Abstract
The notion of the “cryptic species” has recently become an important agenda in biodiversity research. In this study, we show, by applying a periodic-like morphological and molecular framework to the nudibranch genus Cadlina from the world’s least explored locations in the Kuril Islands in the northwestern Pacific, including a description of six new species, that the term “cryptic species” should be removed from biodiversity research terminology. We explicitly show that different species of this complex have various degrees of molecular phylogenetic distances and morphological distinctness, revealing a truly multilevel system of fine-scale differences and similarities. Therefore, to designate any of these species as “cryptic” or “non-cryptic” would be completely arbitrary, non-operational, and generally meaningless. By this, we finally strongly propose to remove the notion of “crypticity” and the term “cryptic species” (in the sense of an “indistinguishable species”) from the arsenal of modern biology, including phylogeny and taxonomy. The importance of fine-scale species differentiation in the multilevel framework is shown for addressing conservation and global warming challenges. Six new species are named after scientists who have not always received the honours they deserve, including two women who did not receive their respective Nobel Prizes.
1. Introduction
Recently, a number of significant contradictions in the use of the term “cryptic species” have been clearly shown [1,2,3,4,5,6,7,8,9]. Despite this, the term “cryptic species” is still widely used in biodiversity studies [10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28]. It is important to note that the apparent “crypticity” has been indicated for the widest range of groups of organisms, including plants, fungi, animals, and unicellular and multicellular groups, as exemplified by the references above.
In this study, we ultimately show, applying a periodic-like morphological and molecular framework (which has recently been coined as phyloperiodic [29]) to the nudibranch genus Cadlina from one of the least explored and hard-to-reach places in the world (the Kuril Islands in the north-western Pacific [30,31]), that the term “cryptic species” should be excluded from the terminology of biodiversity studies. The present Cadlina case, where even phylogenetically distant species exhibit a seemingly “almost indistinguishable” pale body with yellow spots and lines [32], is particularly relevant to show that simply the combination of molecular and morphological data, which has previously been applied under various terms, including “integrative taxonomy” [33,34], is not enough and more consistent approaches need to be developed.
Although the periodic approach has not been in common usage in biological taxonomy previously, compelling interdisciplinary evidence has accumulated over the past two decades showing that periodic processes influence the origin of the biological diversity at the genomic [35,36,37], cellular [38], and morphological levels, including chromatic patterns [39,40]. Thus, the basis of any taxonomic diversity is not only “tree-like” phylogenetic patterns as such, but also periodic processes at different organismal levels, united by the process of ontogenesis [29]. Recently, it has also been shown that the common phylogenetic “lineage approach” is insufficient for understanding biodiversity [41].
The presence of an apparently huge number of “cryptic species” [42,43,44], which presumably cannot be assessed and described other than using exclusively genetic data [45,46,47], greatly hamper the description of biological diversity. It also poses a particular challenge for practitioners, including conservationists, because most of them do not have the ability to conduct molecular analysis in the field. The phyloperiodic approach is particularly relevant for the study of “hidden diversity” and is applied here to facilitate the identification of the particular species within a species complex. It involves the building of phyloperiodic tables according to the central biological periodic process—ontogenesis, which underlies and unites all biological levels from molecular to macromorphological—and therefore provides not just artificially formed, but biologically well founded sets of regularities, “established rules” for the further construction of tables.
Thus, the phyloperiodic approach captures and compares organism-based shared regularities (phyloperiods) at any ontogenetic level (both large-scale, such as earlier ontogenetic patterns, or small-scale, including chromatic variants) with phylogeny-based molecular “lineages” [29]. The molecular phylogenetic data are placed in vertical columns (groups, taxa) and diagnostic morphological data (in a broad ontogenetic sense) are arranged in horizontal rows (periods) in an orderly manner (see below for more details). Due to the multilevel basis of any organisms (=ontogeneses), it is possible to construct many different phyloperiodic tables based on different traits, including commonly used diagnostic morphological characters in a given group.
This study does not simply focus on the “narrow taxonomic group”, but instead uses relevant particular material in order to show that the basic terminology and methodology in the widely important field of biological diversity still needs significant improvement.
2. Materials and Methods
2.1. Sampling
Specimens for this study were collected from the Kuril Islands (NW Pacific) in the course of the two recent expeditions (2021, 2022). Specimens were photographed underwater and measured in the laboratory. Specimens were placed in magnesium sulphate (7%) before preservation in 96% ethanol. The specimens were deposited in the Zoological Museum of Lomonosov Moscow State University (ZMMU).
2.2. Nomenclatural Acts
The electronic version of this article in Portable Document Format (PDF) represents a published work according to the International Commission on Zoological Nomenclature (ICZN), and hence the new names contained in the electronic version are effectively published under that Code from the electronic edition alone. This published work and the nomenclatural acts it contains have been registered in ZooBank, the online registration system for the ICZN. The ZooBank LSIDs (Life Science Identifiers) can be resolved and the associated information viewed through any standard web browser by appending the LSID to the prefix http://zoobank.org/. The LSID for this publication is: [urn:lsid:zoobank.org:pub: B33A0BCE-F0A2-4370-A25D-78821AB80BB2].
2.3. Morphological Analysis
The external and internal morphology was studied using a stereomicroscope and digital cameras (Nikon D-810 and Nikon D-80). An amount of 10% sodium hypochlorite solution was used to extract the radula and the jaws. The dried radulae and jaws were coated with gold or palladium and examined using scanning electron microscopes, SEM (CamScan Series II, JSM 6380, and QuattroS). Reproductive systems were examined using a stereomicroscope.
2.4. Molecular Analysis
Thirty-seven specimens of the genus Cadlina were newly sequenced for the mitochondrial genes cytochrome c oxidase subunit I (COI), 16S rRNA, and the nuclear 28S rDNA (C1-C2 domain). The DNA extraction procedure, primers, PCR amplification options, and sequencing have been previously described in detail [4,32]. The PCR amplification options are presented in the Supplementary Materials, Table S1. DNA sequences of both strands were obtained using the ABI PRISM1Big-Dye™ Terminator v. 3.1. on an automated DNA sequencer (Applied Biosystems Prism 3700). All new sequences were deposited in GenBank. Moreover, molecular data of Cadlina and Aldisa species were obtained from GenBank. GenBank accession numbers and references for all sequences used in this study are presented in the Supplementary Materials, Table S2 (newly sequenced data highlighted in bold). Separate analyses were conducted for COI (658 bp), 16S (457 bp), 28S (331 bp), and the concatenated dataset (1446 bp). Sequences were aligned with the MAFFT algorithm [48].
Evolutionary models for each dataset were selected using MEGA11 [49]. The HKY+I+G model was chosen for the COI dataset and for the combined full dataset, the HKY+G model for the 16S dataset, and the GTR model for the 28S dataset. Two different methods of phylogenetic analyses, Bayesian Inference (BI) and Maximum Likelihood (ML), were used. Bayesian estimation of posterior probability was performed in MrBayes 3.2 [50]. Four Markov chains were sampled at intervals of 500 generations; analysis was started with random starting trees and 10,000,000 generations. Maximum Likelihood-based phylogeny inference was performed in RAxML 7.2.8 [51] with bootstrap in 1000 pseudo-replications. Final phylogenetic tree images were rendered in FigTree 1.4.2 (http://tree.bio.ed.ac.uk, accessed on 3 February 2023). The Assemble Species by Automatic Partitioning (ASAP) [52] analysis (available at https://bioinfo.mnhn.fr/abi/public/asap/asapweb.html, accessed on 3 February 2023) was performed for the COI dataset separately using the models proposed by Jukes–Cantor and Kimura.
The program MEGA11 [49] was used to calculate the uncorrected p-distances. The haplotype networks for the COI molecular data were reconstructed using Population Analysis with Reticulate Trees (PopART, http://popart.otago.ac.nz, accessed on 3 February 2023) with the TCS network method.
3. Results
3.1. Molecular Phylogeny
In this molecular study, a total of 90 specimens from the genus Cadlina were examined combining 102 novel sequences with 133 sequences from GenBank. Taxon sampling including the outgroup species from the genus Aldisa was used according to previous results [32]. Bayesian Inference (BI) and Maximum Likelihood (ML) analyses based on the combined dataset for the mitochondrial COI and 16S and the nuclear 28S genes yielded the same results (Figure 1). All Cadlina taxa are clustered in a highly supported monophyletic clade (PP = 1, BS = 100).
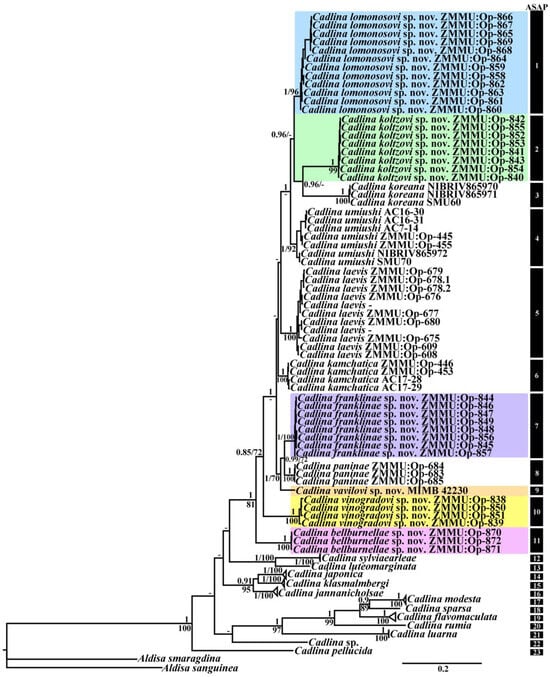
Figure 1.
Phylogenetic tree of the genus Cadlina based on concatenated molecular data (COI + 16S + 28S) represented by Bayesian Inference. The numbers above branches represent posterior probabilities from Bayesian Inference. The numbers below branches indicate bootstrap values for Maximum Likelihood. Summary of species delimitation results are noted by numbered clusters from the ASAP analyses for the cytochrome c oxidase subunit I (COI) dataset.
The ASAP analysis (asap-score 1.0) of the COI dataset run with two different models revealed 23 lineages: C. bellburnellae sp. nov., C. flavomaculata, C. franklinae sp. nov., C. jannanicholsae, C. japonica, C. kamchatica, C. klasmalmbergi, C. koltzovi sp. nov., C. koreana, C. laevis, C. lomonosovi sp. nov., C. luarna, C. luteomarginata, C. modesta, C. paninae, C. pellucida, C. rumia, C. sparsa, C. sylviaearleae, C. umiushi, C. vavilovi sp. nov., C. vinogradovi sp. nov., and Cadlina sp. from South Africa.
All six new species from the Kuril Islands formed separate distinct clades [C. bellburnellae sp. nov. (PP = 1, BS = 100), C. franklinae sp. nov. (PP = 1, BS = 100), C. koltzovi sp. nov. (PP = 1, BS = 99), C. lomonosovi sp. nov. (PP = 1, BS = 96), C. vavilovi sp. nov., and C. vinogradovi sp. nov. (PP = 1, BS = 100)] and clustered in a high supported monophyletic clade (PP = 1, BS = 81) together with four species from the North Pacific Ocean [C. kamchatica (PP = 1, BS = 100), C. koreana (PP = 1, BS = 100), C. paninae (PP = 1, BS = 100), and C. umiushi (PP = 1, BS = 92)], and one species from the North Atlantic [C. laevis (PP = 1, BS = 100)]. Unresolved evolutionary ties were revealed for these species. Whereas C. koltzovi sp. nov. and C. koreana showed close evolutionary relationships, as well as another group C. franklinae sp. nov. and C. paninae.
The results based on COI molecular data obtained by PopART showed a network of haplotypes that clearly clustered into 11 groups (Figure 2A). The haplotype network results show close evolutionary relationships between C. franklinae sp. nov., C. paninae, C. bellburnellae sp. nov., and C. vavilovi sp. nov.
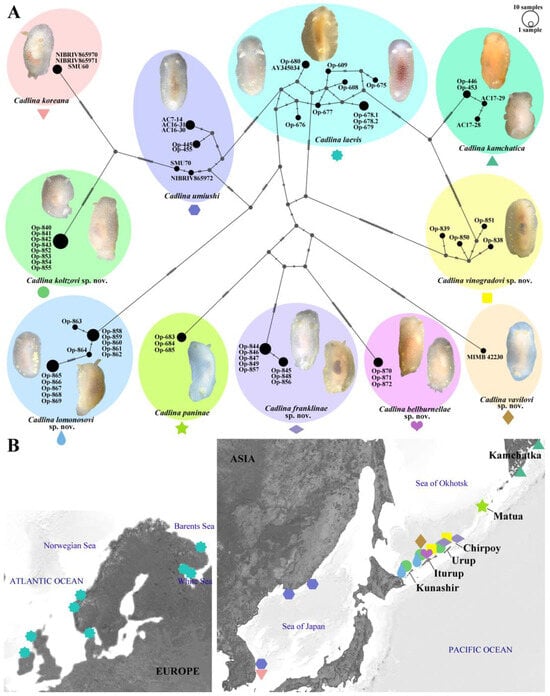
Figure 2.
(A) The haplotype network is based on cytochrome c oxidase subunit I (COI) molecular data and shows genetic mutations occurring within eleven Cadlina species. (B) Coloured symbols placed on the map denote the Cadlina species distribution used in the above analysis.
3.2. Practical Guidelines Proposing How to Perform a Taxonomic Study in the Molecular Era
The following guidelines have been proposed recently [7,53], and we follow them in this study with a little modification: (1) make a selection of a taxonomic group and specimens; (2) conduct a morphological study in a given group, e.g., SEM of previously used diagnostic characters, or application of other methods if available; (3) ensure that ontogenetic information is taken into account during taxonomic assessment (see more details in [29]); (4) perform a relevant bibliographic study of original sources, rather than simply referencing a taxonomic data base; (5) conduct a molecular study of taxonomically checked specimens with appropriate genetic markers; (6) compare the results of morphological (step 2) and taxonomic studies (steps 3, 4) with the molecular (step 5) results; (7) in case inconsistencies are found between commonly used diagnostic characters in a given group and results of molecular phylogeny, respective features should be presented in the phyloperiodic framework, which will enable their detailed comparison and further search for fine-scale differences for each of the closely related taxa; (8) in case difficult to distinguish variants (patterns) are present among the same parallel rows of the phyloperiodic tables, a relevant presentation of diagnostic features should be conducted in order to reveal fine-scale differences between related taxa; (9) a complete study at a given time and using available research possibilities for a given group of taxa should result in fine-scale taxonomic diagnoses for all closely related species in a given taxa/complex (including new taxa); and (10) test the established framework by further investigation of a given group with new materials and data.
3.3. Phyloperiodic Framework for Recognition of Diagnosable Characters in Species Complexes
In the present study, we followed steps 1–6 above using both morphological and molecular data and found that the taxonomic complex represents several apparently difficult-to-distinguish sets of diagnostic characters when the conventional taxonomic comparison format did not work properly. Thus, the phyloperiodic arrangements of several taxonomically reliable characters were used as the next step. The method of the periodic-like presentation includes both tree-like patterns (phylogeny) and ontogenetic (morphological in a broad developmental sense [54]) periodic patterns and was therefore coined phyloperiodic [29]. Accordingly, the general construction of phyloperiodic tables depends, on the one hand, on molecular phylogenetic data ordering the vertical columns (Figure 3, Figure 4 and Figure 5), and data derived from the ontogenetic model [32,54], which, in turn, orders the horizontal rows (Figure 3, Figure 4 and Figure 5). It is important to note that in order to use ontogenetic data in a phyloperiodic framework, it is not necessary to obtain ontogenetic data for all species, but it is necessary to formulate an appropriate ontogenetic model for a given high-ranking group (e.g., for the order Doridida in the present case, see more details on the model formulation in [29,32]).

Figure 3.
Phyloperiodic presentation of the external chromatic patterns (including dorsal views and details of gills and rhinophores) among species Cadlina represented by the horizontal rows. The vertical columns (taxa) are ordered by the molecular phylogenetic tree. Eleven main periods (horizontal rows, Roman numerals) are presented. Spotless body/colourless forms are at the bottom and forms with a maximal number of spots and colour intensity are at the top. Empty cells imply non-observed forms. See text for details.
