classification problems. In particular, with regard to the research identified in this work, in Table 10, all the methods used have been grouped by algorithm type.

Table 10.
Searches grouped by type of ML algorithm applied.
From the table, it can be observed that the simplest and most common classification algorithms, such as random forest and support vector machine, and ensemble algorithms, such as gradient boosting machine, are the most used in these types of studies. However, more complex ML algorithms, such as artificial neural network, and deep neural networks, such as convolutional neural network, autoencoder, and more sophisticated approaches based not only on feature or image analysis, but also on natural language processing and the temporal evolution of features (temporal-based approaches, e.g., recursive neural network) are not missing. This could be due to the lack of very large public databases that would allow better use of the more complex ML techniques [127].
It is also possible to note that the methods applied by the authors differ mainly with respect to the type of the used data and the techniques of analysis and data processing. In particular, in cases where the database is composed exclusively of numerical features, derived from patients’ medical records, classifiers such as support vector machine, random forest, and artificial neural network are the most frequently applied. Whenever diagnostic images are present, instead, the type of ML technique varies according to the preprocessing applied to the data. In the case of minimal or null preprocessing, techniques such as convolutional neural network are used, in which the model directly analyzes the image and finds the most relevant features in order to classify it. Instead, when algorithms are used for the extraction of radiomic features from specific anatomical regions, algorithms generally applied to numerical features are used; in particular, ensemble algorithms are exploited, which typically, in these cases, guarantee a better result in terms of metrics.
Finally, for the evaluation of algorithms’ performance, the authors feel that it could be misleading to compare methods applied to the same objective based on the values obtained from the evaluated metrics computed with different data. However, it is possible offer some considerations about the various metrics used, in order to understand in which cases some metrics are used instead of others. Since the most commonly used metrics are accuracy and AUC, we consider it appropriate to briefly discuss what the differences are: accuracy is a metric that represents the ratio of the number of correctly predicted samples to the total number of samples present; AUC, on the other hand, represents the area under the receiver operating characteristic (ROC) curve that shows, for different probability thresholds, the relationship between the false positive rate (ratio of the number of false positives to the total number of negative cases) and the true positive rate (ratio of the number of true positives to the total number of positive cases). Looking at the two definitions, it may be deduced that the accuracy is a more intuitive metric and therefore more frequently used, but its simplicity has drawbacks, since it cannot be used in all cases—for example, in the case of unbalanced datasets, where it is preferable to use metrics such as the F1 score or AUC, or in case it is desired to take into account the probability associated with the various classes predicted, in which case only AUC takes this aspect into account. With the above in mind, the use of AUC is strongly recommended as it encapsulates increasingly confident information than accuracy alone.
Despite the limits of this work, given the continuous evolution of research in this area, based on what has been analyzed so far, it is possible to conclude that, given the many existing applications of ML in nephrology, AI has great potential and versatility in this field. An example of a possible application for kidney image analysis can be based on the combination of the multiple methodologies that currently exist, such as the use of deep learning to detect kidneys and tumors [38,39,40], followed by the use of other machine learning techniques to classify the nature and/or severity of tumors, or the presence of any kidney disease and/or other possible masses. However, this does not mean that limitations are not still present. Most of the studies identified end before moving to a clinical trial, remaining only single-center retrospective studies, reducing their external validity [128,129]. Consequently, the main and most urgent gap that should be addressed as soon as possible is that of the public availability of data; this will not only allow studies to be compared with each other but will ensure that there are improvements in nephrology itself [2]. To this end, the guidelines for conducting clinical trials in nephrology, reported at the Kidney Disease—Improving Global Outcomes (KDIGO) conference, could be followed [126].
6. Conclusions
In this work, fifty-nine, from a total of 224, studies concerning the application of ML techniques for the segmentation, prediction, and classification of renal diseases were analyzed. First, the studies were divided, analyzed, and presented based on the addressed pathology and the main goal of the research. Then, the existent datasets were analyzed in terms of data typology, size, and public availability; the main concept derived from this analysis is the importance of a large dataset and public availability to allow research to go as far as possible for a specific objective. Finally, the various pathologies were discussed in terms of what does not exist and what can be done to achieve further developments in this specific sector. In conclusion, from the analysis of the literature, it can also be noted how the introduction of modern ML techniques in the nephrological field allows the achievement goals not obtainable with traditional techniques, such as speeding up and automating CT segmentation processes, the possibility to perform non-invasive and reliable diagnosis, and to create predictive models—for example, to evaluate surgical or transplant outcomes and create predictive models to monitor patient’s parameters in order to act promptly.
Among all the works analyzed, it can be seen that the practical purposes of the use of AI in urology range from the diagnosis of a disease, to the analysis of diagnostic images, to the prediction of prognosis, etc., and generally aim to aid doctors in making more accurate decisions, without attempting, in any way, to replace them [130,131,132,133]. The physician’s attendance remains essential both from a human point of view, in establishing a deep doctor–patient bond of trust that can improve the success of any therapies and treatments [134], and from an ethical and accountable point of view for diagnoses [135].
