Abstract
Phylogeographic studies have been conducted on many different mammal species in order to track their recent demographic histories. The climatic fluctuations associated with the Last Glacial Maximum (LGM) appear to have had a profound influence on the geographic patterning of genetic diversity in mammals. However, most phylogenetic studies have focused on single species. Few have used a holistic approach covering multiple taxa to explore common patterns. Here, we conducted meta-analyses of mitochondrial DNA control region sequences, identifying haplotype diversity and private allelic richness patterns in a geographic context. Four general patterns emerged among European mammals: an east–west decline in variation, a Western-Central belt of the highest diversity, southern richness, and homogeneity with no geographic pattern. These patterns likely reflect the refugial origins of modern populations. The east–west variation decline suggests species with eastern LGM refugia; the Western-Central belt of the highest diversity may harbor taxa with cryptic northern refugia, while southern richness may correspond to traditional southern refugia. Species with homogeneity and no geographic pattern may have been panmictic without a specific refugium or may reflect the occurrence of both southern and cryptic northern refugia. Surprisingly, the “no pattern” phenomenon is seldom discussed and may frequently have been discounted. Our study emphasizes the importance of considering multiple taxa, providing valuable insights into the responses of European mammals to past climatic changes.
1. Introduction
In the last three decades, phylogeography has become a major component of biogeography at the within-species level and has helped understand the shaping of contemporary geographic patterning of genetic lineages by climate change during the Pleistocene [1,2]. During glacial periods, the distributions of temperate and cold-adapted species contracted or expanded due to their climate adaptations [3,4,5,6,7,8,9]. Those areas representing the species’ maximum contraction in a geographical range are called refugia [7,10]. The last glacial maximum (LGM), 24,000 to 15,000 years ago, has been of particular importance in shaping the genetic landscape of many species [11].
The data presented in the first phylogeographic studies of Europe suggested that the majority of temperate species analyzed responded with end-glacial/post-glacial colonization of central and northern Europe from southern refugia [3,12,13,14]. These southern refugia have been identified as the Mediterranean peninsulas of Europe (Iberia, Italy and the Balkans) with at least three main latitudinal expansion routes [4,12]. Several studies have suggested that this model is incomplete and may not explain the genetic patterns seen in many temperate species [7,8,15,16,17,18,19]. Therefore, other possibilities have been refined, including the existence of cryptic northern refugia [20], microrefugia [21,22], and refugia within refugia [23]. Understanding the effects of the last glacial and post-glacial period on cold-adapted species is more complex. It has been suggested that the distributions of cold-adapted species are at their minimum in the current interglacial period, and therefore, such species are now in refugia [7]. Continental refugia were also proposed by Stewart et al. [7], with some European species currently occupying eastern refugia. Hewitt [3,4] and Randi [24] earlier posited eastern refugia for temperate species that were active during glacials rather than interglacials.
The evaluation of how populations of species responded to past climate change was initially based on phylogeographic studies solely using contemporary specimens. More recently, ancient DNA (aDNA) has provided a means of testing the likely responses of species to climate changes by adding a time dimension. Carbon dating and aDNA analyses have provided further evidence of the existence of northern refugia for temperate species (e.g., [25]). Cold-adapted species have also been investigated to determine how these taxa have responded to climate warming since the last glacial period, whether by extinction and population turnovers or habitat tracking during the end-glacial/post-glacial period [26,27,28,29,30,31].
Most of the phylogeographic literature is based on single species or limited numbers of related species. Due to differences between studies (discrepancies in genetic markers, geographical areas sampled and theoretical approaches), comparative analyses have not often been attempted, with some exceptions [5,19,32,33,34,35]. It has long been noted that populations in areas that were glacial refugia should show higher genetic diversity than areas colonized by those refugia [2,3]. Thus, patterns of contemporary genetic diversity, using appropriate indices, should be a valuable general approach to infer glacial refugia. However, it is also possible to get high genetic diversity in areas of mixing from different refugia, which may occur far from the refugia [35,36]. Therefore, it is helpful that the locations of glacial refugia are expected to show a high frequency of private alleles [32,37]. Combining the approaches of comparing genetic diversity and private allele frequencies has proved insightful and helpful in identifying common phylogeographic patterns [32,35,37].