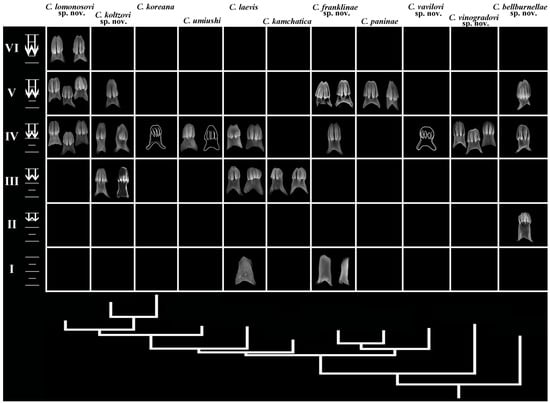
Figure 4.
Phyloperiodic presentation of the central teeth of the radula among species Cadlina, represented by the horizontal rows. The vertical columns (groups, taxa) are ordered by the molecular phylogenetic tree. Six main periods (horizontal rows, Roman numerals) are presented following the direction from non-denticulate and relatively low teeth to higher teeth, where denticles occupy an increasingly greater part of the teeth’s length. Empty cells imply non-observed forms. See text for details.
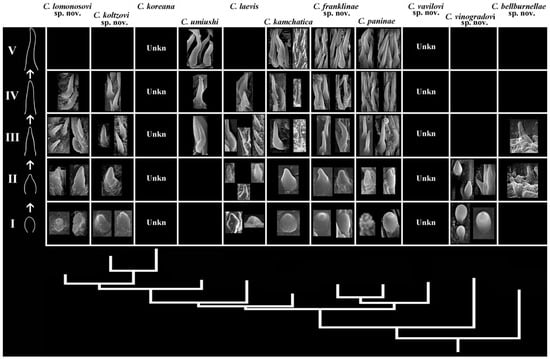
Figure 5.
Phyloperiodic presentation of the copulative spines among species Cadlina, represented by the horizontal rows. The vertical columns (groups, taxa) are ordered by the molecular phylogenetic tree. Five main periods (horizontal rows, Roman numerals) are presented following the direction from short, oval rounded or irregular plates to the elongated spines. Empty cells imply non-observed/unknown forms. See text for details.
The proposed scheme of chromatic patterns has an ontogenetic basis since it has been shown in Cadlina, as well as other disparate members of the nudibranch order Doridida, that during the earlier post-larval ontogenetic development the darker colouration is absent and distinct spots, yellow lines, or darker yellowish or brownish colouration (if any) appear only towards later ontogenetic periods [32,55]. Because any ontogeny encompasses all structures and processes of the organisms (not only those that occurred at the embryonal period), this allows the use of any features for an ontogenetic model, regardless of whether they are earlier embryonal inter-taxon shared nodes (phylotypic periods in a narrow sense) [29,54] or later juvenile–subadult acquisitions of various origin (e.g., chromatic variants, small-scale phyloperiods of the later stages), including potential environmental, food, and other influences. In this regard, the resulting morphology of a particular organism, which has formed during ontogeny and includes both shared and unique features, is important, and any trait of both genetic or epigenetic origin can potentially be represented in phyloperiodic tables.
The central radular teeth and copulative spines, which range from simple non-cuspidate or low cuspidate teeth or rounded plate-like spines to more elongated and elaborated ones, also correspond to the ontogenetic model [32,55]. Therefore, for taxonomic purposes, we present here a phyloperiodic framework for three main sets of commonly used Cadlina diagnostic characters in three different organ systems, including external patterns (colouration, gills, and rhinophoral lamellae patterns), radular details (central teeth have been proven to be the best diagnostic feature in the radula of Cadlina) in the digestive system, and details of copulative spines in the reproductive system [32]. Only specimens for which molecular data have been obtained (Figure 3, Figure 4 and Figure 5) are included in the phyloperiodic tables to ensure that any morphological features can be matched with the molecular phylogenetic tree (Figure 1). Under this approach, diagnostic characters in respective periods are presented not as strict diagnoses, but as a complex range (Figure 3, Figure 4 and Figure 5). The premise of the phyloperiodic approach is precisely that the traditional “rigid” diagnosis will be gradually replaced by multidimensional “soft” or “complex” diagnoses that therefore go beyond the current conventional taxonomic rules and contribute to the formation of future biological nomenclature. The construction of the phyloperiodic tables and the placement of some characters in a certain cell already indicate potential “complex” diagnoses.
This method has been successfully applied to three distantly related groups of nudibranchs [7,29,56]. The shape of the jaw elements [32], traditionally used in taxonomy, is shown here as similar in all species and is therefore not included here in the phyloperiodic table, despite the fact that some small-scale differences could potentially be discovered later. The presentation of the general schemes of reproductive systems required drawings, which may include subjective elements; therefore, we did not use them for the purposes of the phyloperiodic approach. In addition, not all specimens may represent equally similar reproductive systems due to varying degrees of maturity and subsequent preservation. However, we carefully checked the reproductive systems and documented all differences in a more conventional taxonomic way. The molecular phylogenetic component of the phyloperiodic approach helps to clearly confirm the presence of periodic patterns in morphological characters in every available specimen of each species. The vertical columns based on molecular data (Figure 3, Figure 4 and Figure 5) show existing or potentially existing similar morphological characters in each corresponding horizontal row, which is the essence of the periodic approach in science [36,57]. The conjunction of morphology in the broad ontogenetic sense [54] and molecular data thus brings forward truly biological, multilevel dimensions for both traditional and “phylogenetic” systematics.
Building the phyloperiodic rows of chromatic patterns. Similar chromatic patterns within each potential species were aligned using calibration by the degree of light to dark surface pigmentation and transparency of body tissue, including the presence of yellow spots, yellow notal lines, and brownish/dark pigmentation. The present scheme is a further elaboration of a similar scheme that has been used on a practical basis [7].
These similar forms establish horizontal rows of similar-looking specimens within the species (Figure 3). In total, the following eleven chromatic patterns (horizontal rows I–XI) are recognized:
- I
- Body white, yellow spots absent, brownish/dark colouration absent, dispersed white spots may be present, yellow notal line absent, light yellow subepidermal glands distinguishable, and rhinophores and gills white;
- II
- Body white, yellow spots absent, brownish/dark colouration absent, dispersed white spots may be present, yellow notal line absent, yellow subepidermal glands barely distinguishable, and rhinophores and gills light yellow;
- III
- Body white, yellow spots absent to hardly distinguishable, dispersed white spots may be present, brownish/dark colouration absent, yellow notal line absent, yellow subepidermal glands distinguishable, and rhinophores and gills white to light yellow;
- IV
- Body white to creamy, yellow spots absent, dispersed white spots may be present, brownish/dark colouration absent, yellow notal line absent, yellow subepidermal glands distinguishable, and rhinophores and gills yellow to light brownish;
- V
- Body white to creamy, faint yellow spots may be present, dispersed white spots may be present, brownish/dark colouration absent, yellow notal line absent, yellow subepidermal glands barely distinguishable, and rhinophores and gills white to light yellow;
- VI
- Body white to creamy, faint yellow spots present, dispersed white spots may be present, brownish/dark colouration absent, yellow notal line partially present, dispersed, yellow subepidermal glands distinguishable, and rhinophores and gills yellow to light brownish;
- VII
- Body white to creamy, distinct yellow spots present, white spots barely distinguishable, brownish/dark colouration absent, yellow notal line present, moderately distinct, yellow subepidermal glands distinguishable, and rhinophores and gills white to light brownish;
- VIII
- Body white to creamy and light brownish, distinct yellow spots present, white spots may form distinct opaque white covering, brownish/dark colouration absent, yellow notal line present, yellow subepidermal glands distinguishable, and rhinophores and gills yellow to light brownish;
- IX
- Body white to brownish, distinct yellow spots present, white spots barely distinguishable, brownish/dark colouration completely absent, yellow notal line distinct, yellow subepidermal glands distinguishable, and rhinophores and gills white to light yellow;
- X
- Body with patchy brownish to uniform yellowish/dark colouration, white spots barely distinguishable, numerous yellow spots present, yellow notal line distinct, yellow subepidermal glands distinguishable, and rhinophores and gills yellow to brownish;
- XI
- Body with almost completely brownish/dark colouration, white and yellow spots, yellow notal line or subepidermal glands barely distinguishable, and rhinophores and gills brownish.
Building the phyloperiodic rows of the central teeth of the radula. Juveniles have no denticles on the central (rachidian) teeth, and teeth are generally low [32,58]. Therefore, rows here are built following the direction from non-denticulate and relatively low teeth to higher teeth, where denticles occupy an increasingly larger part of the teeth’s length (Figure 4). Six main phyloperiods are presented as follows:
I—central teeth without denticles; II—denticles on central teeth occupy ca. ¼ or are shorter than ¼ of the teeth height; III—denticles on central teeth are longer than ¼ but shorter than ½ of the teeth height; IV—denticles on central teeth occupy ca. ½ of the teeth height; V—denticles on central teeth are longer than ½ but shorter than ¾ of the teeth height; and VI—denticles on central teeth occupy ca. ¾ or are longer than ¾ of the teeth height.
Building the phyloperiodic rows of copulative spines. Juveniles have no elaborated reproductive system [55] or spines of the copulative apparatus; therefore, further development should first involve short spines and then longer spines. The rows were built using direction from short, oval rounded or irregular plates to the elongated spines (Figure 5): I—short, oval to irregularly shaped, and top part not elongated; II—short, oval to irregularly shaped, and top part elongated; III—relatively short, straight, conical, and top part slightly elongated; IV—elongated, partly curved, conical, and top part rather rounded; and V—elongated, significantly convoluted, conical, and top part rather rounded.
3.4. Systematics
Class Gastropoda Cuvier, 1795
Family Cadlinidae Bergh, 1891
Genus Cadlina Bergh, 1879
Cadlina bellburnellae sp. nov.
zoobank no.: urn:lsid:zoobank.org:act: D0B858B6-578A-4310-AC95-9E84F4536C35
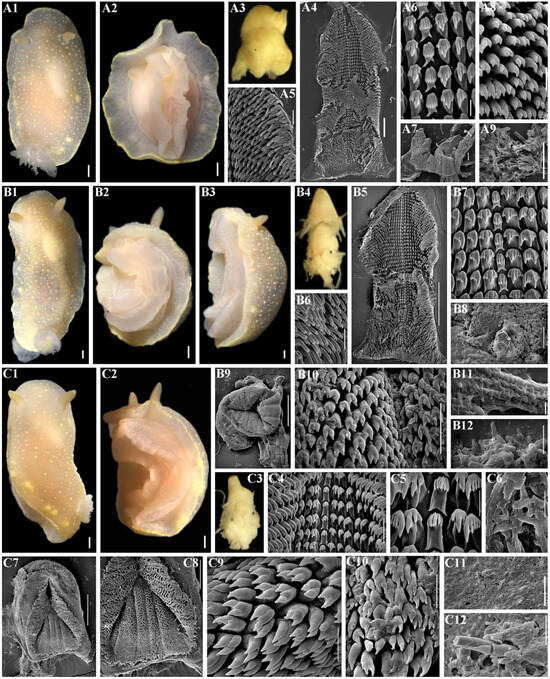
Figure 6.
Cadlina bellburnellae sp. nov., external and internal features. (A) Holotype ZMMU Op-870, length 14 mm live, Iturup Island. (A1) Dorsal view. (A2) Ventral view. (A3) Buccal bulb, light microscopy (LM). (A4) Complete radula, scanning electron microscopy (SEM), scale bar 200 µm. (A5) Outer lateral teeth, SEM, 50 µm. (A6) Central teeth and first lateral teeth, SEM, 20 µm. (A7) Labial cuticle, SEM, 100 µm. (A8) Elements of labial cuticle, SEM, 10 µm. (A9) Spicules of notum, SEM, 500 µm. (B) Paratype ZMMU Op-871, length 20 mm live, Iturup Island. (B1) Dorsal view. (B2) Ventral view. (B3) Lateral view. (B4) Buccal bulb, LM. (B5) Complete radula, SEM, 400 µm. (B6) outer lateral teeth, SEM, 50 µm. (B7) Central teeth and first lateral teeth, SEM, 40 µm. (B8) Notum, SEM, 100 µm. (B9) Complete labial cuticle, SEM, 300 µm. (B10) Elements of labial cuticle, SEM, 20 µm. (B11) Copulative spines, SEM, 5 µm. (B12) Copulative spines, SEM, 10 µm. (C) Paratype ZMMU Op-872, length 15 mm live. (C1) Dorsal view. (C2) Ventrolateral view. (C3) Buccal bulb, LM. (C4) Central part of radula, SEM, 50 µm. (C5) Enlarged central part of radula, central teeth and first lateral teeth, SEM, 20 µm. (C6) Copulative spines, SEM, 10 µm. (C7) Complete labial cuticle, SEM, 200 µm. (C8) Enlarged labial cuticle, SEM, 100 µm. (C9) Elements of labial cuticle, SEM, 10 µm. (C10) Elements of labial cuticle, SEM, 10 µm. (C11) Notum, SEM, 100 µm. (C12) Spicules of notum, SEM, 20 µm. Scale bars for all living specimens are 1 mm. Photos: Tatiana Korshunova and Alexander Martynov.
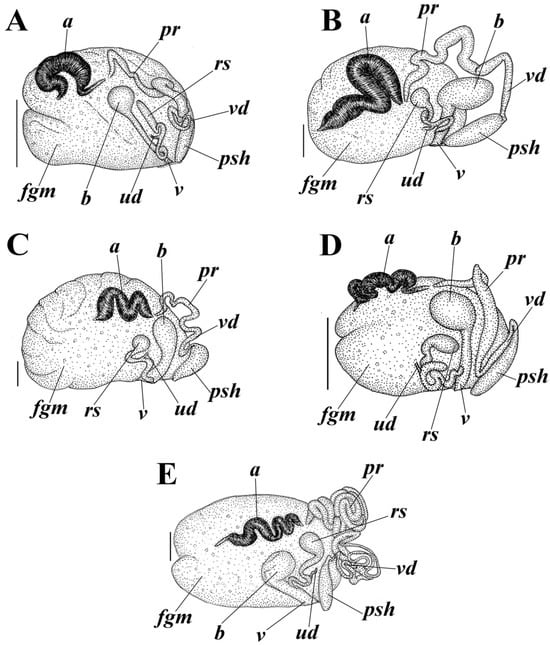
Figure 7.
Reproductive systems of the genus Cadlina. (A) Cadlina bellburnellae sp. nov. (B) Cadlina franklinae sp. nov. (C) Cadlina koltzovi sp. nov. (D) Cadlina lomonosovi sp. nov. (E) Cadlina vinogradovi sp. nov. Abbreviations: a, ampulla; b, bursa; fgm, female gland mass; rs, receptaculum seminis; pr, prostate; psh, penial sheath; ud, uterine duct; v, vaginal duct; and vd, vas deferens. Scale bars are 1 mm. Drawings: Tatiana Korshunova.
Etymology. This species is named after Jocelyn Bell Burnell, who was one of the main contributors to the discovery of pulsars, but did not receive the recognition of a Nobel Prize in Physics in 1974.
Type materials. Holotype ZMMU Op-870, length 14 mm live, Kuril Islands, Iturup Island, 29 August 2022, depth 15–20 m, coll. T.A. Korshunova, A.V. Martynov. Paratype ZMMU Op-871, length 20 mm live, Kuril Islands, Iturup Island, 30 August 2022, depth 15–20 m, coll. T. A. Korshunova, A.V. Martynov. Paratype ZMMU Op-872, length 15 mm live, Kuril Islands, Iturup Island, 3 September 2022, depth 15–20 m, coll. T.A. Korshunova, A.V. Martynov.
External morphology. The notum is broad, and rounded in front and posteriorly. The rhinophores are long and retracted into raised soft sheaths bearing small tubercles (Figure 6(A1,B1–B3,C1,C2)); 9–12 rhinophoral lamellae. The notum is covered with medium- to small-sized, rounded or irregular tubercles (Figure 6(A1,A9,B1,B3,B8,C1,C11,C12)). The spicules form a sparse network in the notum. Eight to ten multipinnate gills united by a common membrane form a circle around the anus. The gills are retractable into the common gill cavity. The border of the gill cavity is moderately raised with a tuberculated edge (Figure 6). The oral veil is small, trapezoid, with oblique notched lateral sides (Figure 6(A2,B2,B3,C2)). The foot is broad, anteriorly rounded, and slightly thickened to form a double edge. The foot appears entirely (Figure 6(A2,B2,C2)); posteriorly, it sometimes projects slightly from notum in crawling animals, forming a rounded tail.