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sutherland, S.M.; Goldstein, S.L.; Bagshaw, S.M. Leveraging Big Data and Electronic Health Records to Enhance Novel Approaches to Acute Kidney Injury Research and Care. Blood Purif. 2017, 44, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Thongprayoon, C.; Kaewput, W.; Kovvuru, K.; Hansrivijit, P.; Kanduri, S.R.; Bathini, T.; Chewcharat, A.; Leeaphorn, N.; Gonzalez-Suarez, M.L.; Cheungpasitporn, W. Promises of Big Data and Artificial Intelligence in Nephrology and Transplantation. J. Clin. Med. 2020, 9, 1107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zitt, E.; Pscheidt, C.; Concin, H.; Kramar, R.; Peter, R.S.; Beyersmann, J.; Lhotta, K.; Nagel, G. Long-Term Risk for End-Stage Kidney Disease and Death in a Large Population-Based Cohort. Sci. Rep. 2018, 8, 7729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, R.; Kanso, A.; Sedor, J.R. Chronic Kidney Disease and Its Complications. Prim. Care-Clin. Off. Pract. 2008, 35, 329–344. [Google Scholar] [CrossRef] [Green Version]
- Thompson, S.; James, M.; Wiebe, N.; Hemmelgarn, B.; Manns, B.; Klarenbach, S.; Tonelli, M. Cause of Death in Patients with Reduced Kidney Function. J. Am. Soc. Nephrol. 2015, 26, 2504–2511. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.K.; Knicely, D.H.; Grams, M.E. Chronic Kidney Disease Diagnosis and Management: A Review. J. Am. Med. Assoc. 2019, 322, 1294–1304. [Google Scholar] [CrossRef]
- Yang, H.C.; Zuo, Y.; Fogo, A.B. Models of Chronic Kidney Disease. Drug Discov. Today Dis. Model. 2010, 7, 13–19. [Google Scholar] [CrossRef] [Green Version]
- Thompson, R.H.; Kurta, J.M.; Kaag, M.; Tickoo, S.K.; Kundu, S.; Katz, D.; Nogueira, L.; Reuter, V.E.; Russo, P. Tumor Size Is Associated With Malignant Potential in Renal Cell Carcinoma Cases. J. Urol. 2009, 181, 2033–2036. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Remulla, D.; Nguyen, J.H.; Aastha, D.; Liu, Y.; Dasgupta, P.; Hung, A.J. Current Status of Artificial Intelligence Applications in Urology and Their Potential to Influence Clinical Practice. BJU Int. 2019, 124, 567–577. [Google Scholar] [CrossRef]
- Rashidi, H.H.; Tran, N.K.; Betts, E.V.; Howell, L.P.; Green, R. Artificial Intelligence and Machine Learning in Pathology: The Present Landscape of Supervised Methods. Acad. Pathol. 2019, 6, 2374289519873088. [Google Scholar] [CrossRef]
- Zaidi, S.S.A.; Ansari, M.S.; Aslam, A.; Kanwal, N.; Asghar, M.; Lee, B. A Survey of Modern Deep Learning Based Object Detection Models. Digit. Signal Process. Rev. J. 2022, 126, 103514. [Google Scholar] [CrossRef]
- Papastratis, I. Speech Recognition: A Review of the Different Deep Learning Approaches. Available online: https://theaisummer.com/speech-recognition/ (accessed on 7 June 2022).
- Magherini, R.; Mussi, E.; Servi, M.; Volpe, Y. Emotion Recognition in the Times of COVID 19: Coping with Face Masks. Intellingent Syst. Appl. 2022, 200094. [Google Scholar] [CrossRef]
- Wankhade, M.; Rao, A.C.S.; Kulkarni, C. A Survey on Sentiment Analysis Methods, Applications, and Challenges; Springer: Amasterdam, The Netherlands, 2022; ISBN 0123456789. [Google Scholar]
- Ligthart, A.; Catal, C.; Tekinerdogan, B. Systematic Reviews in Sentiment Analysis: A Tertiary Study; Springer: Amasterdam, The Netherlands, 2021; Volume 54, ISBN 1046202109973. [Google Scholar]
- Hardy, M.; Harvey, H. Artificial Intelligence in Diagnostic Imaging: Impact on the Radiography Profession. Br. J. Radiol. 2020, 93, 20190840. [Google Scholar] [CrossRef]
- Jvion. Healthcare & Clinical AI Platform. Available online: https://jvion.com/ (accessed on 7 June 2022).
- Wellframe. Digital Health Management. Available online: www.wellframe.com (accessed on 7 June 2022).
- Enlitic. Healthcare Information Technology with Enlitic. Available online: https://www.enlitic.com/ (accessed on 7 June 2022).
- Johnson, K.B.; Wei, W.Q.; Weeraratne, D.; Frisse, M.E.; Misulis, K.; Rhee, K.; Zhao, J.; Snowdon, J.L. Precision Medicine, AI, and the Future of Personalized Health Care. Clin. Transl. Sci. 2021, 14, 86–93. [Google Scholar] [CrossRef] [PubMed]
- GNS Health Care. Available online: www.gnshealthcare.com (accessed on 7 June 2022).
- Oncora Medical: Real World Data to Fight Cancer. Available online: www.oncora.ai (accessed on 7 June 2022).
- Zakipoint Health. Available online: www.zakipointhealth.com (accessed on 7 June 2022).