The present study aims to determine the main phylogeographic patterns of 23 terrestrial mammal species, including modern humans, by combining all available sequences of a fragment of the control region of the mtDNA for these species within Europe. We combine intra-specific and inter-specific approaches in a meta-analysis of haplotype diversity and private haplotype richness to detect potential refugia and possible common phylogeographic patterns among species.
2. Materials and Methods
For our analyses, after an initial literature search generated 225 mammal species in Europe, we limited ourselves to terrestrial mammal species, excluding bats, with a broad sample range (0.5 × 106 km2) within Europe (SI, Supplementary Text). Due to the small sample size and peculiarities in the sequence availability, we restricted consideration to 23 species based on available data up to 2021; all had a sample size of at least 50 individuals. For those species with less than 50 sequences available, the analysis has not been conducted (SI, Supplementary Text, Tables S1 and S2).
The total number of individuals included for each taxon and the number of individuals from each geographic region varied among the studies considered for the meta-analysis. For a comparison of geographic regions, we divided Europe based on biogeographic subdivisions or discontinuities identified from previous phylogeographic and palaeoecological studies. Not assigning a coordinate to each sample is because of insufficient published information [38], and therefore, countries are defined as the minimum geographical unit in the analysis.
This ensured a minimum sample size across each region of more than five individuals for the species analyzed. These regions are represented by Iberia, Western Europe, Central Europe, Italy, the British Isles, the Balkans, Eastern Europe, Scandinavia, Caucasus, and Anatolia (SI, Figure S1). Through this delimitation, the understanding and identification of refugial areas can be addressed with confidence. This is because the main hypotheses suggested for most of the species previously analyzed were southern and northern refugia, which can be readily distinguished from our geographic subdivisions.
The most widely used measures of genetic diversity in population studies are haplotype and nucleotide diversity [39,40]. To calculate those values, we required a full reconstruction of the data sets. We calculated the number of haplotypes and haplotype (hd) diversity using DNAsp v.5.10.01 [41]. Under-sampling can affect diversity indices, as larger sample sizes tend to increase haplotype diversity [40,42]. Estimates of haplotype sampling coverage indicate how much of a population’s genetic diversity is represented by a specific sample size. Achieving high sampling completeness is crucial for ensuring the accuracy of meta-analyses. As recommended by Pedreschi et al. [19], we established a minimum completeness threshold of 75%. To account for differences in the geographical distribution of samples, we employed rarefaction [43]. Specifically, to mitigate the effects of varying sample sizes on genetic diversity estimates, we used the rarefaction method in HP-RARE [44] to calculate haplotype richness and the richness of private alleles. The minimum sample size across species and regions was set according to the number of rarefied haplotypes, following Lumibao et al. [37].
Another commonly used estimation for genetic diversity is the raw number of haplotypes. To exploit private haplotypes as a means to identify refugial populations [32,45], we made use of the expectation that the haplotype frequencies should be higher than the genetic distance between haplotypes [10,35,45]. For standardization of the private allelic richness, we set a rarefaction method to a standard sample size of 10 using HP-Rare v1 [44]. We analyzed and calculated these values for each species and for each geographic region (where sufficient sequences were available) with the aim of determining commonality in patterns between species.
3. Results and Discussion
The present study used a comparative approach to study 23 wild mammal species from across Europe. The taxa studied belong to the Rodentia, Lagomorpha, Eulipotyphla, Carnivora, Primates and Artiodactyla (Table 1). Haplotype and nucleotide diversity values were calculated for each species, and each geographic region analyzed (SI, Figure S2). Rodents and Eulipotyphla show relatively consistent high values for nucleotide diversity (SI, Figure S2). This is in accordance with Pedreschi et al. [19], who revealed high genetic diversity values for small mammals in general. In carnivorans and artiodactyls, the diversity values have been found to be more variable (Figure 1 and Figure S2), and there are species with relatively low haplotype diversity (Lynx lynx, Bison bonasus, and Sus scrofa). Small mammals displayed higher average haplotype diversity (mean ± s.d. = 0.94 ± 0.04) than large mammals (0.78 ± 0.17). The highest values of diversity across all mammals studied were identified in Arvicola amphibius, Lepus timidus, and Mustela nivalis (Figure 1).