Colour. The notum is partly semi-transparent or almost completely opaque, from white to pale creamy to light yellow and brownish (Figure 6(A1,A2,B1–B3,C1,C2)). The gills are somewhat paler or darker than the notum with a patchy light-yellow pigment. The rhinophores are slightly darker than the notum. The digestive gland is black brownish to brownish; the stomach and intestine are partly visible through the notum dorsally (Figure 6(A1,B1,C1)). A whitish area (parts of the reproductive and digestive systems) may shine through the notum in front of the stomach. The internal organs can also be almost completely invisible. A range of 3–8 subepidermal yellow glands are well conspicuous dorsally and less conspicuous ventrally (some of them are placed close to each other) from each body side (Figure 6(A1,B1,B3,C1)). A yellow line appears around the notum (sometimes is less distinct) and yellow pigment in the dorsal tubercles is well defined (Figure 6(A1,A2,B1–B3,C1,C2)). The yellow line around the foot is somewhat more dispersed but also distinct.
Buccal bulb and oral tube. The buccal bulb is relatively short, similar in length to the oral tube (Figure 6(A3,B4,C3)). The salivary glands are relatively long and narrow.
Jaws. The jaws take the form of a rounded labial disk covered by a yellowish-brown cuticle bearing rod-shaped labial elements with double- to triple-pointed, hook-shaped tips (Figure 6(A7,A8,B9,B10,C7–C10)).
Radula. Radular formula ca. 65–80 × 35–40.1.35–40. The central tooth is moderately low to relatively high and bears up to nine cusps (Figure 6(A6,B7,C4,C5)). The inner lateral teeth (including unique teeth with two main cusps and double sets of denticles on lateral edges) have up to five denticles on the outer edge, and up to three on the inner edge (Figure 6(A4,A6,B5,B7,C4,C5)). The middle and outer teeth are comb-shaped, bearing up to ca. seven, which are concentrated at the top of the teeth, making the shape of the denticles quite peculiar (Figure 6(A5,B6)).
Reproductive system. The ampulla comprises at least two thickened compartments (Figure 7A, a). The ampulla bifurcates into moderately long vas deferens and oviduct. The uterine duct emerges some distance from the female gland mass. The prostatic part of the vas deferens is long, narrow, and only moderately distinct (Figure 7A, pr). The prostate transits to narrow vas deferens (Figure 7A, vd), which considerably widen towards the penial sheath that encloses the evertable ejaculatory duct (Figure 7A, psh). The copulative (penial) spines are conical and relatively short according to available data (Figure 5 and Figure 6(B11,B12,C6)). The vagina narrows (Figure 7A, v) and enters a medium-sized rounded bursa copulatrix (Figure 7A, b). The uterine duct is short and narrow (Figure 7A, ud); it begins from the female gland mass and then enters near the base of a relatively large elongate receptaculum seminis (Figure 7A, rs).
Habitat. The species inhabits shallow waters with rocky and stony substrates at depths of c. 15–20 m.
Distribution. Currently, the species is only known to occur in the coastal waters of Iturup Island (Figure 2B); however, potential distribution includes at least the neighbouring Kuril Islands and Hokkaido Island.
Remarks. The species exhibits known chromatic patterns VIII–X (Figure 3), central teeth patterns II, IV, and V (Figure 4), and copulative spine patterns II and III (Figure 5). Due to the combination of a significant number of yellow spots, the distinct to the less distinct yellow line around the notum, the moderately low to relatively high (in that case only pattern II is known) central radular teeth, the moderately long distal part of vas deferens, the relatively short but elongated copulative spines (more elongated ones can be potentially discovered), and the molecular phylogenetic data (Figure 1), this new species Cadlina bellburnellae sp. nov. is distinguished from all known [32] and herein-described species of Cadlina (Figure 3, Figure 4, Figure 5 and Figure 7). The intragroup distance in C. bellburnellae sp. nov. is 0% for the COI. The lowest COI intergroup distance of 6.53% is found between C. bellburnellae sp. nov. and C. paninae (Table 1). See also the details in Section 4.

Table 1.
Uncorrected p-distances for COI (%) estimated within (highlighted in bold) and between species of the genus Cadlina.
Cadlina franklinae sp. nov.
zoobank no.: urn:lsid:zoobank.org:act: 25F3E3DB-9E36-4BE8-A62B-044F61A08F4A
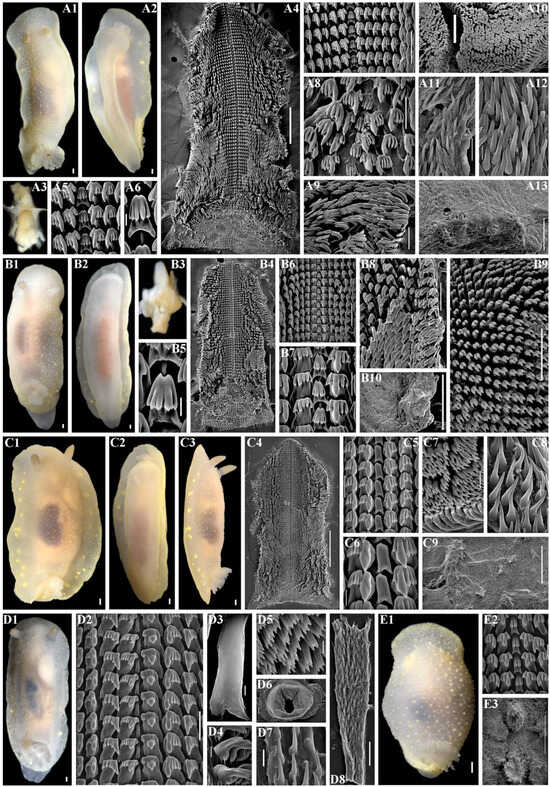
Figure 8.
Cadlina franklinae sp. nov., external and internal features. (A) Holotype ZMMU Op-845, length 39 mm live, Urup Island. (A1) Dorsal view. (A2) Ventral view. (A3) Buccal bulb, light microscopy (LM). (A4) Complete radula, scanning electron microscopy (SEM), scale bar 500 µm. (A5) Central teeth and first lateral teeth, SEM, 40 µm. (A6) Central tooth, SEM, 10 µm. (A7) Central part of radula, SEM, 100 µm. (A8) Central part of radula, SEM, 100 µm. (A9) Outer lateral teeth, SEM, 100 µm. (A10) Labial cuticle, SEM, 100 µm. (A11) Copulative spines, SEM, 30 µm. (A12) Copulative spines, SEM, 10 µm. (A13) Notum and spicules, SEM, 100 µm. (B) Paratype ZMMU Op-844, length 36 mm live, Urup Island. (B1) Dorsal view. (B2) Ventral view. (B3) Buccal bulb, LM. (B4) Complete radula, SEM, 500 µm. (B5) Central tooth, SEM, 10 µm. (B6) Central part of radula, SEM, 100 µm. (B7) Central teeth and first lateral teeth, SEM, 30 µm. (B8) Outer lateral teeth, SEM, 100 µm. (B9) Elements of labial cuticle, SEM, 50 µm. (B10) Notum, SEM, 500 µm. (C) Paratype ZMMU Op-849, length 24 mm live, Urup Island. (C1) Dorsal view. (C2) Ventral view. (C3) Lateral view. (C4) Complete radula, SEM, 500 µm. (C5) Central part of radula, SEM, 50 µm. (C6) Central teeth and first lateral teeth, SEM, 20 µm. (C7) Elements of labial cuticle, SEM, 20 µm. (C8) Copulative spines, SEM, 10 µm. (C9) Notum, SEM, 500 µm. (D) Paratype ZMMU Op-846, length 34 mm, Chirpoy Island. (D1) Dorsal view. (D2) Central part of radula, SEM, 50 µm. (D3) Central tooth, SEM, 2 µm. (D4) Outer lateral teeth, SEM, 20 µm. (D5) Elements of labial cuticle, SEM, 10 µm. (D6) Complete labial cuticle, SEM, 200 µm. (D7) Copulative spines, SEM, 10 µm. (D8) Copulative spines, SEM, 50 µm. (E) Paratype ZMMU Op-857, length 14 mm live, Urup Island. (E1) Dorsal view. (E2) Central part of radula, SEM, 20 µm. (E3) Notum, SEM, 300 µm. Scale bars for all living specimens are 1 mm. Photos: Tatiana Korshunova and Alexander Martynov.
Etymology. This species is named after Rosalind Franklin, co-author of the DNA discovery, who died early (radiation during the study of DNA is considered a factor in her death), not having received the Nobel Prize together with Crick and Watson.
Type materials. Holotype ZMMU Op-845, length 39 mm live, Kuril Islands, Urup Island, 27 August 2021, depth 15–20 m, coll. T.A. Korshunova, A.V. Martynov. Paratype ZMMU Op-844, length 36 mm live, Kuril Islands, Urup Island, 2 September 2021, depth 10–20 m, coll. T.A. Korshunova, A.V. Martynov. Paratype ZMMU Op-846, length 34 mm, Kuril Islands, Chirpoy Island, 24 August 2021, depth 15–20 m, coll. T.A. Korshunova, A.V. Martynov. Paratype ZMMU Op-847, 19 mm live, Kuril Islands, Urup Island, 2 September 2021, depth 20–30 m, coll. T. A. Korshunova, A.V. Martynov. Paratype ZMMU Op-848, length 17 mm live, Kuril Islands, Urup Island, 27 August 2021, depth 15–20 m, coll. T.A. Korshunova, A.V. Martynov. Paratype ZMMU Op-849, 24 mm, Kuril Island, Urup Island, 2 September 2021, depth 20–30 m, coll. T.A. Korshunova, A.V. Martynov. Paratype ZMMU Op-856, length 15 mm live, Kuril Islands, Urup Island, 1 September 2022, depth 15–20 m, coll. T.A. Korshunova, A.V. Martynov. Paratype ZMMU Op-857, length 14 mm live, Kuril Islands, Urup Island, 1 September 2022, depth 15–20 m, coll. T.A. Korshunova, A.V. Martynov.
External morphology. The notum is broad, and rounded in front and posteriorly. The rhinophores are long and retracted into raised soft almost smooth or bearing very small tubercle sheaths (Figure 8(A1,B1,C1,C3,D1,E1)); 10–16 rhinophoral lamellae. The notum is almost smooth or covered with very small, indistinct tubercles (Figure 8(A1,A13,B1,B10,C1,C3,C9,D1,E1,E3)). The spicules form a sparse network in the notum. Seven to nine multipinnate gills united by a common membrane form a circle around the anus. The gills are retractable into a common gill cavity. The border of the gill cavity is moderately raised with a tuberculated edge (Figure 8). The oral veil is small, trapezoid, with oblique notched lateral sides (Figure 8(A2,B2,C2)). The foot is broad, anteriorly rounded and slightly thickened to form a double edge. The foot appears entirely (Figure 8(A2,B2,C2)); posteriorly, it sometimes projects slightly from the notum in crawling animals, forming a rounded tail.
Colour. The notum is commonly white, and additionally may have patchy covers with opaque white dust, partly semi-transparent, more rarely more towards a yellowish uniform colour (Figure 8(A1,B1,C1,C3,D1,E1)). The gills are somewhat paler, darker, or almost the same colour as the notum, without or with a patchy yellow pigment. The rhinophores are slightly darker than notum. The digestive gland is black and brownish; the stomach and intestine are partly visible through the notum dorsally (Figure 8(A1,B1,C1,C3,D1,E1)). The whitish to pinkish area (parts of the reproductive and digestive systems) may shine through the notum in front and behind the stomach. The internal organs can also be almost completely invisible. A range of 2–15 (sometimes 19 if smaller ones are counted) subepidermal yellow glands are barely distinguishable to conspicuous dorsally and ventrally from each body side. The yellow line around the notum is more commonly absent or very indistinct; only rarely is it more distinct. A yellow pigment appears dorsally or within small-sized tubercles (Figure 8(A1,B1,C1,C3,D1,E1)). The yellow lines around the foot are commonly absent or very weak but can also be more distinct.
Buccal bulb and oral tube. The buccal bulb is relatively short, similar in length to the oral tube (Figure 8(A3,B3)). The salivary glands are relatively long and narrow.
Jaws. The jaws take the form of a rounded labial disk covered by a yellowish-brown cuticle bearing rod-shaped labial elements with double-pointed hook-shaped tips (Figure 8(A10,B9,C7,D5,D6)).
Radula. Radular formula ca. 80–120 × 30–50.1.30–50. The central tooth is moderately low, bears up to six cusps, and is rarely completely devoid of cusps (Figure 8(A5–A8,B5–B7,C5,C6,D2,D3,E2)). The inner lateral teeth have up to eight denticles on the outer edge, and up to two on the inner edge (Figure 8(A4,A5,A7,A8,B4,B6,B7,C4,C5,C6,D2,E2)). The middle and outer teeth are comb-shaped, bearing up to 25 denticles (Figure 8(A4,A5,A9,B4,B8,C4,D4)).
Reproductive system. The ampulla comprises at least three thickened compartments (Figure 7B, a). The ampulla bifurcates into moderately long vas deferens and oviduct. The uterine duct emerges some distance from the female gland mass. The prostatic part of the vas deferens is long, narrow, and only moderately distinct (Figure 7B, pr). The prostate transits to narrow vas deferens (Figure 7B, vd), which considerably widens towards the penial sheath that encloses the evertable ejaculatory duct (Figure 7B, psh). The copulative (penial) spines are commonly elongated and convoluted, and some shorter ones may present in addition (Figure 5 and Figure 8(A11,A12,C8,D7,D8)). The vagina narrows (Figure 7B, v) and enters a relatively large oval bursa copulatrix (Figure 7B, b). The uterine duct is short and narrow (Figure 7B, ud); it begins from the female gland mass and then enters near the base of the pear-shaped receptaculum seminis (Figure 7B, rs).
Habitat. The species inhabits shallow waters with rocky and stony substrates at depths of c. 15–30 m.
Distribution. Currently, the species is only known to occur in the coastal waters of the Urup and Chirpoy Islands (Figure 2B); however, potential distribution includes at least the neighbouring Kuril Islands.
Remarks. The species exhibits known chromatic patterns II, V, VIII, and X (Figure 3), central teeth patterns I, IV, and V (Figure 4), and copulative spine patterns I–V (Figure 5). Due to the combination of the moderate number of yellow spots, the usual absence of a distinct yellow line around the notum, the notum commonly with opaque white sprinkled small spots, the relatively low central radular teeth, the moderate distal part of vas deferens and commonly elongate copulative spines, and the molecular phylogenetic data (Figure 1), this new species Cadlina franklinae sp. nov. is distinguished from all known [32] and herein-described species of Cadlina (Figure 3, Figure 4, Figure 5 and Figure 7B). The intragroup distances in C. franklinae sp. nov. are 0–0.30% for the COI. The lowest COI intergroup distance of 2.74% is found between C. franklinae sp. nov. and C. paninae (Table 1). See also the details in Section 4.
Cadlina koltzovi sp. nov.
zoobank no.: urn:lsid:zoobank.org:act: 31006AD5-3604-4574-8A4B-66E277DA5441
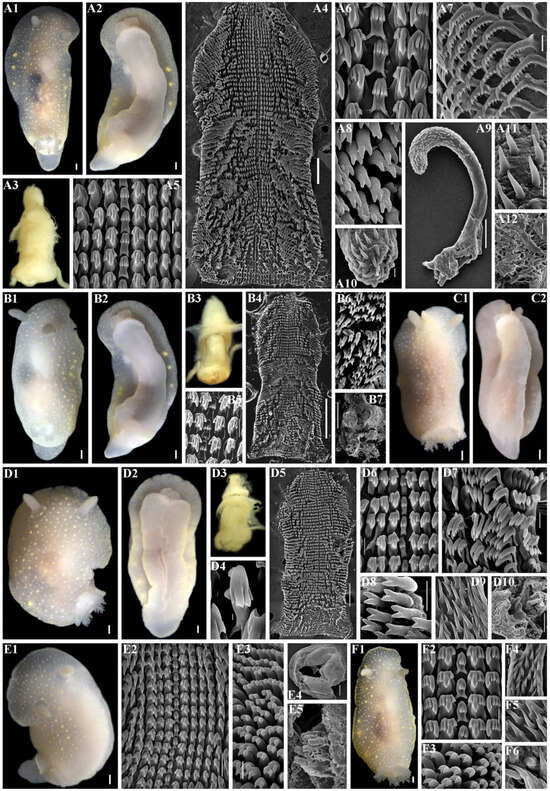
Figure 9.