- Khan, Z.F.; Alotaibi, S.R. Applications of Artificial Intelligence and Big Data Analytics in M-Health: A Healthcare System Perspective. J. Healthc. Eng. 2020, 2020, 8894694. [Google Scholar] [CrossRef] [PubMed]
- Ballard, B.D.; Guzman, N. Renal Mass. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Chen, C.J.; Pai, T.W.; Fujita, H.; Lee, C.H.; Chen, Y.T.; Chen, K.S.; Chen, Y.C. Stage diagnosis for chronic kidney disease based on ultrasonography. In Proceedings of the International Conference on Fuzzy Systems and Knowledge Discovery (FSKD), Xiamen, China, 19–21 August 2014; pp. 525–530. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, N.; Cho, K.S.; Kang, S.H.; Dae, Y.K.; Yoon, Y.J.; Kim, J.K. Bayesian Classifier for Predicting Malignant Renal Cysts on MDCT: Early Clinical Experience. Am. J. Roentgenol. 2009, 193, 106–111. [Google Scholar] [CrossRef]
- Kunapuli, G.; Varghese, B.A.; Ganapathy, P.; Desai, B.; Cen, S.; Aron, M.; Gill, I.; Duddalwar, V. A Decision-Support Tool for Renal Mass Classification. J. Digit. Imaging 2018, 31, 929–939. [Google Scholar] [CrossRef]
- Erdim, C.; Yardimci, A.H.; Bektas, C.T.; Kocak, B.; Koca, S.B.; Demir, H.; Kilickesmez, O. Prediction of Benign and Malignant Solid Renal Masses: Machine Learning-Based CT Texture Analysis. Acad. Radiol. 2020, 27, 1422–1429. [Google Scholar] [CrossRef]
- Kocak, B.; Ates, E.; Durmaz, E.S.; Ulusan, M.B.; Kilickesmez, O. Influence of Segmentation Margin on Machine Learning–Based High-Dimensional Quantitative CT Texture Analysis: A Reproducibility Study on Renal Clear Cell Carcinomas. Eur. Radiol. 2019, 29, 4765–4775. [Google Scholar] [CrossRef]
- Kocak, B.; Durmaz, E.S.; Kaya, O.K.; Kilickesmez, O. Machine Learning-Based Unenhanced CT Texture Analysis for Predicting BAP1 Mutation Status of Clear Cell Renal Cell Carcinomas. Acta Radiol. 2019, 61, 856–864. [Google Scholar] [CrossRef]
- Cui, E.; Li, Z.; Ma, C.; Li, Q.; Lei, Y.; Lan, Y.; Yu, J.; Zhou, Z.; Li, R.; Long, W.; et al. Predicting the ISUP Grade of Clear Cell Renal Cell Carcinoma with Multiparametric MR and Multiphase CT Radiomics. Eur. Radiol. 2020, 30, 2912–2921. [Google Scholar] [CrossRef]
- Bektas, C.T.; Kocak, B.; Yardimci, A.H.; Turkcanoglu, M.H.; Yucetas, U.; Koca, S.B.; Erdim, C.; Kilickesmez, O. Clear Cell Renal Cell Carcinoma: Machine Learning-Based Quantitative Computed Tomography Texture Analysis for Prediction of Fuhrman Nuclear Grade. Eur. Radiol. 2018, 29, 1153–1163. [Google Scholar] [CrossRef] [PubMed]
- Azuaje, F.; Kim, S.-Y.; Perez Hernandez, D.; Dittmar, G. Connecting Histopathology Imaging and Proteomics in Kidney Cancer through Machine Learning. J. Clin. Med. 2019, 8, 1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, B.; Mukherjee, S.; Jain, M. A New Method Using Multiphoton Imaging and Morphometric Analysis for Differentiating Chromophobe Renal Cell Carcinoma and Oncocytoma Kidney Tumors. Multiphot. Microsc. Biomed. Sci. XVI 2016, 9712, 179–186. [Google Scholar] [CrossRef]
- Singh, N.P.; Bapi, R.S.; Vinod, P.K. Machine Learning Models to Predict the Progression from Early to Late Stages of Papillary Renal Cell Carcinoma. Comput. Biol. Med. 2018, 100, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Betanzos, A.; Bolón-Canedo, V.; Morán-Fernández, L.; Sánchez-Maroño, N. A Review of Microarray Datasets: Where to Find Them and Specific Characteristics; Methods in Molecular Biology Series; Humana Press: Totowa, NJ, USA, 2019; Volume 1986, pp. 65–85. ISBN 9781493994427. [Google Scholar]
- Isensee, F.; Maier-Hein, K.H. An Attempt at Beating the 3D U-Net. arXiv 2019, arXiv:1908.02182. [Google Scholar] [CrossRef] [Green Version]
- Hou, X.; Xie, C.; Li, F.; Nan, Y. Cascaded Semantic Segmentation for Kidney and Tumor. Submiss. 2019 Kidney Tumor Segm. Chall. KiTS19 2019, 2–6. [Google Scholar] [CrossRef] [Green Version]
- Mu, G.; Lin, Z.; Han, M.; Yao, G.; Gao, Y. Segmentation of Kidney Tumor by Multi-Resolution VB-Nets. Submiss. 2019 Kidney Tumor Segm. Chall. KiTS19 2019, 1–5. [Google Scholar] [CrossRef] [Green Version]
- National Cancer Institute. Clinical Proteomic Tumor Analysis Consortium (CPTAC) Radiology Data from the Clinical Proteomic Tumor Analysis Consortium Clear Cell Renal Cell Carcinoma CPTAC-CCRCC Collection. 2018. Available online: https://wiki.cancerimagingarchive.net/display/Public/CPTAC-CCRCC (accessed on 7 June 2022).
- National Cancer Institute GDC Data Portal. Genes. Available online: https://portal.gdc.cancer.gov/genes/ENSG00000143294 (accessed on 7 June 2022).
- Yang, X.J.; Tan, M.-H.; Kim, H.L.; Ditlev, J.A.; Betten, M.W.; Png, C.E.; Kort, E.J.; Futami, K.; Furge, K.A.; Takahashi, M.; et al. A Molecular Classification of Papillary Renal Cell Carcinoma. Cancer Res. 2005, 65, 5628–5637. [Google Scholar] [CrossRef] [Green Version]
- KiTS19—Grand Challenge. Available online: https://kits19.grand-challenge.org/home (accessed on 7 June 2022).