Table 1.
Classification of mammalian species included in the analysis. Number of individuals, sequence length, and number of haplotypes.
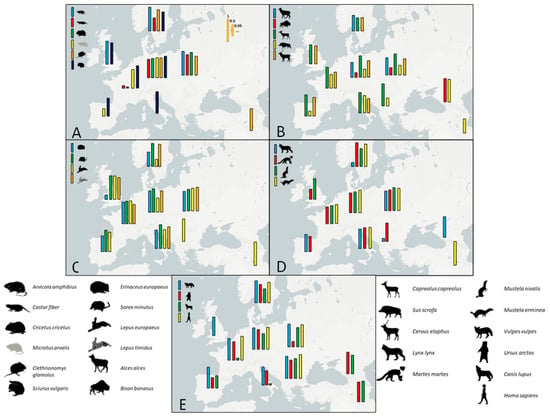
Figure 1.
Maps displaying the contemporary D-loop haplotype diversities for each species among ten predefined geographic regions. (A) Rodentia, (B) Artiodactyla, (C) Lagomorpha and Eulipotyphla, (D) Carnivora (Felidae, Mustelidae), and (E) Carnivora (Canidae, Ursidae) and Primates. The contemporary human haplotype diversity shown is that during the Mesolithic and not modern times.
The haplotype diversity values are displayed in Figure 1, where species are grouped by taxonomy. The results vary considerably by species and region analyzed. This reinforces the view that haplotype diversity, taken alone, provides misleading inferences for identifying refugia, even when diversity is shown on maps on a species-by-species and region-by-region basis [32]. The high haplotype diversity found, for example, across modern humans in the Palaeolithic and Mesolithic complicates any identification of possible refugia from the haplotype diversity values (Figure 1).
The results of this study underscore the complexity inherent in using genetic diversity in mtDNA to identify glacial refugia in mammals. Our findings suggest that common genetic signals are often elusive, with species-specific responses playing a crucial role in reconstructing post-glacial recolonization patterns. Although we focused on a single taxonomic group (mammals) compared to previous comparative phylogeographic studies (e.g., [4,5]), the inclusion of a large number of species with substantial sample sizes has provided a robust framework for addressing these key phylogeographic questions.
However, it is important to acknowledge that relying solely on genetic diversity for the identification of refugia presents significant challenges and potential biases. These limitations must be carefully considered in future studies. For instance, the concept of southern richness, often interpreted within the southern refugial paradigm [3,12], may represent a simplistic view and might reflect areas of endemism rather than true source populations for post-glacial recolonization of northern regions [7,15,19,20]. In this context, the allelic richness observed in mid-latitudinal regions could provide insights into the significance of western and central European areas as “hubs of diversity” rather than merely zones of contact and hybridization.