Cadlina koltzovi sp. nov., external and internal features. (A) Holotype ZMMU Op-842, length 24 mm live, Urup Island. (A1) Dorsal view. (A2) Ventral view. (A3) Buccal bulb, light microscopy (LM). (A4) Complete radula, scanning electron microscopy (SEM), scale bar 200 µm. (A5) Central part of radula, SEM, 20 µm. (A6) Central teeth and first lateral teeth, SEM, 10 µm. (A7) Outer lateral teeth, SEM, 20 µm. (A8) Elements of labial cuticle, SEM, 5 µm. (A9) Everted copulative apparatus, SEM, 100 µm. (A10) Copulative spines, SEM, 10 µm. (A11) Copulative spines, SEM, 10 µm. (A12) Notum, SEM, 50 µm. (B) Paratype ZMMU Op-840, length 17 mm live, Urup Island. (B1) Dorsal view. (B2) Ventral view. (B3) Buccal bulb, LM. (B4) Complete radula, SEM, 500 µm. (B5) Central teeth and first lateral teeth, SEM, 30 µm. (B6) Elements of labial cuticle, SEM, 20 µm. (B7) Notum, SEM, 500 µm. (C) Paratype ZMMU Op-841, length 15 mm live, Urup Island. (C1) Dorsal view. (C2) Ventral view. (D) Paratype ZMMU Op-843, length 18 mm live, Urup Island. (D1) Dorsal view. (D2) Ventral view. (D3) Buccal bulb, LM. (D4) Central tooth, SEM, 3 µm. (D5) Complete radula, SEM, 300 µm. (D6) Central part of radula, SEM, 30 µm. (D7) Outer lateral teeth, SEM, 30 µm. (D8) Elements of labial cuticle, SEM, 10 µm. (D9) Copulative spines, SEM, 10 µm. (D10) Notum and spicules, SEM, 100 µm. (E) Paratype ZMMU Op-852, length 17 mm live, Kunashir Island. (E1) Dorsal view. (E2) Central part of radula, SEM, 50 µm. (E3) Elements of labial cuticle, SEM, 10 µm. (E4) Complete labial cuticle, SEM, 200 µm. (E5) Spicules of notum, SEM, 20 µm. (F) Paratype ZMMU Op-853, length 22 mm live, Iturup Island. (F1) Dorsal view. (F2) Central part of radula, SEM, 20 µm. (F3) Elements of labial cuticle, SEM, 10 µm. (F4) Copulative spines, SEM, 10 µm. (F5) Copulative spines, SEM, 10 µm. (F6) Copulative spines, SEM, 3 µm. Scale bars for all living specimens are 1 mm. Photos: Tatiana Korshunova and Alexander Martynov.
Etymology. This species is named after Nikolai Koltzov, founder of the Institute of Experimental Biology (which has now transitioned to the Institute of Developmental Biology RAS), who pioneered the idea of a cytoskeleton and anticipated many concepts of modern genetics (including the structure of DNA and epigenetics), but died unexpectedly following persecution after refusal to support false testimonies against Nikolai Vavilov.
Type materials. Holotype ZMMU Op-842, length 24 mm live, Kuril Islands, Urup Island, 23 August 2021, depth 10–20, coll. T.A. Korshunova, A. V. Martynov. Paratype ZMMU Op-840, length 17 mm live, Kuril Islands, Urup Island, 2 September 2021, depth 10–40 m, coll. T.A. Korshunova, A.V. Martynov. Paratype ZMMU Op-841, length 15 mm live, Kuril Islands, Urup Island, 30 August 2021, depth 10–20 m, coll. T.A. Korshunova, A. V. Martynov. Paratype ZMMU Op-843, length 18 mm live, Kuril Island, Urup Island, 23 August 2021, coll. T. A. Korshunova, A.V. Martynov. Paratype ZMMU Op-852, length 17 mm live, Kuril Islands, Kunashir Island, 29 August 2022, depth 5–10 m, coll. T.A. Korshunova, A. V. Martynov. Paratype ZMMU Op-853, length 22 mm live, Kuril Islands, Iturup Island, 4 September 2022, depth 10–15 m, coll. T.A. Korshunova, A.V. Martynov. Paratype ZMMU Op-854, length 25 mm live, Kuril Island, Iturup Island, 4 September 2022, depth 10–15 m, coll. T.A. Korshunova, A.V. Martynov. ZMMU Op-855, length 24 mm live, Kuril Islands, Iturup Island, 4 September 2022, depth 10–15 m, coll. T.A. Korshunova, A.V. Martynov.
External morphology. The notum is broad, and rounded in front and posteriorly. The rhinophores are long and retracted into raised soft almost smooth or bearing very small tubercle sheaths (Figure 9(A1,B1,C1,D1,E1,F1)); 8–13 rhinophoral lamellae. The notum is almost smooth or covered with very small, indistinct tubercles (Figure 9(A1,A12,B1,B7,C1,D1,D10,E1,E5,F1)). The spicules form a sparse network in the notum. Seven to nine multipinnate gills united by a common membrane form a circle around the anus. The gills are retractable into a common gill cavity. The border of the gill cavity is moderately raised with a tuberculated edge (Figure 9(A1,B1,C1,D1,E1,F1)). The oral veil is small, trapezoid, with oblique notched lateral sides (Figure 9(A2,B2,C2,D2)). The foot is broad, anteriorly rounded, and slightly thickened to form a double edge. The foot appears entirely (Figure 9(A2,B2,C2,D2)); posteriorly, it sometimes projects slightly from the notum in crawling animals, forming a rounded tail.
Colour. The notum is partly semi-transparent to more opaque, white to creamy, and patchy brownish to uniform yellowish in colour (Figure 9(A1,A2,B1,B2,C1,C2,D1,D2,E1,F1)). The gills are almost the same colour as the notum, without or with a patchy yellow pigment. The rhinophores are slightly darker than the notum. The digestive gland is black brownish to creamy; the stomach and intestine are partly visible through the notum dorsally (Figure 9(A1,B1,C1,D1,E1,F1)). The whitish area (parts of the reproductive and digestive systems) may shine through the notum in front and behind the stomach. The internal organs can also be almost completely invisible. A range of 3–12 subepidermal yellow glands are barely distinguishable to conspicuous dorsally and ventrally from each body side. The yellow line around the notum is usually absent or dispersed, or, rarely, present (Figure 9(A1,A2,B1,B2,C1,C2,D1,D2,E1,F1)). A yellow pigment appears dorsally or within small- to medium-sized tubercles (Figure 9(A1,B1,C1,D1,E1,F1)). The yellow line around the foot is absent.
Buccal bulb and oral tube. The buccal bulb is relatively short, similar in length to the oral tube (Figure 9(A3,B3,D3)). The salivary glands are relatively long and narrow.
Jaws. The jaws take the form of a rounded labial disk covered by a yellowish-brown cuticle bearing rod-shaped labial elements with double hook-shaped tips (Figure 9(A8,B6,D8,E3,E4,F3)).
Radula. Radular formula ca. 60–75 × 25–45.1.25–45. The central tooth is moderately high and bears up to seven cusps (Figure 9(A5,A6,B5,D4,D6,E2,F2)). The inner lateral teeth have up to five denticles on the outer edge, and up to three on the inner edge (Figure 9(A4–A6,B4,B5,D5–D7,E2,F2)). The middle and outer teeth are comb-shaped, bearing up to 12 denticles (Figure 9(A4,A7,D5,D7)).
Reproductive system. The ampulla comprises at least three thickened compartments (Figure 7C, a). The ampulla bifurcates into moderately long vas deferens and oviduct. The uterine duct emerges some distance from the female gland mass. The prostatic part of the vas deferens is long, narrow, and not distinct (Figure 7C, pr). The prostate transits to narrow vas deferens (Figure 7C, vd), which considerably widens towards the penial sheath that encloses the evertable ejaculatory duct (Figure 7C, psh). The copulative (penial) spines are commonly elongated and curved, and some shorter ones may present in addition (Figure 5 and Figure 9(A9–A11,D9,F4–F6)). The vagina narrows (Figure 7C, v) and enters a medium-sized oval bursa copulatrix (Figure 7C, b). The uterine duct is short and narrow (Figure 7C, ud); it begins from the female gland mass and then enters near the base of the pear-shaped receptaculum seminis (Figure 7C, rs).
Habitat. The species inhabits shallow waters with rocky and stony substrates at depths of c. 5–40 m.
Distribution. Currently, the species is only known to occur in the coastal waters of the Urup, Iturup, and Kunashir Islands (Figure 2B); however, potential distribution includes at least the neighbouring Kuril Islands and Hokkaido Island.
Remarks. The species exhibits known chromatic patterns V, VIII–X (Figure 3), central teeth patterns III–V (Figure 4), and copulative spine patterns I–IV (Figure 5). Due to the combination of a moderate number of yellow spots, the presence or absence of a distinct yellow line around the notum, the relatively high central radular teeth, the moderate distal part of vas deferens and commonly elongate copulative spines, and the molecular phylogenetic data (Figure 1), this new species Cadlina koltzovi sp. nov. is distinguished from all known [32] and herein-described species of Cadlina (Figure 3, Figure 4 and Figure 5). The intragroup distances in C. koltzovi sp. nov. are 0–0.15% for the COI. The lowest COI intergroup distance of 5.63% is found between C. koltzovi sp. nov. and C. umiushi (Table 1). See also the details in Section 4.
Cadlina lomonosovi sp. nov.
zoobank no.: urn:lsid:zoobank.org:act: DA87EC75-2E17-45B3-8117-51DBF6960402
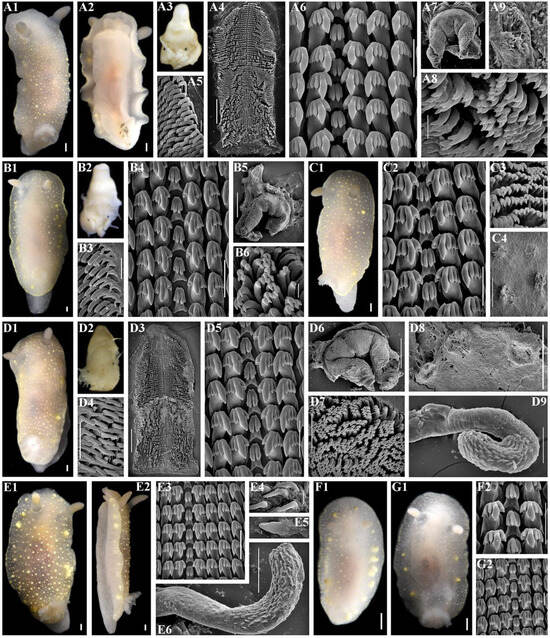
Figure 10.
Cadlina lomonosovi sp. nov., external and internal features. (A) Holotype ZMMU Op-868, length 17 mm live, Kunashir Island. (A1) Dorsal view. (A2) Ventral view. (A3) Buccal bulb, light micros-copy (LM). (A4) Complete radula, scanning electron microscopy (SEM), scale bar, 300 µm. (A5) Outer lateral teeth, SEM, 30 µm. (A6) Central part of radula, 30 µm. (A7) Complete labial cuticle, SEM, 100 µm. (A8) Elements of labial cuticle, SEM, 10 µm. (A9) Notum, SEM, 30 µm. (B) Paratype ZMMU Op-866, length 24 mm live, Kunashir Island. (B1) Dorsal view. (B2) Buccal bulb, LM. (B3) Outer lateral teeth, SEM, 50 µm. (B4) Central part of radula, SEM, 40 µm. (B5) Complete labial cuticle, SEM, 500 µm. (B6) Elements of labial cuticle, SEM, 10 µm. (C) Paratype ZMMU Op-869, length 15 mm live, Kunashir Island. (C1) Dorsal view. (C2) Central part of radula, SEM, 30 µm. (C3) Elements of labial cuticle, SEM, 30 µm. (C4) Notum and spicules, SEM, 300 µm. (D) Paratype ZMMU Op-861, length 19 mm live, Iturup Island. (D1) Dorsal view. (D2) Buccal bulb, LM. (D3) Complete radula, SEM, 500 µm. (D4) Outer laterals, SEM, 100 µm. (D5) Central part of radula, SEM, 50 µm. (D6) Complete labial cuticle, SEM, 500 µm. (D7) Elements of labial cuticle, SEM, 50 µm. (D8) Notum and spicules, SEM, 500 µm. (D9) Everted copulative apparatus, SEM, 100 µm. (E) Paratype ZMMU Op-859, length 20 mm live, Iturup Island. (E1) Dorsal view. (E2) Lateral view. (E3) Central part of radula, SEM, 50 µm. (E4) Copulative spines, SEM, 10 µm. (E5) Copulative spines, SEM, 1 µm. (E6) Everted copulative apparatus, SEM, 100 µm. (F) Paratype ZMMU Op-864, length 8 mm live, Iturup Islands. (F1) Dorsal view. (F2) Central teeth and first lateral teeth, SEM, 30 µm. (G) Paratype ZMMU Op-863, length 11 mm live, Iturup Island. (G1) Dorsal view. (G2) Central part of the radula, SEM, 20 µm. Scale bars for all living specimens are 1 mm. Photos: Tatiana Korshunova and Alexander Martynov.
Etymology. This species is named after Mikhail Lomonosov, founder of the Moscow State University. Despite his fame, Lomonosov struggled with scientific bureaucrats and obscurantists in Russia throughout most of his career.
Type materials. Holotype ZMMU Op-868, length 17 mm live, Kuril Islands, Kunashir Island, 29 August 2022, depth 5–10 m, coll. T.A. Korshunova, A.V. Martynov. Paratype ZMMU Op-858, length 15 mm live, Kuril Islands, Iturup Island, 4 September 2022, depth 15–20 m, coll. T.A. Korshunova, A.V. Martynov. Paratype ZMMU Op-859, length 20 mm live, Kuril Islands, Iturup Island, 4 September 2022, depth 15–20 m, coll. T.A. Korshunova, A.V. Martynov. Paratype ZMMU Op-860, length 14 mm live, Kuril Islands, Iturup Island, 4 September 2022, depth 15–20 m, coll. T. A. Korshunova, A.V. Martynov. Paratype ZMMU Op-861, length 19 mm live, Kuril Islands, Iturup Island, 17 August 2022, depth 10–15 m, coll. T.A. Korshunova, A.V. Martynov. Paratype ZMMU Op-862, length 12 mm live, Kuril Islands, Iturup Island, 9 September 2022, depth 15–20 m, coll. T. A. Korshunova, A.V. Martynov. Paratype ZMMU Op-863, length 11 mm live, Kuril Islands, Iturup Island, 17 August 2022, depth 10–15 m, coll. T. A. Korshunova, A. V. Martynov. Paratype ZMMU Op-864, length 8 mm live, Kuril Islands, Iturup Islands, 5 September 2022, depth 10–15 m, coll. T. A. Korshunova, A.V. Martynov. Paratype ZMMU Op-865, length 10 mm live, Kuril Islands, Iturup Island, 30 August 2022, depth 15–20 m, coll. T.A. Korshunova, A.V. Martynov. Paratype ZMMU Op-866, length 24 mm live, Kuril Islands, Kunashir Island, 29 August 2022, depth 5–10 m, coll. T.A. Korshunova, A.V. Martynov. Paratype ZMMU Op-867, length 7 mm live, Kuril Islands, Iturup Island, 29 August 2022, depth 10–15 m, coll. T.A. Korshunova, A.V. Martynov. Paratype ZMMU Op-869, length 15 mm live, Kuril Islands, Kunashir Island, 29 August 2022, depth 5–10, coll. T. A. Korshunova, A.V. Martynov.
External morphology. The notum is broad, and rounded in front and posteriorly. The rhinophores are long and retracted into raised soft sheaths bearing small tubercles (Figure 10(A1,A2,B1,C1,D1,E1,E2,F1,G1)); 8–15 rhinophoral lamellae. The notum is covered with medium- to small-sized, rounded or irregular tubercles (Figure 10(A1,A9,B1,C1,C4,D1,D8,E1,E2,F1,G1)). The spicules form a sparse network in the notum. Eight to nine multipinnate gills united by a common membrane form a circle around the anus. The gills are retractable into a common gill cavity. The border of the gill cavity is moderately raised with a tuberculated edge (Figure 10(A1,B1,C1,D1,E1,E2,F1,G1)). The oral veil is small, trapezoid, with oblique notched lateral sides (Figure 10(A2,E2)). The foot is broad, anteriorly rounded, and slightly thickened to form a double edge. The foot appears entirely (Figure 10(A2,E2)); posteriorly, it sometimes projects slightly from the notum in crawling animals, forming a rounded tail.