- Sanchez-Pinto, L.N.; Venable, L.R.; Fahrenbach, J.; Churpek, M.M. Comparison of Variable Selection Methods for Clinical Predictive Modeling. Int. J. Med. Inform. 2018, 116, 10–17. [Google Scholar] [CrossRef]
- Penny-Dimri, J.C.; Bergmeir, C.; Reid, C.M.; Williams-Spence, J.; Cochrane, A.D.; Smith, J.A. Machine Learning Algorithms for Predicting and Risk Profiling of Cardiac Surgery-Associated Acute Kidney Injury. Semin. Thorac. Cardiovasc. Surg. 2021, 33, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, D.; Liu, Z.; Chen, C.; Ge, M.; Li, X.; Luo, T.; Wu, Z.; Shi, C.; Wang, B.; et al. An Explainable Supervised Machine Learning Predictor of Acute Kidney Injury after Adult Deceased Donor Liver Transplantation. J. Transl. Med. 2021, 19, 321. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.K.; Sen, S.; Palmieri, T.L.; Lima, K.; Falwell, S.; Wajda, J.; Rashidi, H.H. Artificial Intelligence and Machine Learning for Predicting Acute Kidney Injury in Severely Burned Patients: A Proof of Concept. Burns 2019, 45, 1350–1358. [Google Scholar] [CrossRef] [PubMed]
- Fix, E.; Hodges, J.L. Discriminatory Analysis. Nonparametric Discrimination: Consistency Properties. Int. Stat. Rev. Rev. Int. Stat. 1989, 57, 238. [Google Scholar] [CrossRef]
- Ibrahim, N.E.; McCarthy, C.P.; Shrestha, S.; Gaggin, H.K.; Mukai, R.; Magaret, C.A.; Rhyne, R.F.; Januzzi, J.L. A Clinical, Proteomics, and Artificial Intelligence-Driven Model to Predict Acute Kidney Injury in Patients Undergoing Coronary Angiography. Clin. Cardiol. 2019, 42, 292–298. [Google Scholar] [CrossRef] [Green Version]
- Tseng, P.Y.; Chen, Y.T.; Wang, C.H.; Chiu, K.M.; Peng, Y.S.; Hsu, S.P.; Chen, K.L.; Yang, C.Y.; Lee, O.K.S. Prediction of the Development of Acute Kidney Injury Following Cardiac Surgery by Machine Learning. Crit. Care 2020, 24, 478. [Google Scholar] [CrossRef]
- Azzalini, L.; Candilio, L.; McCullough, P.A.; Colombo, A. Current Risk of Contrast-Induced Acute Kidney Injury After Coronary Angiography and Intervention: A Reappraisal of the Literature. Can. J. Cardiol. 2017, 33, 1225–1228. [Google Scholar] [CrossRef]
- Connell, A.; Montgomery, H.; Martin, P.; Nightingale, C.; Sadeghi-Alavijeh, O.; King, D.; Karthikesalingam, A.; Hughes, C.; Back, T.; Ayoub, K.; et al. Evaluation of a Digitally-Enabled Care Pathway for Acute Kidney Injury Management in Hospital Emergency Admissions. NPJ Digit. Med. 2019, 2, 67. [Google Scholar] [CrossRef] [Green Version]
- Scanlon, L.A.; O’hara, C.; Garbett, A.; Barker-Hewitt, M.; Barriuso, J. Developing an Agnostic Risk Prediction Model for Early Aki Detection in Cancer Patients. Cancers 2021, 13, 4182. [Google Scholar] [CrossRef]
- Song, X.; Yu, A.S.L.; Kellum, J.A.; Waitman, L.R.; Matheny, M.E.; Simpson, S.Q.; Hu, Y.; Liu, M. Cross-Site Transportability of an Explainable Artificial Intelligence Model for Acute Kidney Injury Prediction. Nat. Commun. 2020, 11, 5668. [Google Scholar] [CrossRef]
- Kursa, M.B.; Jankowski, A.; Rudnicki, W.R. Boruta—A System for Feature Selection. Fundam. Inform. 2010, 101, 271–285. [Google Scholar] [CrossRef]
- Churpek, M.M.; Yuen, T.C.; Winslow, C.; Robicsek, A.A.; Meltzer, D.O.; Gibbons, R.D.; Edelson, D.P. Multicenter Development and Validation of a Risk Stratification Tool for Ward Patients. Am. J. Respir. Crit. Care Med. 2014, 190, 649–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Pinto, L.N.; Khemani, R.G. Development of a Prediction Model of Early Acute Kidney Injury in Critically Ill Children Using Electronic Health Record Data. Pediatr. Crit. Care Med. 2016, 17, 508–515. [Google Scholar] [CrossRef] [PubMed]
- National Database|ANZSCTS. Available online: https://anzscts.org/database (accessed on 17 February 2022).
- ClinicalTrials.Gov. The CASABLANCA Study: Catheter Sampled Blood Archive in Cardiovascular Diseases—Full Text View—ClinicalTrials.Gov. Available online: https://clinicaltrials.gov/ct2/show/study/NCT00842868 (accessed on 16 March 2022).
- Waitman, L.R.; Aaronson, L.S.; Nadkarni, P.M.; Connolly, D.W.; Campbell, J.R. The Greater Plains Collaborative: A PCORnet Clinical Research Data Network. J. Am. Med. Inform. Assoc. 2014, 21, 637–641. [Google Scholar] [CrossRef] [Green Version]
- Facts about Chronic Kidney Disease. Available online: https://www.kidney.org/news/newsroom/fsindex (accessed on 9 May 2022).