The haplotype diversity values have more value when combined with calculations of private allelic richness after rarefraction to S = 10. The values for each species by region are shown in Figure 2. Four different patterns can be inferred based on the private allelic richness values complemented with the haplotype diversity values (Figure 1 and Figure 2). The first pattern (Figure 2A) shows an east-to-west cline with the highest values of private allelic richness in the east and the lowest in the west and is seen in Bison bonasus, Alces alces, Lynx lynx, Castor fiber, Cricetus cricetus, Mustela nivalis, and Mustela erminea (excluding the British Isles). This pattern is also reinforced by the haplotype diversity values found in Castor fiber, Cricetus cricetus, Alces alces, and Mustela nivalis but not for Mustela erminea, where the haplotype diversity values appear more homogeneous over the geographical distribution. The results for these species may reflect more than a single historical process. Lynx lynx and Alces alces include a strong association with boreal forests today [34,46], which is most prominent in northern and eastern Europe and may have had LGM refugia in the east [46]. The same may also apply to Mustela erminea [47]. Cricetus cricetus is, however, a species that is adapted to more continental steppe habitats [48] and may well have contracted since the LGM only to re-expand during the Neolithic as cereal cultivation spread west across Europe, as has been suggested for the common vole (Microtus arvalis) [49]. Cricetus cricetus would otherwise be in an Eastern refugium today [7]. In Castor fiber, Mustela nivalis, and Bison bonasus, the situation is more complex due to the substantial reduction of populations and the low diversity in recent times seen in Bison bonasus and Castor fiber [50,51] and uncertainty about the subspecies affinities of different populations previously described in Mustela nivalis [52].
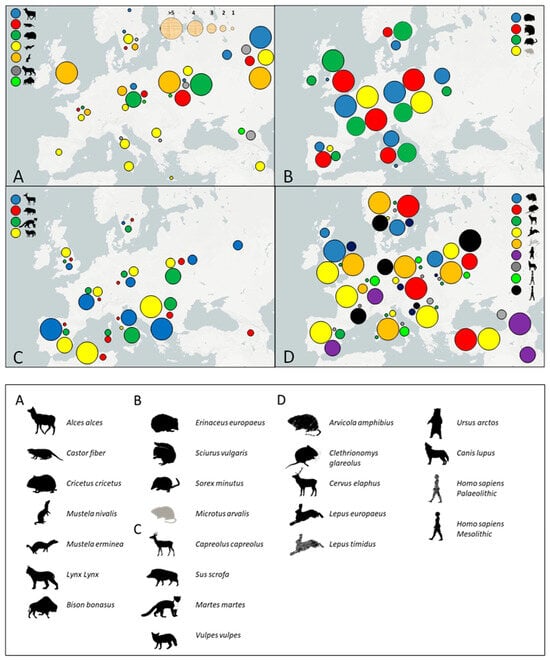
Figure 2.
Maps displaying the four main patterns of distribution identified for private allelic richness identified after rarefaction to S = 10. (A) East–west decline in variation; (B) Western-Central belt of highest diversity; (C) Southern richness; and (D) Homogeneous with no geographic pattern.
The second pattern identified (Figure 2B) relates to a Western-Central belt that comprises the highest values of private allelic richness for Erinaceus europaeus, Sciurus vulgaris, Sorex minutus, and Microtus arvalis. It is interesting that the species showing this second pattern are all small mammals, suggesting some association with low dispersal rates, although not all small mammals conform to the pattern. Erinaceus europaeus and Microtus arvalis have a similar haplotype diversity pattern as found with private alleles [53], although that was not the case for Sciurus vulgaris and Sorex minutus, further emphasizing species-by-species variation [54] (Figure 1). Although haplotype diversity did not reveal a pattern for the red squirrel (Sciurus vulgaris), private allelic richness indicates Western and Central Europe as possible refugial areas for the species. For Sorex minutus, the pattern is not as clear as for the other species and the high values for private allelic richness identified in the whole range of the species complicate the resolution. However, the importance of areas in France as possible refugia has been suggested [55], which is consistent with the results here. Therefore, these species may be the ones that were in cryptic northern refugia during the LGM in Europe, as has been previously suggested [20,56,57].
The third pattern (Figure 2C) is one where the greatest diversity is in the south of Europe. Capreolus capreolus, Sus scrofa, Martes martes, and Vulpes vulpes display this pattern of high private allelic richness in the south (east and west) and low values in northern latitudes. Capreolus capreolus, Sus scrofa, and Martes martes appear to have had southern refugia apparently coupled with complex demographic histories. The human actions of the introduction of domestic pigs, conspecific with wild boar (Sus scrofa), and the translocation of roe deer (Capreolus capreolus) may have affected the genetics of these species [58,59,60], although our analyses have not detected that. A previously described lack of phylogeographic structure and homogeneous distribution in the red fox (Vulpes vulpes), suggested that there may have been a constant occupation of the region by this species [61]. In contrast, the results here show Vulpes vulpes to have a southern refugial pattern where Iberia, and especially the Balkans, represent important areas for private allelic richness. The pattern observed here for Vulpes vulpes is in accordance with the recent study from McDevitt et al. [62]. Overall, the pattern of southern richness in diversity accords with species that resided in southern refugia during the LGM, conforming to the traditional scenario for European species [12,13].