Colour. The notum is partly semi-transparent or almost completely opaque, from white to pale creamy to light yellow and brownish (Figure 10(A1,A2,B1,C1,D1,E1,E2,F1,G1)). The gills are somewhat paler or darker than the notum with a patchy light-yellow pigment. The rhinophores are slightly darker than the notum. The digestive gland is black brownish to pinkish; the stomach and intestine are partly visible through the notum dorsally (Figure 10(A1,B1,C1,D1,E1,F1,G1)). A pinkish area (parts of the reproductive and digestive systems) may shine through the notum in front of the stomach. The internal organs can also be almost completely invisible. A range of 2–12 subepidermal yellow glands are well conspicuous dorsally and less conspicuous ventrally (some of them are placed close to each other) from each body side. The yellow lines around the notum and the yellow pigment in the dorsal tubercles are usually distinct and numerous (Figure 10(A1,A2,B1,C1,D1,E1)), although specimens without distinct yellow spots and dispersed notal lines are also present, which is consistent with the ontogenetic model since these specimens are commonly smaller in size (Figure 10(F1,G1)). The yellow line around the foot is dispersed and indistinct.
Buccal bulb and oral tube. The buccal bulb is relatively short, similar in length to the oral tube (Figure 10(A3,B2,D2)). The salivary glands are relatively long and narrow.
Jaws. The jaws take the form of a rounded labial disk covered by a yellowish-brown cuticle bearing rod-shaped labial elements with double-pointed hook-shaped tips (Figure 10(A7,A8,B6,C3,D6,D7)).
Radula. Radular formula ca. 100–110 × 30–50.1.30–50. The central tooth is moderately low and bears up to eight cusps (Figure 10(A6,B4,C2,D5,E3,F2,G2)). The inner lateral teeth have up to five denticles on the outer edge, and up to three on the inner edge (Figure 10(A4,A6,B4,C2,D3,D5,E3,F2,G2)). The middle and outer teeth are comb-shaped, bearing up to ca. 15 denticles (Figure 10(A4,A5,B3,D3,D4)).
Reproductive system. The ampulla comprises at least three thickened and convoluted compartments (Figure 7D, a). The ampulla bifurcates into moderately long vas deferens and oviduct. The uterine duct emerges some distance from the female gland mass (Figure 7D). The prostatic part of the vas deferens is long, narrow, and moderately distinct (Figure 7D, pr). The prostate transits to narrow vas deferens (Figure 7D, vd), which considerably widens towards the penial sheath that encloses the evertable ejaculatory duct (Figure 7D, psh). The copulative (penial) spines are commonly elongated and curved, and some shorter ones may present in addition (Figure 5 and Figure 10(D9,E4–E6)). The vagina narrows (Figure 7D, v) and enters a relatively large rounded bursa copulatrix (Figure 7D, b). The uterine duct is short and narrow (Figure 7D, ud); it begins from the female gland mass and then enters near the base of the oval receptaculum seminis (Figure 7D, rs).
Habitat. The species inhabits shallow waters with rocky and stony substrates at depths of c. 5–20 m.
Distribution. Currently, the species is only known to occur in the coastal waters of the Iturup and Kunashir Islands (Figure 2B); however, potential distribution includes at least the neighbouring Kuril Islands and Hokkaido Island.
Remarks. The species features known chromatic patterns VI–X (Figure 3), central teeth patterns IV–VI (Figure 4), and copulative spine patterns I–IV (Figure 5). Due to the combination of an insignificant to significant number of yellow spots, the distinct or less distinct but still present yellow line around the notum, the moderately low central radular teeth (in this case, pattern II is not known, only patterns IV–VI), the moderately long distal part of vas deferens, the commonly elongated copulative spines, and the molecular phylogenetic data (Figure 1), this new species Cadlina lomonosovi sp. nov. is distinguished from all known [32] and herein-described species of Cadlina (Figure 3, Figure 4 and Figure 5). The intragroup distances in C. lomonosovi sp. nov. are 0–1.52% for the COI. The lowest COI intergroup distance of 3.28% is found between C. lomonosovi sp. nov. and C. umiushi (Table 1). See also the details in Section 4.
Cadlina vavilovi sp. nov.
zoobank no.: urn:lsid:zoobank.org:act: C2A62183-1931-4F61-8CAA-381302E047D3
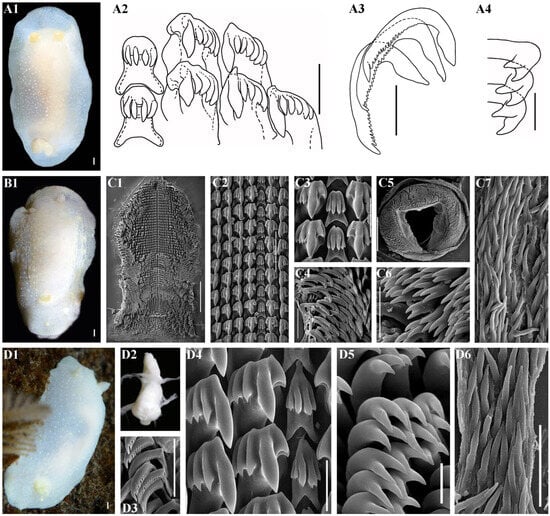
Figure 11.
Cadlina vavilovi sp. nov. (A) and Cadlina paninae Korshunova et al., 2020 (B–D), external and internal features. (A) Cadlina vavilovi sp. nov., Holotype MIMB 42230, length 24 mm live, Iturup Island. (A1) Dorsal view, drawing. (A2) Central teeth and first lateral teeth, drawing, scale bar 30 µm. (A3) Outer lateral teeth, drawing, 30 µm. (A4) Elements of labial cuticle, drawing, 10 µm (the drawings based on data [59]). (B–D) Cadlina paninae Korshunova et al., 2020. (B1) Holotype ZMMU Op-683, Matua Island, dorsal view. (C) Paratype ZMMU Op-684, Matua Island. (C1) Complete radula, scanning electron microscopy (SEM), 500 µm. (C2) Central part of the radula, SEM, 100 µm. (C3) Central teeth with first lateral teeth, SEM, 40 µm. (C4) Outer lateral teeth, 100 µm. (C5) Complete labial cuticle, SEM, 500 µm. (C6) Elements of labial cuticle, SEM, 20 µm. (C7) Copulative spines, SEM, 40 µm. (D) Paratype ZMMU Op-685, Matua Island. (D1) Dorso-lateral view. (D2) Buccal bulb, light microscopy (LM). (D3) Outer lateral teeth, SEM, 100 µm. (D4) Central teeth with first lateral teeth, SEM, 30 µm. (D5) Elements of labial cuticle, SEM, 10 µm. (D6) Copulative spines, SEM, 40 µm. Scale bars for all living specimens are 1 mm. Photos and drawings: Tatiana Korshunova, Nadezhda Sanamyan, and Alexander Martynov.
Etymology. This species is named after Nikolai Vavilov. Famous for his studies on cultivated plants, Vavilov was one of the advocates of the periodic approach in taxonomy. He died in Stalin’s prison due to falsehood allegations made by agronomist Trofim Lysenko, whose work was initially supported by Nikolai Vavilov.
Type material. Holotype MIMB 42230, length 24 mm live, Kuril Islands, Iturup Island, 19 August 2019, depth 263–273 m, coll. unspecified.
External morphology. The notum is broad, and rounded in front and posteriorly. The rhinophores are retracted into raised soft sheaths bearing small tubercles (Figure 11(A1)); 14 rhinophoral lamellae. The notum is covered with small, indistinct tubercles (Figure 11(A1)). Eight multipinnate gills united by a common membrane form a circle around the anus. The gills are retractable into a common gill cavity. The border of the gill cavity is moderately raised with a tuberculated edge (Figure 11(A1)). The foot is broad.
Colour. The notum is partly semi-transparent, white, without a conspicuous yellow pigment (Figure 11(A1)). The gills are light creamy. The rhinophores are slightly darker than the notum. The digestive system is light brownish and pinkish, partly visible through the notum dorsally (Figure 11(A1)). The subepidermal glands are barely visible. The yellow line around the notum is absent. The yellow pigment dorsally or within small tubercles is barely distinguishable (Figure 11(A1)). The yellow line around the foot is absent.
Buccal bulb and oral tube. The buccal bulb and oral tube are present. The salivary glands are narrow.
Jaws. The jaws take the form of an oval labial disk covered by cuticle-bearing rod-shaped labial elements with double-hook-shaped tips (Figure 11(A4)).
Radula. Radular formula 79 × 34.1.34. The central tooth is moderately low and bears up to six cusps (Figure 11(A2)). The inner lateral teeth have up to six denticles on the outer edge, and up to three on the inner edge (Figure 11(A2)). The middle and outer teeth are comb-shaped, bearing up to 20 denticles (Figure 11(A3)).
Reproductive system. The ampulla is folded and broad; it bifurcates into moderately long vas deferens and oviduct. The prostate transits to narrow vas deferens that enclose the ejaculatory duct with copulative (penial) spines. The vagina is narrow, and the bursa copulatrix and receptaculum seminis are oval.
Habitat. The species inhabits relatively deeper waters with gravel sand substrates at depths of c. 263–273 m.
Distribution. Currently, the species is only known to occur in the coastal waters of Iturup Island (Figure 2B); however, potential distribution includes at least the neighbouring Kuril Islands.
Remarks. The species exhibits known chromatic pattern II and central teeth pattern IV. Due to the combination of indistinguishable yellow spots, the absence of the yellow line around the notum, the uniformly coloured ground notum, the relatively low central radular teeth, and the molecular phylogenetic data (Figure 1, Figure 2, Figure 3, Figure 4, Figure 5 and Figure 11), this new species Cadlina vavilovi sp. nov. is distinguished from all known [32] and herein-described species of Cadlina (Figure 3, Figure 4 and Figure 5). The species is mentioned as Cadlina sp. in [59]. The lowest COI intergroup distance of 4.82% is found between C. vavilovi sp. nov. and C. franklinae sp. nov. as well as between C. vavilovi sp. nov. and C. paninae (Table 1). Extended morphological data for C. paninae are provided here (Figure 3, Figure 4, Figure 5 and Figure 11(B1,C1–C7,D1–D6)). See also the details in Section 4.
Cadlina vinogradovi sp. nov.
zoobank no.: urn:lsid:zoobank.org:act: 1C45ED81-E24B-487C-95C8-986D6C53ABF2
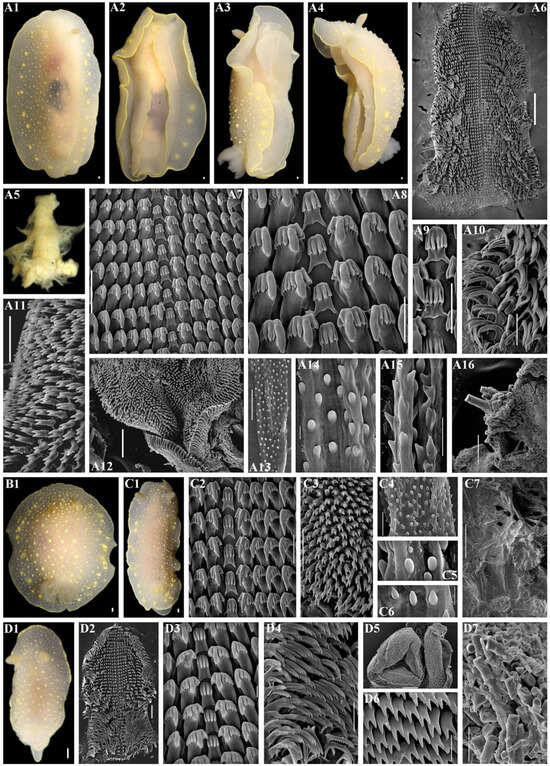
Figure 12.
Cadlina vinogradovi sp. nov., external and internal features. (A) Holotype ZMMU Op-839, length 60 mm live, Urup Island. (A1) Dorsal view. (A2) Ventral view. (A3) Lateral view. (A4) Lateral view. (A5) Buccal bulb, light microscopy (LM). (A6) Complete radula, scanning electron microscopy (SEM), scale bar, 500 µm. (A7) Central part of radula, SEM, 100 µm. (A8) Central teeth with first lateral teeth, SEM, 50 µm. (A9) Central teeth, SEM, 50 µm. (A10) Outer lateral teeth, SEM, 100 µm. (A11) Elements of labial cuticle, SEM, 40 µm. (A12) Labial cuticle, SEM, 100 µm. (A13) Copulative spines, SEM, 30 µm. (A14) Copulative spines, SEM, 3 µm. (A15) Copulative spines, SEM, 10 µm. (A16) Notum and spicules, SEM, 100 µm. (B1) Paratype ZMMU Op-838, length 32 mm live, Urup Island, dorsal view. (C) Paratype ZMMU Op-850, length 33 mm live, Iturup Island. (C1) Dorsal view. (C2) Central part of radula, SEM, 50 µm. (C3) Elements of labial cuticle, SEM, 40 µm. (C4) Copulative spines, SEM, 10 µm. (C5) Copulative spines, SEM, 1 µm. (C6) Copulative spines, SEM, 3 µm. (C7) Notum and spicules, SEM, 400 µm. (D). Paratype ZMMU Op-851, length 12 mm live, Iturup Island. (D1) Dorsal view. (D2) Complete radula, SEM, 100 µm. (D3) Central part of radula, SEM, 10 µm. (D4) Outer lateral teeth, SEM, 50 µm. (D5) Complete labial cuticle, SEM, 100 µm. (D6) Elements of labial cuticle, SEM, 10 µm. (D7) Spicules of notum, SEM, 50 µm. Scale bars for all living specimens are 1 mm. Photos: Tatiana Korshunova and Alexander Martynov.
Etymology. This species is named after Dmitry Vinogradov, one of the first re-inventors of porcelain in Europe, who developed Russian hard-paste porcelain. Vinogradov died at the age of 38 without receiving an honour for his important contributions.
Type materials. Holotype ZMMU Op-839, length 60 mm live, Kuril Islands, Urup Island, 23 August 2021, depth 10–20 m, coll. T.A. Korshunova, A.V. Martynov. Paratype ZMMU Op-838, length 32 mm live, Kuril Islands, Urup Island, 2 September 2021, depth 10–40 m, coll. T.A. Korshunova, A.V. Martynov. Paratype ZMMU Op-850, length 33 mm live, Kuril Islands, Iturup Island, 5 September 2022, 10–15 m, coll. T.A. Korshunova, A.V. Martynov. Paratype ZMMU Op-851, length 12 mm live, Kuril Islands, Iturup Island, 3 September 2022, 20–30 m, coll. T.A. Korshunova, A.V. Martynov.
External morphology. The notum is broad, and rounded in front and posteriorly. The rhinophores are long and retracted into raised soft sheaths bearing small tubercles (Figure 12(A1,A3,A4,B1,C1,D1)); 12–20 rhinophoral lamellae. The notum is covered with very distinct, medium-sized, rounded or irregular tubercles (Figure 12(A1,A16,A3,A4,B1,C1,C7,D1,D7)). The spicules form a sparse network in the notum. Eight to nine multipinnate gills united by a common membrane form a circle around the anus. The gills are retractable into a common gill cavity. The border of the gill cavity is moderately raised with a tuberculated edge (Figure 12(A1,A3,A4,B1,C1,D1)). The oral veil is small, trapezoid, with oblique notched lateral sides (Figure 12(A2–A4)). The foot is broad, anteriorly rounded and slightly thickened to form a double edge. The foot appears entirely (Figure 12(A2–A4)); posteriorly, it sometimes projects slightly from the notum in crawling animals, forming a rounded tail.
Colour. The notum is partly semi-transparent or almost completely opaque, from white to pale creamy to light yellow and brownish (Figure 12(A1–A4,B1,C1,D1)). The gills are somewhat paler or darker than the notum with a patchy light-yellow pigment. The rhinophores are slightly darker than the notum. The digestive gland is black-to-brownish; the stomach and intestine are partly visible through the notum dorsally (Figure 12(A1,A3,A4,B1,C1,D1)). A pinkish area (parts of the reproductive and digestive systems) may shine through the notum in front of the stomach. The internal organs can also be almost completely invisible. A range of 14–23 subepidermal yellow glands are well conspicuous dorsally and less conspicuous ventrally (some of them are placed close to each other) from each body side. The yellow lines around the notum and the yellow pigment in the dorsal tubercles are well defined in both larger and smaller specimens (Figure 12(A1–A4,B1,C1,D1)). Together with the tubercles, the yellow spots form an extensive field of yellow pigmentation. The yellow line around the foot is somewhat more dispersed but also distinct.