- Botev, R.; Mallié, J.-P. Reporting the EGFR and Its Implication for CKD Diagnosis. Clin. J. Am. Soc. Nephrol. 2008, 3, 1606–1607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dua, D.; Graff, C. UCI Machine Learning Repository. Available online: http://archive.ics.uci.edu/ml (accessed on 7 June 2022).
- Salekin, A.; Stankovic, J. Detection of chronic kidney disease and selecting important predictive attributes. In Proceedings of the 2016 IEEE International Conference on Healthcare Informatics (ICHI), Chicago, IL, USA, 4–7 October 2016; pp. 262–270. [Google Scholar] [CrossRef]
- Boukenze, B.; Haqiq, A.; Mousannif, H. Predicting Chronic Kidney Failure Disease Using Data Mining Techniques. Lect. Notes Electr. Eng. 2017, 397, 701–712. [Google Scholar] [CrossRef]
- Charleonnan, A.; Fufaung, T.; Niyomwong, T.; Chokchueypattanakit, W.; Suwannawach, S.; Ninchawee, N. Predictive analytics for chronic kidney disease using machine learning techniques. In Proceedings of the Management and Innovation Technology International Conference (MITicon 2016), Bang-Saen, Thailand, 12–14 October 2016; pp. MIT80–MIT83. [Google Scholar] [CrossRef]
- Wibawa, M.S.; Maysanjaya, I.M.D.; Putra, I.M.A.W. Boosted classifier and features selection for enhancing chronic kidney disease diagnose. In Proceedings of the International Conference on Cyber and IT Service Management (CITSM), Denpasar, Indonesia, 8–10 August 2017. [Google Scholar] [CrossRef]
- Subasi, A.; Alickovic, E.; Kevric, J. Diagnosis of Chronic Kidney Disease by Using Random Forest; IFMBE Proceedings Book Series; Springer: Berlin/Heidelberg, Germany, 2017; pp. 589–594. [Google Scholar]
- Aljaaf, A.J.; Al-Jumeily, D.; Haglan, H.M.; Alloghani, M.; Baker, T.; Hussain, A.J.; Mustafina, J. Early prediction of chronic kidney disease using machine learning supported by predictive analytics. In Proceedings of the 2018 IEEE Congress on Evolutionary Computation (CEC 2018), Rio de Janeiro, Brazil, 8–13 July 2018. [Google Scholar] [CrossRef]
- Vanaja, R.; Mukherjee, S. Novel Wrapper-Based Feature Selection for Efficient Clinical Decision Support System. Commun. Comput. Inf. Sci. 2018, 941, 113–129. [Google Scholar] [CrossRef]
- Rady, E.H.A.; Anwar, A.S. Prediction of Kidney Disease Stages Using Data Mining Algorithms. Inform. Med. Unlocked 2019, 15, 100178. [Google Scholar] [CrossRef]
- Senan, E.M.; Al-Adhaileh, M.H.; Alsaade, F.W.; Aldhyani, T.H.H.; Alqarni, A.A.; Alsharif, N.; Uddin, M.I.; Alahmadi, A.H.; Jadhav, M.E.; Alzahrani, M.Y. Diagnosis of Chronic Kidney Disease Using Effective Classification Algorithms and Recursive Feature Elimination Techniques. J. Healthc. Eng. 2021, 2021, 1004767. [Google Scholar] [CrossRef]
- Wickramasinghe, M.P.N.M.; Perera, D.M.; Kahandawaarachchi, K.A.D.C.P. Dietary prediction for patients with chronic kidney disease (CKD) by considering blood potassium level using machine learning algorithms. In Proceedings of the IEEE Lifesciences Conference—IEEE LSC 2018, Sydney, Australia, 13–15 December 2017; pp. 300–303. [Google Scholar] [CrossRef]
- Mitch, W.E.; Remuzzi, G. Diets for Patients with Chronic Kidney Disease, Should We Reconsider? BMC Nephrol. 2016, 17, 80. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, Y.; Nakajima, K.; Nakajima, K. A Rule Extraction Approach to Explore the Upper Limit of Hemoglobin during Anemia Treatment in Patients with Predialysis Chronic Kidney Disease. Inform. Med. Unlocked 2019, 17, 100262. [Google Scholar] [CrossRef]
- Liu, K.; Xu, S.; Feng, N. A Radial Basis Probabilistic Process Neural Network Model and Corresponding Classification Algorithm. Appl. Intell. 2019, 49, 2256–2265. [Google Scholar] [CrossRef]
- Han, H.; Segal, A.M.; Seifter, J.L.; Dwyer, J.T. Nutritional Management of Kidney Stones (Nephrolithiasis). Clin. Nutr. Res. 2015, 4, 137. [Google Scholar] [CrossRef] [Green Version]
- Yarnell, J.; O’Reilly, D. Epidemiology and Disease Prevention, 2nd ed.; Oxford University Press: London, UK, 2013. [Google Scholar]
- Miernik, A.; Hein, S.; Wilhelm, K.; Schoenthaler, M. Harnsteindiagnostik—Was Bringt Uns Die Zukunft? Aktuelle Urol. 2017, 48, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Große Hokamp, N.; Lennartz, S.; Salem, J.; Pinto dos Santos, D.; Heidenreich, A.; Maintz, D.; Haneder, S. Dose Independent Characterization of Renal Stones by Means of Dual Energy Computed Tomography and Machine Learning: An Ex-Vivo Study. Eur. Radiol. 2019, 30, 1397–1404. [Google Scholar] [CrossRef]
- De Perrot, T.; Hofmeister, J.; Burgermeister, S.; Martin, S.P.; Feutry, G.; Klein, J.; Montet, X. Differentiating Kidney Stones from Phleboliths in Unenhanced Low-Dose Computed Tomography Using Radiomics and Machine Learning. Eur. Radiol. 2019, 29, 4776–4782. [Google Scholar] [CrossRef]
- Aminsharifi, A.; Irani, D.; Pooyesh, S.; Parvin, H.; Dehghani, S.; Yousofi, K.; Fazel, E.; Zibaie, F. Artificial Neural Network System to Predict the Postoperative Outcome of Percutaneous Nephrolithotomy. J. Endourol. 2017, 31, 461–467. [Google Scholar] [CrossRef]
- Shabaniyan, T.; Parsaei, H.; Aminsharifi, A.; Movahedi, M.M.; Jahromi, A.T.; Pouyesh, S.; Parvin, H. An Artificial Intelligence-Based Clinical Decision Support System for Large Kidney Stone Treatment. Australas. Phys. Eng. Sci. Med. 2019, 42, 771–779. [Google Scholar] [CrossRef]
- Yang, S.W.; Hyon, Y.K.; Na, H.S.; Jin, L.; Lee, J.G.; Park, J.M.; Lee, J.Y.; Shin, J.H.; Lim, J.S.; Na, Y.G.; et al. Machine Learning Prediction of Stone-Free Success in Patients with Urinary Stone after Treatment of Shock Wave Lithotripsy. BMC Urol. 2020, 20, 88. [Google Scholar] [CrossRef]
- National Kidney Foundation. Understanding Glomerular Diseases. Available online: https://www.kidney.org/atoz/content/understanding-glomerular-diseases (accessed on 20 April 2022).