The homogeneity in genetic variation identified in some species, the fourth pattern, suggests there is no clear, structured pattern of distribution of private alleles (Figure 2D). This is superficially similar to that of the northern refugium pattern; however, the distinction is seen in the results for each individual species (Figure 2). This is in accordance with the findings of Hofreiter et al. [63] for the Late Pleistocene genetic landscape but extended to the present day. For eight different species, a homogeneous distribution of private alleles was found, and areas with much higher diversity than others were not identified (Figure 1 and Figure 2). Modern humans from the Palaeolithic and the Mesolithic are amongst the species with this pattern.
The body size of a species does not appear to be associated with a homogeneous distribution of private alleles, as four of the species displaying this are small mammals (Arvicola amphibius, Clethrionomys glareolus, Lepus europaeus, and Lepus timidus), and four are large (Cervus elaphus, Canis lupus, Ursus arctos, and Homo sapiens). In general, these species are characterized by a widespread range and homogeneous distribution, and a relative lack of ecological restrictions may help explain this private allele distribution. The absence of phylogeographic patterns in some species has been explained by their high migration rate, such as Canis lupus [64]. It is interesting to note that two of these species (water vole Arvicola amphibius and brown bear Ursus arctos) were described by Hewitt [3,4] as having had southern refugia exclusively. The latter has been shown to be incorrect for the brown bear and has been questioned for Arvicola amphibius, as well as Cervus elaphus, based on aDNA and other studies [65,66,67,68]. The Spanish brown bear haplotype of Hewitt [4] has been found in Belgium during the LGM, and the red deer and water vole are represented on either side of the LGM by the same populations in northern Europe. These data may be consistent with the homogeneity described here. Cervus elaphus should perhaps be regarded with some caution as they are known to have been translocated in historical times, which may have homogenized their geographic pattern of diversity [68]. Kotlík et al. [36], on the other hand, have inferred that Clethrionomys glareolus had cryptic northern refugia as well as southern refugia—and this might be another explanation for the pattern described here. Homo sapiens populations from the Palaeolithic as well as the Mesolithic do not show any strong pattern, with only a slightly higher observed occurrence of private alleles in the west during the Mesolithic. In general, the private allelic richness is higher in the Mesolithic than the Palaeolithic, either indicating fragmentation of populations in the continent during the LGM and/or major re-expansions [69,70], leading to an increased number of regionally private haplotypes.
The four disparate phylogeographic patterns identified here are likely to be dominantly influenced by the historical biogeography of the individual component species and their constituent populations. The general explanation for such patterns is that the species’ populations were restricted to refugia during the adverse conditions of the last glacial maximum, followed by a re-expansion during the Holocene [3,4]. While it is likely that the most recent dramatic climate changes have affected these species, it is also possible that some geographic patterns have an older legacy. It should be kept in mind that the Milankovitch glacial/interglacial cycles have affected taxa during the entirety of the Quaternary.