Buccal bulb and oral tube. The buccal bulb is relatively short, similar in length to the oral tube (Figure 12(A5)). The salivary glands are relatively long and narrow.
Jaws. The jaws take the form of a rounded labial disk covered by a yellowish-brown cuticle bearing rod-shaped labial elements with double- to triple-pointed hook-shaped tips (Figure 12(A11,C3,D5,D6)).
Radula. Radular formula ca. 60–90 × 20–40.1.20–40. The central tooth is moderately low and bears up to seven cusps (Figure 12(A7–A9,C2,D3)). The inner lateral teeth have up to 12 denticles on the outer edge, and up to four on the inner edge (Figure 12(A6–A8,C2,D2,D3)). The middle and outer teeth are comb-shaped, bearing up to ca. 40 denticles (Figure 12(A6,A10,D2,D4)).
Reproductive system. The ampulla comprises at least four thickened and convoluted compartments (Figure 7E, a). The ampulla bifurcates into very long vas deferens and oviduct. The uterine duct emerges some distance from the female gland mass. The prostatic part of the vas deferens is long, narrow, and only moderately distinct (Figure 7E, pr). The prostate transits to narrow vas deferens (Figure 7E, vd), which considerably widens towards the penial sheath that encloses the evertable ejaculatory duct (Figure 7E, psh). The copulative (penial) spines are significantly short, blunt, oval or slightly more triangular; elongated ones are absent (Figure 5 and Figure 12(A13–A15,C4–C6)). The vagina narrows (Figure 7E, v) and enters a medium-sized rounded bursa copulatrix (Figure 7E, b). The uterine duct is short and narrow (Figure 7E, ud); it begins from the female gland mass and then enters near the base of the pear-shaped receptaculum seminis (Figure 7E, rs).
Habitat. The species inhabits shallow waters with rocky and stony substrates at depths of c. 10–40 m.
Distribution. Currently, the species is only known to occur in the coastal waters of the Urup and Iturup Islands (Figure 2B); however, potential distribution includes at least the neighbouring Kuril Islands and Hokkaido Island.
Remarks. The species exhibits known chromatic patterns IX–X (Figure 3), central teeth pattern IV (Figure 4), and copulative spine patterns I and II (Figure 5). Due to the combination of a significant number of yellow spots, the consistently present distinct yellow line around the notum, the moderately low central radular teeth, the very long vas deferens, the peculiar short, rounded, with occasionally partly elongated top, copulative spines (Figure 12(A13–A15,C4–C6)), and the molecular phylogenetic data (Figure 1 and Figure 2), this new species Cadlina vinogradovi sp. nov. is distinguished from all known [32] and herein-described species of Cadlina (Figure 3, Figure 4 and Figure 5). Cadlina vinogradovi sp. nov. has a remarkable pattern of short copulative spines in the adult state, whereas most other Cadlina species of this complex commonly have elaborated, elongated, curved spines, with only a few shorter spines in addition (Figure 5). In this regard, the copulative spines of C. vinogradovi sp. nov. could correspond to an earlier ontogenetic state (and, hence, evolutionary [54]) in other Cadlina species. The intragroup distances in C. vinogradovi sp. nov. are 0.61–1.06% for the COI. The lowest COI intergroup distance of 7.75% is found between C. vinogradovi sp. nov. and C. laevis from the North Atlantic (Table 1). See also the details in Section 4 below.
4. Discussion
There are several definitions of “cryptic species” that are widely circulated in the current literature. For example, Sáez et al. [42] indicated that “These molecular differences have been taken as evidence of reproductive isolation between morphologically indistinguishable (cryptic), or only a posteriori distinguished species (pseudocryptic)”. Bickford et al. [43] outlined that “We consider two or more species to be ‘cryptic’ if they are, or have been, classified as a single nominal species because they are at least superficially morphologically indistinguishable”. The latter definition is more straightforwardly emphasized in the glossary of the same work [43]: “Cryptic species: two or more distinct species that are erroneously classified (and hidden) under one species name”. Gill et al. [60] concluded that “Cryptic species are taxa that are morphologically indistinguishable and consequently often incorrectly considered as a single nominal species when in fact constituent taxa are genetically divergent and reproductively isolated from each other”.
It is immediately apparent that all these different authors strongly referred to an “indistinguishable” morphology as the main definition. Notably, while Bickford et al.’s [43] consideration of “cryptic species” was presented in a relatively balanced way (though still with a central reference to “indistinguishability”), instead, there has been a recent trend towards a too straightforward definition of the term “cryptic species”. For example, Walters et al. [61] define the term as “those that lack distinguishing morphological characters”); Kramina et al. [11] state that “Bickford et al. (2007) defined cryptic species as two or more species which are sufficiently distinct based on molecular or other evidence, but classified as a single nominal species, because they are at least superficially morphologically indistinguishable”; and in Yin et al. [28] writes that “…cryptic species are morphologically indistinguishable…”. The tendency to overuse the term “cryptic species” is notable in that studies highly critical of the “cryptic species concept” have been cited inaccurately, without mentioning the underlying criticism of the “cryptic concept” [61,62].
Recently, the combination of molecular analysis with a phyloperiodic framework as a set of practical methods [7,29,56] has been used in order to break with those notorious indications of the “impossibility of morphological distinguishing” in closely related species complexes [43]. This approach is considerably simpler in practical use than, for example, the complex, although otherwise very useful, method of geometric morphometrics [2,63] because commonly used taxonomic characters in a given group (which are usually declared “indistinguishable”) are presented explicitly, allowing fine morphological details to be searched by eye (Figure 3, Figure 4 and Figure 5). Thus, such a phyloperiodic framework can be used by both scientists and various practitioners (for example, citizen scientists) when it is necessary to distinguish species of a given complex/group in the field. Although this is still a complicated task, we have already successfully applied the phyloperiodic approach to another complex case: the dorids of the genus Polycera [7].
This is very evident in the case of Cadlina, when the putative “chaos” of external colouration (typically presented for nudibranchs [64]) represented in ordered rows (in combination with the molecular phylogeny, Figure 1, Figure 3, Figure 4 and Figure 5), in most cases it reveals fine-scale differences, as well as the presence or absence of some particular patterns (variants) within a species (Figure 3, Figure 4 and Figure 5). For example, Cadlina vinogradovi sp. nov. has a significant number of yellow spots on the notum (Figure 12(A1,B1,C1)) compared to other species of the complex (Figure 3), although partly similar in that respect to Cadlina lomonosovi sp. nov., which commonly includes significantly more pale variants (Figure 10(A1,B1,D1,F1,G1)), but also more intensively coloured (Figure 10(E1)). Cadlina koltzovi sp. nov. has a more translucent notum (Figure 9(A1,B1,C1,D1,E1,F1)) compared to the more opaque notum in C. franklinae sp. nov. (Figure 8(A1,B1,C1,D1,E1)). Moreover, there are clear differences between C. vinogradovi sp. nov., C. koltzovi sp. nov., C. franklinae sp. nov., C. vavilovi sp. nov., as well as in Cadlina laevis, C. kamchatica, C. paninae, and C. umiushi regarding the amount and distinctness of the yellow notal line (Figure 3). Cadlina laevis, a type species of the genus Cadlina, despite showing a wide range of chromatic patterns (I, III, IV, VI, and VIII–X, Figure 3), it only presumably overlaps with several North Pacific Ocean species because forms with distinct yellow notal line are rare [32] (Figure 3). Whereas in species like C. vinogradovi sp. nov., C. lomonosovi sp. nov., C. bellburnellae sp. nov., and C. umiushi instead, a distinct yellow notal line is very common (Figure 3). In its turn, Cadlina lomonosovi sp. nov. and C. bellburnellae sp. nov. are more similar to C. umiushi, but the latter species invariably has a very distinct yellow notal line, whereas C. lomonosovi sp. nov. and Cadlina bellburnellae sp. nov. sp. may include forms with a less distinct notal yellow line, and also generally have more yellow spots on the notum than C. umiushi (Figure 3). Cadlina franklinae sp. nov. and Cadlina paninae so far have not been revealed as commonly having distinct yellow notal lines [32] (Figure 3). These apparently subjective qualitative characteristics can be objectively represented within a phyloperiodic framework (Figure 3) and can then significantly facilitate species identification.
We conclude that the incorporation of morphological characters in the PhyloPeriodic Tables is a practical method for detecting diagnosable characters within apparently “cryptic species”. A molecular phylogeny allows ordering the morphological characters in a multilevel framework [7,29,65] (Figure 3, Figure 4 and Figure 5). While the resolved phylogeny provides supported hypotheses of monophyly (Figure 1), the use of the phyloperiodic framework provides a more rigorous study of the morphology (Figure 3, Figure 4, Figure 5, Figure 6, Figure 7, Figure 8, Figure 9, Figure 10, Figure 11 and Figure 12). Thus, the periodic framework is not just a theoretical addition to existing practical methods, but also a key element for truly integrative study. Currently, in applying the controversial concept of “cryptic species”, a substantial part of the diversity remains unnamed [66], whereas the phyloperiodic approach and multilevel organismal diversity (MOD) method allow us to name each particular small unit of diversity. This is a very important practical achievement (including conservation biology) that immediately distinguishes the phyloperiodic and MOD approaches from the notion of “integrative taxonomy” since the latter does not necessarily recognize morphodiversity and does not necessarily allow all molecularly distinct groups to be named [34,44]. Even taking into account all biological irregularities, the phyloperiodic representation greatly streamlines any further potential variations that may be discovered and significantly facilitates species identification. These features are directly related to predictability, which is an essential characteristic of any periodic system [57].
Furthermore, naming as small units of biodiversity as possible using the phyloperiodic approach is very important for other fields such as biogeography and climatology, and directly relevant to a key recent issue such as global warming [67,68,69]. For example, in this study we discovered very interesting patterns associated with the overall distribution of sea temperatures: Cadlina franklinae sp. nov. is restricted so far only to the Urup and Chirpoy Islands, the Middle Kuril Islands (Figure 2), where at the depth of 10 m the mean annual temperature is +3.64 °C … +5.64 °C [70], whereas C. lomonosovi sp. nov. sp. and C. bellburnellae sp. nov. were found only at the most southern Kuril Islands, Iturup and Kunashir (Figure 2), where at the depth of 10 m the mean annual temperature is +5.62 °C …+8.88 °C [70], thus, the average ca. is 1.5 times higher than in Urup Island. Cadlina vinogradovi and Cadlina koltozvi sp. nov. in turn occur both in cold water (Urup, Chirpoy) and temperate water (Iturup, Kunashir) in the Kuril Islands (Figure 2). We have detailed sampling for two years, so we can say with confidence that these patterns are not due to insufficient collection efforts since, given the diversity of locations around the islands and the number of specimens, we have never found C. franklinae sp. nov. further south than Urup Island, and we have never found C. lomonosovi sp. nov. and C. bellburnellae sp. nov. further north than the Iturup and Kunashir Islands. Cadlina vavilovi sp. nov. occurs at Iturup Island (Figure 2) in a deeper location, c. 250 m, with mean annual temperature at this depth considerably low, +1.0 °C …+1.93 °C [70]. The +12 °C isotherm in the south direction was indicated precisely at the borders of the Urup and Iturup Islands [71].
Thus, the fine-scale morphological, molecular, and biogeographical differences in this species complex identified in the present study are in broad agreement with the well established geophysical data. All these environmental peculiarities, coupled with the direct development known in the related species C. laevis [32], may imply similar modes of development in other species in this complex and therefore limited dispersal ability. These processes may contribute to the formation of a significant amount of fine-scale species-level diversity. These interesting patterns can be used in planning conservation activities since the natural reserve on the Kuril Islands is planned to be expanded to all the islands [31]. Furthermore, due to the ongoing global warming, it will be important to check if the ranges of colder water species in this complex, such as C. franklinae sp. nov., will shift southward, whereas ranges of more temperate species, like C. lomonosovi sp. nov. sp. and C. bellburnellae sp. nov., will shift northward instead. Without the coherent morphological and molecular framework presented here using the phyloperiodic approach, species-related information of such importance to biogeography, climatology, and conservation would simply be lost in the “cryptic species” approach, which lumps all the diversity into a single “undistinguishable” assemblage.
Ultimately, the phyloperiodic framework removes the term “cryptic species”. In this study, we show this explicitly: within the uncovered diversity of the genus Cadlina, which is supposedly “completely cryptic”, we can indicate, for example, that C. vinogradovi sp. nov. readily distinguishes from all other new species described here (C. koltzovi sp. nov., C. franklinae sp. nov. sp., C. vavilovi sp. nov., C. lomonosovi sp. nov., and C. bellburnellae sp. nov.), as well as the previously described Cadlina laevis, C. kamchatica [32], C. koreana (external and radular drawings on Figure 2, Figure 3 and Figure 4 for the latter species are based on [72]), C. paninae, and C. umiushi [32] based on the shape of copulative spines (Figure 5 and Figure 12(A13–A15)). However, the radular morphology (central teeth) and external features of C. vinogradovi sp. nov. remain “cryptic” (in the currently accepted erroneous meaning) compared to the above-listed species (Figure 4 and Figure 12(A8,C2,D3))). In turn, Cadlina koltzovi sp. nov. is well distinguished from all of the species listed, with the exception of C. koreana, by relatively high central teeth of radula (Figure 4 and Figure 9(A5,A6,D6,F2)), but remains “cryptic” by the external features and copulative spines (Figure 3, Figure 5 and Figure 9(A1,B1,D1,E1,F1)). Several more species, like Cadlina bellburnellae sp. nov., may also include relatively high central teeth, but they differ from other species by the fine-scale details of the central teeth (Figure 4). The type species of the genus Cadlina, C. laevis, which also belongs to the studied clade (Figure 1), externally includes almost all colour varieties, which can be found in any other member of this clade (Figure 1), even such distinct species such as C. umiushi and C. kamchatica (Figure 3), and according to the radula and copulative spines, C. laevis is “cryptic” to the majority of these species except Cadlina vinogradovi sp. nov., C. koltzovi sp. nov., and C. koreana (Figure 3 and Figure 4). Cadlina franklinae sp. nov., C. paninae, and C. vavilovi sp. nov. show more subtle differences in external, radular, and copulative spine morphology (Figure 3, Figure 4 and Figure 5), and compared to some variants of Cadlina laevis, but not all of them (Figure 3). The North Atlantic Cadlina laevis clearly does not overlap the range of the central radular teeth and copulative spines with the North Pacific Ocean C. vinogradovi sp. nov., C. koltzovi sp. nov., C. vavilovi sp. nov., C. lomonosovi sp. nov., and C. bellburnellae sp. nov. (Figure 4 and Figure 5).
This series of comparisons can be continued, but it is already possible to point out a key consideration for the crucial problem of “cryptic species”. We can designate this complex as “cryptic”, but then some of these species exhibit distinct internal features (Figure 4, Figure 5, Figure 9(A5,A6,D6,F2) and Figure 12(A13–A15)). We may attempt to designate some of these species as “cryptic” by external features, e.g., Cadlina franklinae sp. nov. and C. vavilovi sp. nov., but the type species Cadlina laevis will include some “cryptic” specimens and some “non-cryptic” specimens (Figure 3). Then, we can designate several species in this complex as “cryptic” in terms of internal features, whereas Cadlina vinogradovi sp. nov. and C. koltzovi sp. nov. can be designated as “non-cryptic”. However, according to external features, Cadlina vinogradovi sp. nov. and C. koltzovi sp. nov. will remain “cryptic”, and C. koltzovi sp. nov. will also remain “cryptic” by radular morphology. In addition, most of the species newly described here, as well as Cadlina umiushi, can be externally considered as cryptic to Cadlina luteomarginata and Cadlina klasmalmbergi, which definitely belong to other clades and are easily distinguished by radular and reproductive characters [32]. Considering that different species from this complex have various degrees of molecular–phylogenetic distances (Figure 1 and Figure 2) and morphological distinctness (Figure 4 and Figure 5), a truly multilevel system of fine-scale differences and similarities is revealed (Figure 3, Figure 4 and Figure 5). This is very instructive because when comparing the horizontal rows of the phyloperodic tables IX–XI containing the larger, darker, and ontogenetically more advanced specimens, the fine-scale differences in the colouration patterns between most of the species become evident (Figure 3).