- Leung, R.K.K.; Wang, Y.; Ma, R.C.W.; Luk, A.O.Y.; Lam, V.; Ng, M.; So, W.Y.; Tsui, S.K.W.; Chan, J.C.N. Using a Multi-Staged Strategy Based on Machine Learning and Mathematical Modeling to Predict Genotype-Phenotype Risk Patterns in Diabetic Kidney Disease: A Prospective Case-Control Cohort Analysis. BMC Nephrol. 2013, 14, 162. [Google Scholar] [CrossRef]
- Dagliati, A.; Marini, S.; Sacchi, L.; Cogni, G.; Teliti, M.; Tibollo, V.; De Cata, P.; Chiovato, L.; Bellazzi, R. Machine Learning Methods to Predict Diabetes Complications. J. Diabetes Sci. Technol. 2018, 12, 295–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennett, C.C. Artificial Intelligence for Diabetes Case Management: The Intersection of Physical and Mental Health. Inform. Med. Unlocked 2019, 16, 100191. [Google Scholar] [CrossRef]
- Schena, F.P.; Nistor, I. Epidemiology of IgA Nephropathy: A Global Perspective. Semin. Nephrol. 2018, 38, 435–442. [Google Scholar] [CrossRef] [PubMed]
- National Kidney Foundation. IgA Nephropathy. Available online: https://www.kidney.org/atoz/content/iganeph (accessed on 7 June 2022).
- Takahashi, K.; Kitamura, S.; Fukushima, K.; Sang, Y.; Tsuji, K.; Wada, J. The Resolution of Immunofluorescent Pathological Images Affects Diagnosis for Not Only Artificial Intelligence but Also Human. J. Nephropathol. 2021, 10, e26. [Google Scholar] [CrossRef]
- Schena, F.P.; Anelli, V.W.; Trotta, J.; Di Noia, T.; Manno, C.; Tripepi, G.; D’Arrigo, G.; Chesnaye, N.C.; Russo, M.L.; Stangou, M.; et al. Development and Testing of an Artificial Intelligence Tool for Predicting End-Stage Kidney Disease in Patients with Immunoglobulin A Nephropathy. Kidney Int. 2021, 99, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- Konieczny, A.; Stojanowski, J.; Krajewska, M.; Kusztal, M. Machine Learning in Prediction of Iga Nephropathy Outcome: A Comparative Approach. J. Pers. Med. 2021, 11, 312. [Google Scholar] [CrossRef] [PubMed]
- Agar, J.W.M.; Webb, G.I. Application of Machine Learning to a Renal Biopsy Database. Nephrol. Dial. Transplant. 1992, 7, 472–478. [Google Scholar] [CrossRef]
- Iakovidis, D.K.; Goudas, T.; Smailis, C.; Maglogiannis, I. Ratsnake: A Versatile Image Annotation Tool with Application to Computer-Aided Diagnosis. Sci. World J. 2014, 2014, 312. [Google Scholar] [CrossRef]
- Aldeman, N.L.S.; de Sá Urtiga Aita, K.M.; Machado, V.P.; da Mata Sousa, L.C.D.; Coelho, A.G.B.; da Silva, A.S.; da Silva Mendes, A.P.; de Oliveira Neres, F.J.; do Monte, S.J.H. Smartpathk: A Platform for Teaching Glomerulopathies Using Machine Learning. BMC Med. Educ. 2021, 21, 248. [Google Scholar] [CrossRef]
- Niel, O.; Bastard, P.; Boussard, C.; Hogan, J.; Kwon, T.; Deschênes, G. Artificial Intelligence Outperforms Experienced Nephrologists to Assess Dry Weight in Pediatric Patients on Chronic Hemodialysis. Pediatr. Nephrol. 2018, 33, 1799–1803. [Google Scholar] [CrossRef]
- Higgins, R.; Hathaway, M.; Lowe, D.; Lam, F.; Kashi, H.; Tan, L.C.; Imray, C.; Fletcher, S.; Zehnder, D.; Chen, K.; et al. Blood Levels of Donor-Specific Human Leukocyte Antigen Antibodies After Renal Transplantation: Resolution of Rejection in the Presence of Circulating Donor-Specific Antibody. Transplantation 2007, 84, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Gloor, J.M.; Stegall, M.D. ABO Incompatible Kidney Transplantation. Curr. Opin. Nephrol. Hypertens. 2007, 16, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Khovanova, N.; Daga, S.; Shaikhina, T.; Krishnan, N.; Jones, J.; Zehnder, D.; Mitchell, D.; Higgins, R.; Briggs, D.; Lowe, D. Subclass Analysis of Donor HLA-Specific IgG in Antibody-Incompatible Renal Transplantation Reveals a Significant Association of IgG4 with Rejection and Graft Failure. Transpl. Int. 2015, 28, 1405–1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greco, R.; Papalia, T.; Lofaro, D.; Maestripieri, S.; Mancuso, D.; Bonofiglio, R. Decisional Trees in Renal Transplant Follow-Up. Transplant. Proc. 2010, 42, 1134–1136. [Google Scholar] [CrossRef]
- Brown, T.S.; Elster, E.A.; Stevens, K.; Graybill, J.C.; Gillern, S.; Phinney, S.; Salifu, M.O.; Jindal, R.M. Bayesian Modeling of Pretransplant Variables Accurately Predicts Kidney Graft Survival. Am. J. Nephrol. 2012, 36, 561–569. [Google Scholar] [CrossRef]