The four patterns identified include two that correspond to a refugial source recognized by Hewitt [3,4], namely the southern and eastern refugia, although, since that time, there has been greater acceptance of the third pattern, the extra-Mediterranean northern refugia [24]. The taxa that had a Western-Central diversity increase may represent species that had cryptic northern refugia, and all are relatively small species that have previously had such claims [20,56]. It should be noted, however, that for Sorex minutus, Vega et al. [71] (and previous studies referenced therein) posit southern refugia in addition to the northern refugia that we have highlighted thus far—a similar situation as for the other species with Western-Central diversity [14,53,54]. However, the signal of refugia, in terms of the presence of private alleles, is stronger for the northern refugia. The species with an apparent lack of pattern are ones that have not previously been widely recognized and indicate species that either had a panmictic distribution during the LGM or had refugia both in the Mediterranean and in the extra-Mediterranean region (with an equally strong signal for refugia in the southern and northern refugia). Interestingly, these taxa include several (the water vole and the brown bear) that had been identified by Hewitt [3,4] as having had southern LGM refugia. Furthermore, some of these had also led to the suggestion of cryptic northern refugia, namely Cervus elaphus and Arvicola amphibius [66,67] and Ursus arctos was also implicated in a model for the evolution of Ursus maritimus in a cryptic maritime northern refugium [69]. The idea that Ursus arctos had been in one or more cryptic northern refugium could be interpreted from the work of Ersmark et al. [65], although this had not been specifically claimed by them (they used the more general phrase, “northern survival”). This may suggest that the concept of cryptic northern refugia may have been overstated and that some of the species proposed as having had such refugia had instead panmictic populations. The alternative is that some or all of these species had both northern and southern refugia, as has been proposed for the bank vole [36].
A possibility is that the more dispersive larger mammals like Canis lupus and Homo sapiens were panmictic while the smaller mammals had more isolated northern and southern refugia. Both archaeological [72] and whole mtDNA and whole-genome aDNA data [73,74] suggest that southwest Europe served as a human refuge during the LGM and that there was substantial genetic turnover in Europe between 25,000 and 11,500 years ago but with the continuity from the Aurignacian to the Magdalenian and the Solutrean followed by new ancestries spreading from a likely refuge in the southeast after 14,500 years ago [75,76,77]. Nevertheless, the pattern is complex, and in our analyses, modern humans from the Palaeolithic and the Mesolithic conform to the lack of phylogeographic structure, indicating that contrary to some suggestions [69,78], they were not restricted to a southern refugium during the LGM [70,79]. Some of the other mammalian species with a lack of pattern for private allele richness may be similar to birds in the Palaearctic, for whom it was suggested there had been panmixia during the LGM [80]. The lack of pattern is interesting because this may be viewed as a negative result and/or suffer from publication bias. Negative results are often difficult to publish [81], which can lead to non-representative perceptions.
4. Conclusions
The species-specific analysis of the 23 mammals investigated here has identified a number of patterns in the diversity of mitochondrial D-loop sequences. This meta-analysis is unusually rich in terms of the number of species using the same genetic marker and contributes to a better understanding of mammalian phylogeography in Europe specifically and the complexity of biogeographical processes more widely.
A new approach to locating potential refugial areas in the continent has been established based on identifying areas where private allelic richness displayed high values and was complemented by haplotype diversity indices. This research builds on the legacy of the comparative approach [5,13,32,35], although it provides a novel method to recognize similarities between species.
A weakness of the analysis performed here is that it does not take into account the phylogenetic distribution of genetic lineages. In the future, when it becomes possible for taxa with good sampling, it would be beneficial to study diversity and private allele distribution on a lineage-by-lineage basis. It would also be of interest to see a replication of the meta-analysis with whole mitochondrial genomes, and indeed whole genomes, on large numbers of specimens over large areas to interpret phylogeography. It is also desirable to analyze numerous samples over time using DNA analysis. Such approaches would help identify confounding factors, such as recent bottlenecking, but have as yet only been possible for a few taxa.