Therefore, to designate any of these species as “cryptic”, “pseudocryptic”, or non-cryptic would be completely arbitrary, non-operational, and generally meaningless. By this, we finally strongly propose that the concept of “crypticity” and the term “cryptic species” be removed from the arsenal of modern biology, including phylogeny and taxonomy. These terms no longer help to reveal the enormous biological diversity, but, on the contrary, significantly hinder its further exploration. Prospects for other groups of organisms, which do not always have such complex morphological features as nudibranchs, e.g., protists [42], should not be used as a justification of the rationale for the term “cryptic species”. This is also due to the fact that any statements about the “morphologically indistinguishable species” have no biological basis since there cannot be two completely identical biological organisms, even twins [5,7,73], so “cryptic twin species” should not be excluded from that strict biological rule. There is growing dissatisfaction with the concept of “cryptic species” in various studies of disparate organismal groups, such as plants and animals [3,6,8,26,74,75], and more recently species for which morphological data can be provided have been highlighted as “non-cryptic” [76]. There are also a number of studies [77,78,79,80,81,82,83,84] in which the more neutral term “hidden diversity” has already been used, which does not overlap semantically with “cryptic species” in its original meaning: “well camouflaged on a substrate and in an environment” [7,9,85,86,87]. However, there is also a confusing mixed use of both “cryptic” and “hidden” terms [88,89,90,91,92].
Rather than continuing to rely on the straightforward notion of “crypticity”, the proposed multilevel periodic-like framework is able to provide a reliable method for displaying and distinguishing even very subtle differences, both external and internal, within a given Cadlina species complex (Figure 1, Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 7, Figure 8, Figure 9, Figure 10, Figure 11 and Figure 12). The phyloperiodic approach makes it possible to perform a “granular”, fine-scale comparison of every specimen, whereas the “cryptic species approach” instead generally asserts that some species morphologically are completely “indistinguishable” [42]. This is consistent with the growing understanding that the conventional “species concept” represents a considerable overgeneralization and that more organism-focused approaches need to be implemented [29]. Potentially, the phyloperiodic approach can be expanded to more features, but this is definitely a task for the next studies, as even the current framework is quite complex to implement. If new material becomes available, future studies will test the framework developed here, and, for example, more periodic rows and columns can be recognized. This approach can be universally applied to any other groups of organisms, using appropriate diagnostic characters in the ontogenetic framework of each given group in combination with molecular phylogenetic data. The periodic approach in various forms opens up a new avenue for the further development of various practical applications in biology and has begun to be applied, for example, in the fields of ecology [93,94] or biomolecules [95]. Significant efforts remain to be made in the taxonomy to ensure widespread acceptance of the periodic approach. This is especially important since systematics is the inevitable basis for all other areas of biology.
The traditional, still widely used, rigid taxonomic diagnoses need to be gradually replaced with the fine-scale multilevel presentation (Figure 3, Figure 4 and Figure 5), and the most recent studies are in line with this emerging agenda [74,96]. The patterns of natural biodiversity have not been established according to pre-evolutionary, strict diagnosis-based “box-thinking”, but instead reflect the most complex, dynamic, constantly evolved biological organisms.
Thus, one of the most important messages of this study for any taxonomic, phylogenetic, and biodiversity studies is the following: without the phyloperiodic framework and multilevel understanding of biological diversity, all numerous and existing, fine-scale diversity units will be just considered as a chaotic “cryptic species”.
5. Conclusions
A morphologically difficult-to-distinguish species complex of dorid of the genus Cadlina was discovered in the little-studied Kuril Islands in the North Pacific Ocean and used as a model system to show persistent pitfalls in the common usage of the “cryptic species” concept.
Analysing the diagnostic characters among the newly described and related species in the phyloperiodic framework, it is clearly shown that dividing them into “cryptic” and “non-cryptic” taxa is completely arbitrary and meaningless.
Applying our novel data and analysing the current literature on the widely cited problem of “cryptic species” for a wide range of different groups of organisms, we strongly propose that the notion of “crypticity” and the term “cryptic species” be excluded from the arsenal of modern biology. This is because the concept misleadingly divides overwhelming biological diversity into “cryptic” and “non-cryptic” components and, rather than emphasizing the multilevel nature of organismal diversity, artificially reduces it to two simple, supposedly contrasting modes.
The implications of the present study are directly relevant to studying the diversity, phylogeny, biogeography, conservation, and many other aspects of any group of organisms.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d16040220/s1, Table S1. PCR amplification options. Table S2. GenBank accession numbers and references for all sequences used in this study.
Author Contributions
T.K. and A.M. worked on the study conceptualization and fieldwork, contributed resources, conducted analyses, and wrote the original draft. T.K. and A.M. contributed to discussions and reviewed and edited the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The work of AM was performed under a research project of the MSU Zoological Museum (18-1-21 No. 121032300105-0). The work of TK was conducted under the Koltzov Institute of Developmental Biology RAS (IDB) basic research program in 2024 (No. 0088-2024-0011). The research was conducted using equipment of the Core Centrum of IDB RAS. This study was supported by the project “ReDNAcarnation of the museum collections” from the Vladimir Potanin Philanthropic Foundation (the program “Museum without borders”, “Museum 4.0” contest).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Data from this study are available in the main text and the Supplementary Materials.
Acknowledgments
We are sincerely grateful to the organizers and participants of the expeditions of the Russian Geographical Society to the Kuril Islands in 2021 and 2022 and, particularly, to Anton Iurmanov and Anatoly Kalemberg. Three anonymous reviewers helped to improve the manuscript. The Electron Microscopy Laboratory, Moscow State University, is gratefully acknowledged for providing scanning electron microscopy support. The SEM work was conducted in the Shared Research Facility “Electron microscopy in life sciences” using Unique Equipment “Three-dimensional electron microscopy and spectroscopy” (instruments CamScan Series II, JSM 6380, and QuattroS).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Karanovic, T.; Djurakic, M.; Eberhard, S.M. Cryptic species or inadequate taxonomy? Implementation of 2D geometric morphometrics based on integumental organs as landmarks for delimitation and description of copepod taxa. Syst. Biol. 2016, 65, 304–327. [Google Scholar] [CrossRef] [PubMed]
- Zúñiga-Reinoso, A.; Benítez, H.A. The overrated use of the morphological cryptic species concept: An example with Nyctelia darkbeetles (Coleoptera: Tenebrionidae) using geometric morphometrics. Zool. Anz. 2015, 255, 47–53. [Google Scholar] [CrossRef]
- Heethoff, M. Cryptic species—Conceptual or terminological chaos? A response to Struck et al. Trends Ecol. Evol. 2018, 33, 310. [Google Scholar] [CrossRef]
- Korshunova, T.A.; Martynov, A.V.; Bakken, T.; Picton, B.E. External diversity is restrained by internal conservatism: New nudibranch mollusc contributes to the cryptic species problem. Zool. Scr. 2017, 46, 683–692. [Google Scholar] [CrossRef]
- Korshunova, T.A.; Picton, B.; Furfaro, G.; Mariottini, P.; Pontes, M.; Prkić, J.; Fletcher, K.; Malmberg, K.; Lundin, K.; Martynov, A. Multilevel fine scale diversity challenges the ‘cryptic species’ concept. Sci. Rep. 2019, 9, 6732. [Google Scholar] [CrossRef] [PubMed]
- Horsáková, V.; Nekola, J.C.; Horsák, M. When is a “cryptic” species not a cryptic species: A consideration from the Holarctic micro-landsnail genus Euconulus (Gastropoda: Stylommatophora) deciphering “cryptic” pyramidula species. Mol. Phylogenetics Evol. 2019, 132, 307–320. [Google Scholar] [CrossRef]
- Korshunova, T.A.; Driessen, F.; Picton, B.; Martynov, A.V. The multilevel organismal diversity approach deciphers difficult to distinguish nudibranch species complex. Sci. Rep. 2021, 11, 18323. [Google Scholar] [CrossRef]
- Horsáková, V.; Líznarová, E.; Razkin, O.; Nekola, J.C.; Horsák, M. Deciphering “cryptic” nature of European rock-dwelling Pyramidula snails (Gastropoda: Stylommatophora). Contrib. Zool. 2022, 91, 233–260. [Google Scholar] [CrossRef]
- Minelli, A. Two-way exchanges between animal and plant biology, with focus on evo-devo. Front. Ecol. Evol. 2022, 10, 1057355. [Google Scholar] [CrossRef]
- Macleod, N.; Canty, R.J.; Polaszek, A. Morphology-based identification of Bemisia tabaci cryptic species puparia via embedded group-contrast convolution neural network analysis. Syst. Biol. 2022, 71, 1095–1109. [Google Scholar] [CrossRef]
- Kramina, T.E.; Samigullin, T.H.; Meschersky, I.G. Two cryptic species of Lotus (Fabaceae) from the Iberian Peninsula. Wulfenia 2020, 27, 21–45. [Google Scholar]
- Brandão-Dias, P.F.P.; Zhang, Y.M.; Pirro, S.; Vinson, C.C.; Weinersmith, K.L.; Ward, A.K.G.; Forbes, A.A.; Egan, S.P. Describing biodiversity in the genomics era: A new species of Nearctic Cynipidae gall wasp and its genome. Syst. Entomol. 2022, 47, 94–112. [Google Scholar] [CrossRef]
- Christmas, M.J.; Jones, J.C.; Olsson, A.; Wallerman, O.; Bunikis, I.; Kierczak, M.; Peona, V.; Whitley, K.M.; Larva, T.; Suh, A.; et al. Genetic barriers to historical gene flow between cryptic species of alpine bumblebees revealed by comparative population genomics. Mol. Biol. Evol. 2021, 38, 3126–3143. [Google Scholar] [CrossRef] [PubMed]
- Calahorra-Oliart, A.; Ospina-Garcés, S.M.; León-Paniagua, L. Cryptic species in Glossophaga soricina (Chiroptera: Phyllostomidae): Do morphological data support molecular evidence? J. Mammal. 2021, 102, 54–68. [Google Scholar] [CrossRef]
- Chan, K.O.; Hutter, C.R.; Wood, P.L.; Su, Y.C.; Brown, R.M. Gene flow increases phylogenetic structure and inflates cryptic species estimations: A case study on widespread Philippine puddle frogs (Occidozyga laevis). Syst. Biol. 2021, 71, 40–57. [Google Scholar] [CrossRef] [PubMed]
- Clavero-Camacho, I.; Palomares-Rius, J.E.; Cantalapiedra-Navarrete, C.; León-Ropero, G.; Martín-Barbarroja, J.; Archidona-Yuste, A.; Castillo, P. Integrative taxonomy reveals hidden cryptic diversity within pin nematodes of the genus Paratylenchus (Nematoda: Tylenchulidae). Plants 2021, 10, 1454. [Google Scholar] [CrossRef] [PubMed]
- Connolly, J.B.; Romeis, J.; Devos, Y.; Glandorf, D.C.; Turner, G.; Coulibaly, M.B. Gene drive in species complexes: Defining target organisms. Trends Biotechnol. 2022, 41, 154–164. [Google Scholar] [CrossRef]
- Costa-Araújo, R.; Silva, J.S.; Boubli, J.P.; Rossi, R.V.; Canale, G.R.; Melo, F.R.; Bertuol, F.; Silva, F.E.; Silva, D.A.; Nash, S.D.; et al. An integrative analysis uncovers a new, pseudo-cryptic species of Amazonian marmoset (Primates: Callitrichidae: Mico) from the arc of deforestation. Sci. Rep. 2021, 11, 15665. [Google Scholar] [CrossRef] [PubMed]
- Irisarri, I.; Darienko, T.; Pröschold, T.; Fürst-Jansen, J.M.R.; Jamy, M.; de Vries, J. Unexpected cryptic species among streptophyte algae most distant to land plants. Proc. R. Soc. B Biol. Sci. 2021, 288, 20212168. [Google Scholar] [CrossRef]
- Maggia, M.E.; Decaëns, T.; Lapied, E.; Dupont, L.; Roy, V.; Schimann, H.; Orivel, J.; Murienne, J.; Baraloto, C.; Cottenie, K.; et al. At each site its diversity: DNA barcoding reveals remarkable earthworm diversity in neotropical rainforests of French Guiana. Appl. Soil Ecol. 2021, 164, 103932. [Google Scholar] [CrossRef]
- Majtyka, T.; Borczyk, B.; Ogielska, M.; Stöck, M. Morphometry of two cryptic tree frog species at their hybrid zone reveals neither intermediate nor transgressive morphotypes. Ecol. Evol. 2022, 12, e8527. [Google Scholar] [CrossRef] [PubMed]
- Parsons, D.J.; Pelletier, T.A.; Wieringa, J.G.; Duckett, D.J.; Carstens, B.C. Analysis of biodiversity data suggest that mammal species are hidden in predictable places. Proc. Natl. Acad. Sci. USA 2022, 119, e2103400119. [Google Scholar] [CrossRef] [PubMed]
- Pérez, M.; Fernández-Míguez, M.; Matallanas, J.; Lloris, D.; Presa, P. Phylogenetic prospecting for cryptic species of the genus Merluccius (Actinopterygii: Merlucciidae). Sci. Rep. 2021, 11, 5929. [Google Scholar] [CrossRef] [PubMed]
- Yuzefovich, A.P.; Artyushin, I.V.; Kruskop, S.V. Not the cryptic species: Diversity of Hipposideros gentilis (Chiroptera: Hipposideridae) in Indochina. Diversity 2021, 13, 218. [Google Scholar] [CrossRef]
- Pfingstl, T.; Schäffer, S.; Bardel-Kahr, I.; Baumann, J. A closer look reveals hidden diversity in the intertidal Caribbean Fortuyniidae (Acari, Oribatida). PLoS ONE 2022, 17, e0268964. [Google Scholar] [CrossRef] [PubMed]
- Pyron, R.A.; Beamer, D.A. Nomenclatural solutions for diagnosing ‘cryptic’ species using molecular and morphological data facilitate a taxonomic revision of the Black-bellied Salamanders (Urodela, Desmognathus ‘quadramaculatus’) from the southern Appalachian Mountains. Bionomina 2022, 27, 1–43. [Google Scholar] [CrossRef]
- Semedo, T.B.F.; Da Silva, M.N.F.; Carmignotto, A.P.; Rossi, R.V. Three new species of spiny mice, genus Neacomys Thomas, 1900 (Rodentia: Cricetidae), from Brazilian Amazonia. Syst. Biodivers. 2021, 19, 1113–1134. [Google Scholar] [CrossRef]
- Yin, Y.; Yao, L.; Hu, Y.; Shao, Z.; Hong, X.; Hebert, P.D.N.; Xue, X. DNA barcoding uncovers cryptic diversity in minute herbivorous mites (Acari, Eriophyoidea). Mol. Ecol. Res. 2022, 22, 1986–1998. [Google Scholar] [CrossRef] [PubMed]
- Martynov, A.V.; Korshunova, T.A. Multilevel Organismal diversity in an Ontogenetic Framework as a Solution for the Species Concept. In Cryptic Species Morphological Stasis, Circumscription, and Hidden Diversity; Monro, A.K., Mayo, S.J., Eds.; Cambridge University Press: Cambridge, UK, 2022; pp. 78–129. [Google Scholar] [CrossRef]