- Topuz, K.; Zengul, F.D.; Dag, A.; Almehmi, A.; Yildirim, M.B. Predicting Graft Survival among Kidney Transplant Recipients: A Bayesian Decision Support Model. Decis. Support Syst. 2017, 106, 97–109. [Google Scholar] [CrossRef]
- Shaikhina, T.; Lowe, D.; Daga, S.; Briggs, D.; Higgins, R.; Khovanova, N. Decision Tree and Random Forest Models for Outcome Prediction in Antibody Incompatible Kidney Transplantation. Biomed. Signal Process. Control 2017, 52, 456–462. [Google Scholar] [CrossRef]
- Elihimas Júnior, U.F.; Couto, J.P.; Pereira, W.; Barros De Oliveira Sá, M.P.; Tenório De França, E.E.; Aguiar, F.C.; Cabral, D.B.C.; Alencar, S.B.V.; Feitosa, S.J.D.C.; Claizoni Dos Santos, T.O.; et al. Logistic Regression Model in a Machine Learning Application to Predict Elderly Kidney Transplant Recipients with Worse Renal Function One Year after Kidney Transplant: Elderly KTbot. J. Aging Res. 2020, 2020, 118–128. [Google Scholar] [CrossRef]
- Shih, D.T.; Kim, S.B.; Chen, V.C.P.; Rosenberger, J.M.; Pilla, V.L. Efficient Computer Experiment-Based Optimization through Variable Selection. Ann. Oper. Res. 2014, 216, 287–305. [Google Scholar] [CrossRef]
- Martínez-Martínez, J.M.; Escandell-Montero, P.; Barbieri, C.; Soria-Olivas, E.; Mari, F.; Martínez-Sober, M.; Amato, C.; Serrano López, A.J.; Bassi, M.; Magdalena-Benedito, R.; et al. Prediction of the Hemoglobin Level in Hemodialysis Patients Using Machine Learning Techniques. Comput. Methods Programs Biomed. 2014, 117, 208–217. [Google Scholar] [CrossRef]
- Stopper, A.; Amato, C.; Gioberge, S.; Giordana, G.; Marcelli, D.; Gatti, E. Managing Complexity at Dialysis Service Centers across Europe. Blood Purif. 2007, 25, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Jia, Z.; Zeng, X.; Feng, C.; Lu, X.; Duan, H.; Li, H. Renal Biopsy Recommendation Based on Text Understanding. Stud. Health Technol. Inform. 2019, 264, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Kanda, E.; Epureanu, B.I.; Adachi, T.; Tsuruta, Y.; Kikuchi, K.; Kashihara, N.; Abe, M.; Masakane, I.; Nitta, K. Application of Explainable Ensemble Artificial Intelligence Model to Categorization of Hemodialysis-Patient and Treatment Using Nationwide-Real-World Data in Japan. PLoS ONE 2020, 15, e0233491. [Google Scholar] [CrossRef] [PubMed]
- Aalamifar, F.; Rivaz, H.; Cerrolaza, J.J.; Jago, J.; Safdar, N.; Boctor, E.M.; Linguraru, M.G. Classification of Kidney and Liver Tissue Using Ultrasound Backscatter Data. Med. Imaging 2015 Ultrason. Imaging Tomogr. 2015, 9419, 192–199. [Google Scholar] [CrossRef]
- Singh, A.; Nadkarni, G.; Gottesman, O.; Ellis, S.B.; Bottinger, E.P.; Guttag, J.V. Incorporating Temporal EHR Data in Predictive Models for Risk Stratification of Renal Function Deterioration. J. Biomed. Inform. 2015, 53, 220–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [Green Version]
- Higgins, J.C.; Fitzgerald, J.M. Evaluation of Incidental Renal and Adrenal Masses. Am. Fam. Physician 2001, 63, 288–294, 299. [Google Scholar]
- Martin-Isla, C.; Campello, V.M.; Izquierdo, C.; Raisi-Estabragh, Z.; Baeßler, B.; Petersen, S.E.; Lekadir, K. Image-Based Cardiac Diagnosis With Machine Learning: A Review. Front. Cardiovasc. Med. 2020, 7, 1. [Google Scholar] [CrossRef] [Green Version]
- Ng, F.; Kozarski, R.; Ganeshan, B.; Goh, V. Assessment of Tumor Heterogeneity by CT Texture Analysis: Can the Largest Cross-Sectional Area Be Used as an Alternative to Whole Tumor Analysis? Eur. J. Radiol. 2013, 82, 342–348. [Google Scholar] [CrossRef]
- Starmans, M.P.A.; van der Voort, S.R.; Castillo Tovar, J.M.; Veenland, J.F.; Klein, S.; Niessen, W.J. Radiomics. In Handbook of Medical Image Computing and Computer Assisted Intervention; Zhou, S.K., Rueckert, D., Fichtinger, G., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 429–456. ISBN 978-0-12-816176-0. [Google Scholar]
- Raschka, S. Model Evaluation, Model Selection, and Algorithm Selection in Machine Learning. arXiv 2018, arXiv:1811.12808. [Google Scholar] [CrossRef]
- Alnazer, I.; Bourdon, P.; Urruty, T.; Falou, O.; Khalil, M.; Shahin, A.; Fernandez-Maloigne, C. Recent Advances in Medical Image Processing for the Evaluation of Chronic Kidney Disease. Med. Image Anal. 2021, 69, 101960. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute. Computed Tomography (CT) Scans and Cancer Fact Sheet. Available online: https://www.cancer.gov/about-cancer/diagnosis-staging/ct-scans-fact-sheet (accessed on 7 April 2022).