- Korshunova, T.A.; Sanamyan, N.P.; Sanamyan, K.E.; Bakken, T.; Lundin, K.; Fletcher, K.; Martynov, A.V. Biodiversity hotspot in cold waters: A review of the genus Cuthonella with descriptions of seven new species (Mollusca, Nudibranchia). Contrib. Zool. 2021, 90, 216–283. [Google Scholar] [CrossRef]
- Korshunova, T.A.; Martynov, A.V. Increased information on biodiversity from the neglected part of the North Pacific contributes to the understanding of phylogeny and taxonomy of nudibranch molluscs. Can. J. Zool. 2022, 100, 436–451. [Google Scholar] [CrossRef]
- Korshunova, T.A.; Fletcher, K.; Picton, B.; Lundin, K.; Kashio, S.; Sanamyan, N.; Sanamyan, K.; Padula, V.; Schroedl, M.; Martynov, A. The Emperor Cadlina, hidden diversity and gill cavity evolution: New insights for taxonomy and phylogeny of dorid nudibranchs (Mollusca: Gastropoda). Zool. J. Linn. Soc. 2020, 189, 762–827. [Google Scholar] [CrossRef]
- Gómez Daglio, L.; Dawson, M.N. Integrative taxonomy: Ghosts of past, present and future. J. Mar. Biol. Assoc. UK 2019, 99, 1237–1246. [Google Scholar] [CrossRef]
- Korshunova, T.; Grøtan, V.V.; Johnson, K.B.; Bakken, T.; Picton, B.E.; Martynov, A. Similar ones are not related and vice versa—New Dendronotus taxa (Nudibranchia: Dendronotidae) from the North Atlantic ocean provide a platform for discussion of global marine biodiversity patterns. Diversity 2023, 15, 504. [Google Scholar] [CrossRef]
- Taylor, W.R. A ‘periodic table’ for protein structures. Nature 2002, 416, 657–660. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.J.; Hassan, M.; Jernigan, R.L.; Jia, K.; Kihara, D.; Kloczkowski, A.; Kotelnikov, S.; Kozakov, D.; Liang, J.; Liwo, A.; et al. Protein folds vs. protein folding: Differing questions, different challenges. Proc. Natl. Acad. Sci. USA 2023, 120, e2214423119. [Google Scholar] [CrossRef]
- Wang, S.S.; Ellington, A.D. Pattern generation with nucleic acid chemical reaction networks. Chem. Rev. 2019, 119, 6370–6383. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Yanai, I. A periodic table of cell types. Development 2019, 146, dev169854. [Google Scholar] [CrossRef]
- Gante, H.F. How fish get their stripes again and again. Science 2018, 362, 396–397. [Google Scholar] [CrossRef]
- Inaba, M.; Chuong, C.M. Avian pigment pattern formation: Developmental control of macro- (across the body) and micro- (within a feather) level of pigment patterns. Front. Cell Dev. Biol. 2020, 8, 620. [Google Scholar] [CrossRef]
- Freudstein, J.V.; Broe, M.B.; Folk, R.A.; Sinn, B.T. Biodiversity and the species concept—Lineages are not enough. Syst. Biol. 2017, 66, 644–656. [Google Scholar] [CrossRef]
- Sáez, A.G.; Probert, I.; Geisen, M.; Quinn, P.; Young, J.R.; Medlin, L.K. Pseudo-cryptic speciation in coccolithophores. Proc. Natl. Acad. Sci. USA 2003, 100, 7163–7168. [Google Scholar] [CrossRef] [PubMed]
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 2007, 22, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Fišer, C.; Robinson, C.T.; Malard, F. Cryptic species as a window into the paradigm shift of the species concept. Mol. Ecol. 2018, 27, 613–635. [Google Scholar] [CrossRef] [PubMed]
- Jörger, K.M.; Schrödl, M. How to describe a cryptic species? Practical challenges of molecular taxonomy. Front. Zool. 2013, 10, 59. [Google Scholar] [CrossRef] [PubMed]
- Cerca, J.; Meyer, C.; Purschke, G.; Struck, T.H. Delimitation of cryptic species drastically reduces the geographical ranges of marine interstitial ghost-worms (Stygocapitella; Annelida, Sedentaria). Mol. Phyl. Evol. 2020, 143, 106663. [Google Scholar] [CrossRef] [PubMed]
- Minelli, A. Taxonomy needs pluralism, but a controlled and manageable one. Megataxa 2020, 1, 9–18. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis Version 11. Mol. Biol. Evol. 2012, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008, 75, 758–771. [Google Scholar] [CrossRef]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble species by automatic partitioning. Mol. Ecol. Resour. 2021, 21, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Korshunova, T.A.; Martynov, A.; Bakken, T.; Evertsen, J.; Fletcher, K.; Mudianta, I.W.; Saito, H.; Lundin, K.; Schrödl, M.; Picton, B. Polyphyly of the traditional family Flabellinidae affects a major group of Nudibranchia: Aeolidacean taxonomic reassessment with descriptions of several new families, genera, and species (Mollusca, Gastropoda). ZooKeys 2017, 717, 1–139. [Google Scholar] [CrossRef] [PubMed]
- Martynov, A.V.; Lundin, K.; Korshunova, T.A. Ontogeny, phylotypic periods, paedomorphosis, and ontogenetic systematics. Front. Ecol. Evol. 2022, 10, 806414. [Google Scholar] [CrossRef]
- Martynov, A.V.; Korshunova, T.A. A new deep-sea genus of the family Polyceridae (Nudibranchia) possesses a gill cavity, with implications for cryptobranch condition and a ‘Periodic Table’ approach to taxonomy. J. Molluscan Stud. 2015, 81, 365–379. [Google Scholar] [CrossRef]
- Korshunova, T.A.; Malmberg, K.; Prkić, J.; Petani, A.; Fletcher, K.; Lundin, K.; Martynov, A. Fine-scale species delimitation: Speciation in process and periodic patterns in nudibranch diversity. ZooKeys 2020, 917, 15–50. [Google Scholar] [CrossRef] [PubMed]
- Scerri, E. Recent attempts to change the periodic table. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2020, 378, 20190300. [Google Scholar] [CrossRef] [PubMed]
- Martynov, A.V. From ‘tree-thinking’ to ‘cycle-thinking’: Ontogenetic systematics of nudibranch molluscs. Thalassas 2011, 27, 193–224. [Google Scholar]
- Ekimova, I.; Valdés, Á.; Stanovova, M.; Mikhlina, A.; Antokhina, T.; Neretina, T.; Chichvarkhina, O.; Schepetov, D. Connected across the ocean: Taxonomy and biogeography of deep water Nudibranchia from the Northwest Pacific reveal trans Pacific links and two undescribed species. Org. Divers. Evol. 2021, 21, 753–782. [Google Scholar] [CrossRef]
- Gill, B.A.; Kondratieff, B.C.; Casner, K.L.; Encalada, A.C.; Flecker, A.S.; Gannon, D.G.; Ghalambor, C.K.; Guayasamin, J.M.; Poff, N.L.; Simmons, M.P.; et al. Cryptic species diversity reveals biogeographic support for the “mountain passes are higher in the tropics” hypothesis. Proc. R. Soc. B Biol. Sci. 2016, 283, 20160553. [Google Scholar] [CrossRef] [PubMed]
- Walters, A.D.; Cannizzaro, A.G.; Trujillo, D.A.; Berg, D.J. Addressing the Linnean shortfall in a cryptic species complex. Zool. J. Linn. Soc. 2021, 192, 277–305. [Google Scholar] [CrossRef]
- Golo, R.; Vergés, A.; Díaz-Tapia, P.; Cebrian, E. Implications of taxonomic misidentification for future invasion predictions: Evidence from one of the most harmful invasive marine algae. Mar. Pollut. Bull. 2023, 191, 114970. [Google Scholar] [CrossRef] [PubMed]
- Karanovic, T.; Bláha, M. Taming extreme morphological variability through coupling of molecular phylogeny and quantitative phenotype analysis as a new avenue for taxonomy. Sci. Rep. 2019, 9, 2429. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, C.G.; Rauch, C.; Pola, M.; Malaquias, M. Integrative taxonomy reveals a cryptic species of the nudibranch genus Polycera (Polyceridae) in European waters. J. Biol. Assoc. UK. 2020, 100, 733–752. [Google Scholar] [CrossRef]
- Korshunova, T.A.; Bakken, T.; Grøtan, V.; Johnson, K.; Lundin, K.; Martynov, A.V. A synoptic review of the family Dendronotidae (Mollusca: Nudibranchia): A multilevel organismal diversity approach. Contrib. Zool. 2020, 90, 93–153. [Google Scholar] [CrossRef]
- Maroni, P.J.; Wilson, N.G. Multiple Doris “kerguelenensis” (Nudibranchia) species span the Antarctic Polar Front. Ecol. Evol. 2022, 12, e9333. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.; Sato, M.; Ruedy, R.; Lo, K.; Lea, D.W.; Medina-Elizade, M. Global temperature change. Proc. Natl. Acad. Sci. USA 2006, 103, 14288–14293. [Google Scholar] [CrossRef] [PubMed]
- Eyring, V.; Cox, P.M.; Flato, G.M.; Gleckler, P.J.; Abramowitz, G.; Caldwell, P.; Collins, W.D.; Gier, B.K.; Hall, A.D.; Hoffman, F.M.; et al. Taking climate model evaluation to the next level. Nat. Clim. Change 2019, 9, 102–110. [Google Scholar] [CrossRef]
- Supran, G.; Rahmstorf, S.; Oreskes, N. Assessing ExxonMobil’s global warming projections. Science 2023, 379, eabk0063. [Google Scholar] [CrossRef] [PubMed]
- Japan Oceanographic Data Center Portal. 2023. Available online: https://jdoss1.jodc.go.jp/vpage/bts.html (accessed on 9 February 2023).
- Harada, N.; Katsuki, K.; Nakagawa, M.; Matsumoto, A.; Seki, O.; Addison, J.A.; Finney, B.P.; Sato, M. Holocene sea surface temperature and sea ice extent in the Okhotsk and Bering Seas. Prog. Oceanogr. 2014, 126, 242–253. [Google Scholar] [CrossRef]
- Do, T.D.; Jung, D.-W.; Kil, H.-J.; Kim, C.-B. A report of a new species and new record of Cadlina (Nudibranchia, Cadlinidae) from South Korea. ZooKeys 2020, 996, 1–18. [Google Scholar] [CrossRef]
- Fraga, M.F.; Ballestar, E.; Paz, M.F.; Ropero, S.; Setien, F.; Ballestar, M.L.; Heine-Sucer, D.; Cigudosa, J.C.; Urioste, M.; Benitez, J.; et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. USA 2005, 26, 10604–10609. [Google Scholar] [CrossRef] [PubMed]
- Gibert, A.; Louty, F.; Buscail, R.; Baguette, M.; Schatz, B.; Bertrand, J.A.M. Extracting quantitative information from images taken in the wild: A case study of two vicariants of the Ophrys aveyronensis species complex. Diversity 2022, 14, 400. [Google Scholar] [CrossRef]
- Silva, T.S.R.; Hamer, M.T.; Guénard, B. A checklist of Nylanderia (Hymenoptera: Formicidae: Formicinae) from Hong Kong and Macao SARs, with an illustrated identification key for species in Southeast China and Taiwan. Zootaxa 2023, 5301, 501–539. [Google Scholar] [CrossRef] [PubMed]
- González-Wevar, C.A.; Segovia, N.I.; Rosenfeld, S.; Maturana, C.S.; Jeldres, V.; Pinochet, R.; Saucède, T.; Morley, S.A.; Brickle, P.; Wilson, N.G.; et al. Seven snail species hidden in one: Biogeographic diversity in an apparently widespread periwinkle in the Southern Ocean. J. Biogeogr. 2022, 49, 1521–1534. [Google Scholar] [CrossRef]
- Lindeque, P.K.; Parry, H.E.; Harmer, R.A.; Somerfield, P.J.; Atkinson, A. Next Generation sequencing reveals the hidden diversity of zooplankton assemblages. PLoS ONE 2013, 8, e81327. [Google Scholar] [CrossRef] [PubMed]
- Korshunova, T.A.; Martynov, A.V. Consolidated data on the phylogeny and evolution of the family Tritoniidae (Gastropoda: Nudibranchia) contribute to genera reassessment and clarify the taxonomic status of the neuroscience models Tritonia and Tochuina. PLoS ONE 2020, 15, e0242103. [Google Scholar] [CrossRef]
- Edwards, D.; Morris, J.L.; Richardson, J.B.; Kenrick, P. Cryptospores and cryptophytes reveal hidden diversity in early land floras. New Phytol. 2014, 202, 50–78. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Stepanauskas, R.; Moran, N.A. Hidden diversity in honey bee gut symbionts detected by single-cell genomics. PLoS Genet. 2014, 10, e1004596. [Google Scholar] [CrossRef]
- Muggia, L.; Leavitt, S.; Barreno, E. The hidden diversity of lichenised Trebouxiophyceae (Chlorophyta). Phycologia 2018, 57, 503–524. [Google Scholar] [CrossRef]
- Schulz, F.; Alteio, L.; Goudeau, D.; Ryan, E.M.; Yu, F.B.; Malmstrom, R.R.; Blanchard, J.; Woyke, T. Hidden diversity of soil giant viruses. Nature Comm. 2018, 9, 4881. [Google Scholar] [CrossRef]
- Santamaría, R.I.; Bustos, P.; Van Cauwenberghe, J.; González, V. Hidden diversity of double-stranded DNA phages in symbiotic Rhizobium species. Philos. Trans. R. Soc. B Biol. Sci. 2021, 377, 20200468. [Google Scholar] [CrossRef] [PubMed]
- Ampai, N.; Rujirawan, A.; Yodthong, S.; Termprayoon, K.; Stuart, B.L.; Wood, P.L., Jr.; Aowphol, A. Hidden diversity of rock geckos within the Cnemaspis siamensis species group (Gekkonidae, Squamata): Genetic and morphological data from southern Thailand reveal two new insular species and verify the phylogenetic affinities of C. chanardi and C. kamolnorranathi. ZooKeys 2022, 1125, 115–158. [Google Scholar] [CrossRef] [PubMed]
- Claridge, A.W.; Mifsud, G.; Dawson, J.; Saxon, M.J. Use of infrared digital cameras to investigate the behaviour of cryptic species. Wildl. Res. 2004, 31, 645–650. [Google Scholar] [CrossRef]
- Willan, R.C. The taxonomy of two host-specific, cryptic dendronotoid nudibranch species (Mollusca: Gastropoda) from Australia including a new species description. Zool. J. Linn. Soc. 1988, 94, 39–63. [Google Scholar] [CrossRef]
- Karp, D. Detecting small and cryptic animals by combining thermography and a wildlife detection dog. Sci. Rep. 2020, 10, 5220. [Google Scholar] [CrossRef] [PubMed]
- Jaklitsch, W.M.; Voglmayr, H. Hidden diversity in Thyridaria and a new circumscription of the Thyridariaceae. Stud. Mycol. 2016, 85, 35–64. [Google Scholar] [CrossRef] [PubMed]
- Padró, J.; Esteven, A.S.; Soto, I.M. DNA barcodes reveal the hidden arthropod diversity in a threatened cactus forest of the central Andes. Biodivers. Conserv. 2022, 32, 567–587. [Google Scholar] [CrossRef]
- Villalobos-Barrantes, H.M.; Meriño, B.M.; Walter, H.E.; Guerrero, P.C. Independent evolutionary lineages in a globular cactus species complex reveals hidden diversity in a central Chile biodiversity hotspot. Genes 2022, 13, 240. [Google Scholar] [CrossRef]
- Ferreira, V.S.; Ivie, M.A. Lessons from a museum’s cabinet: DNA barcoding and collections-based life stage associations reveals a hidden diversity in the Puerto Rican Bank paedomorphic Lycidae (Coleoptera: Elateroidea: Leptolycini). Insect Syst. Divers. 2022, 6, 1–36. [Google Scholar] [CrossRef]
- Jiang, L.; Zhou, M.; Sarker, K.K.; Huang, J.; Chen, W.; Li, C. Mitochondrial genome uncovered hidden genetic diversity in Microdous chalmersi (Teleostei: Odontobutidae). Fishes 2023, 8, 228. [Google Scholar] [CrossRef]
- Winemiller, K.O.; Fitzgerald, D.B.; Bower, L.M.; Pianka, E.R. Functional traits, convergent evolution, and periodic tables of niches. Ecol. Lett. 2015, 18, 737–751. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves-Souza, T.; Chaves, L.S.; Boldorini, G.X.; Reginaldo, N.F.; Gusmão, A.F.; Perônico, P.B.; Sanders, N.J.; Teresa, F.B. Bringing light onto the Raunkiæran shortfall: A comprehensive review of traits used in functional animal ecology. Ecol. Evol. 2023, 13, e10016. [Google Scholar] [CrossRef] [PubMed]
- Cummings, R.D. A periodic table of monosaccharides. Glycobiology 2024, 34, cwad088. [Google Scholar] [CrossRef]
- Poveda-Cuellar, J.L.; Conde-Saldaña, C.C.; Villa-Navarro, F.A.; Lujan, N.K.; Dergam dos Santos, J.A. Phylogenetic revision of whisker-cheeked suckermouth catfishes (Loricariidae: Lasiancistrus) from east of the Andes: Five species where once there were two. Zool. J. Linn. Soc. 2023, 199, 1–25. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).