- Khan, S.R.; Pearle, M.S.; Robertson, W.G.; Gambaro, G.; Canales, B.K.; Doizi, S.; Traxer, O.; Tiselius, H.-G. Kidney Stones. Nat. Rev. Dis. Prim. 2016, 2, 16008. [Google Scholar] [CrossRef] [PubMed]
- Burlacu, A.; Iftene, A.; Jugrin, D.; Popa, I.V.; Lupu, P.M.; Vlad, C.; Covic, A. Using Artificial Intelligence Resources in Dialysis and Kidney Transplant Patients: A Literature Review. Biomed Res. Int. 2020, 2020, 9867872. [Google Scholar] [CrossRef] [PubMed]
- Thishya, K.; Vattam, K.K.; Naushad, S.M.; Raju, S.B.; Kutala, V.K. Artificial Neural Network Model for Predicting the Bioavailability of Tacrolimus in Patients with Renal Transplantation. PLoS ONE 2018, 13, e0191921. [Google Scholar] [CrossRef] [Green Version]
- Stachowska, E.; Gutowska, I.; Strzelczak, A.; Wesołowska, T.; Safranow, K.; Chlubek, D. The Use of Neural Networks in Evaluation of the Direction and Dynamics of Changes in Lipid Parameters in Kidney Transplant Patients on the Mediterranean Diet. J. Ren. Nutr. 2006, 16, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Baigent, C.; Herrington, W.G.; Coresh, J.; Landray, M.J.; Levin, A.; Perkovic, V.; Pfeffer, M.A.; Rossing, P.; Walsh, M.; Wanner, C.; et al. Challenges in Conducting Clinical Trials in Nephrology: Conclusions from a Kidney Disease—Improving Global Outcomes (KDIGO) Controversies Conference. Kidney Int. 2017, 92, 297–305. [Google Scholar] [CrossRef] [Green Version]
- Ghassemi, M.; Naumann, T.; Schulam, P.; Beam, A.L.; Chen, I.Y.; Ranganath, R. A Review of Challenges and Opportunities in Machine Learning for Health. AMIA Jt. Summits Transl. Sci. Proc. 2020, 2020, 191–200. [Google Scholar]
- Yuan, Q.; Zhang, H.; Deng, T.; Tang, S.; Yuan, X.; Tang, W.; Xie, Y.; Ge, H.; Wang, X.; Zhou, Q.; et al. Role of Artificial Intelligence in Kidney Disease. Int. J. Med. Sci. 2020, 17, 970–984. [Google Scholar] [CrossRef] [Green Version]
- Gameiro, J.; Branco, T.; Lopes, J.A. Artificial Intelligence in Acute Kidney Injury Risk Prediction. J. Clin. Med. 2020, 9, 678. [Google Scholar] [CrossRef] [Green Version]
- Shortliffe, E.H.; Sepúlveda, M.J. Clinical Decision Support in the Era of Artificial Intelligence. JAMA 2018, 320, 2199. [Google Scholar] [CrossRef]
- Obermeyer, Z.; Lee, T.H. Lost in Thought—The Limits of the Human Mind and the Future of Medicine. N. Engl. J. Med. 2017, 377, 1209–1211. [Google Scholar] [CrossRef] [Green Version]
- Gampala, S.; Vankeshwaram, V.; Gadula, S.S.P. Is Artificial Intelligence the New Friend for Radiologists? A Review Article. Cureus 2020, 12, e11137. [Google Scholar] [CrossRef] [PubMed]
- Mistry, N.S.; Koyner, J.L. Artificial Intelligence in Acute Kidney Injury: From Static to Dynamic Models. Adv. Chronic Kidney Dis. 2021, 28, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Chipidza, F.E.; Wallwork, R.S.; Stern, T.A. Impact of the Doctor-Patient Relationship. Prim. Care Companion CNS Disord. 2015, 17, 27354. [Google Scholar] [CrossRef] [Green Version]
- Neri, E.; Coppola, F.; Miele, V.; Bibbolino, C.; Grassi, R. Artificial Intelligence: Who Is Responsible for the Diagnosis? Radiol. Med. 2020, 125, 517–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).