Four different patterns are revealed, consisting of species that have an east-west decline in variation, ones with a Western-Central belt of highest diversity, another with southern richness, and finally, a pattern of homogeneity where no geographic pattern was identified. These patterns would seem to reflect the refugial origins (or lack thereof) of the modern populations. The east–west cline appears to mostly describe the species with eastern refugia during the LGM, albeit differing ones according to the adaptations of the species, and the southern richness pattern seems to correspond to species with southern European refugia. These were the two patterns that had been described by Hewitt [3,4] and Randi [24]. However, one of the species (Cricetus cricetus) may correspond to a continental species that is actually in a refugium today [7], although it may have expanded westwards during the Neolithic in response to anthropogenic changes in the landscape. The species with Western-Central belt richness may describe species that had been in cryptic northern refugia [20]. Finally, the species with a more homogeneous pattern may have been widely distributed in the mid-latitudes of Europe during the LGM and effectively had a large refugium at the time or could be described as having no refugia, or they were located in separate refugia both in the northern extra-Mediterranean as well as Mediterranean areas [36].
The homogeneous pattern seen in some species suggests that large taxa like the brown bear had not been in a southern or a cryptic northern refugium but were widely distributed at the time of the LGM in the European mid-latitudes. The latter may suggest that the concept of cryptic northern refugia may have been overstated and that some of the species proposed as having had such refugia had instead panmictic populations. It may be, however, that other, especially smaller, mammalian taxa, such as the bank vole, had isolated refugia in the south and the north, although distinguishing between these explanations for the homogeneous patterns may be challenging. In conclusion, the meta-analysis of existing phylogeographic data has suggested that four patterns exist: species with eastern, northern and southern refugia, as well as mammals that may not have substantially contracted over non-glaciated areas during the LGM.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/d16100611/s1. Figure S1: Map representing the geographical regions considered for the analysis. Each color represents one area; Figure S2: Maps displaying the nucleotide diversity values for each species per region analyzed. Bars represent nucleotide diversity values, and each color matches with the species that are classified by orders: Artiodactyla, Lagomorpha/Eulipotyphla, Rodentia, Carnivora, and Carnivora/Primates; Supplementary Text: Choice of samples; Table S1: Two-hundred and twenty-five European mammal species considered for the analysis at the beginning of this project with an indication of the number of entries found on GenBank described as control region and D-loop, and for CytB. Species with more than 50 D-loop and control region sequences on GenBank are indicated, and the reasons for excluding the species with adequate data are also given. Inclusion for analysis in green, and exclusion with reason in pink; Table S2: Mammal species chosen for the meta-analysis and the studies that provided the sequences included in the analysis. The sequences were retrieved from GenBank from the associated references. Unpublished means that the sequences were found in GenBank, but the paper was not published by the time this study was finished.
Author Contributions
J.R.S. and O.G.-R. designed research; O.G.-R. performed research; O.G.-R. and E.A.H. analyzed data; and O.G.-R., J.B.S. and J.R.S. wrote the paper with assistance from E.A.H., R.S., M.B.R. and. D.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
This work was supported by Bournemouth University as part of an Institute for the Study of Landscapes and Human Evolution (ISLHE) Ph.D. studentship to O.G.-R. We would also like to thank all the work done by many researchers who produced the data on GenBank. This research would have been impossible without it. We would also like to acknowledge the authors who shared data that were not available in GenBank.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Avise, J.C. Phylogeography: The History and Formation of Species; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Avise, J.C.; Arnold, J.; Ball, R.M.; Bermingham, E.; Lamb, T.; E Neigel, J.; A Reeb, C.; Saunders, N.C. Intraspecific phylogeography: The mitochondrial DNA bridge between population genetics and systematics. Ann. Rev. Ecol. Syst. 1987, 18, 489–522. [Google Scholar] [CrossRef]
- Hewitt, G.M. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 1996, 58, 247–276. [Google Scholar] [CrossRef]
- Hewitt, G.M. The genetic legacy of the quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Fumagalli, L.; Wust-Saucy, A.-G.; Cosson, J.-F. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 1998, 7, 453–464. [Google Scholar] [CrossRef]
- Lister, A.M. The impact of Quaternary Ice Ages on mammalian evolution. Philos. Trans. R. Soc. B Biol. Sci. 2004, 359, 221–241. [Google Scholar] [CrossRef]
- Stewart, J.R.; Lister, A.M.; Barnes, I.; Dalén, L. Refugia revisited: Individualistic responses of species in space and time. Proc. R. Soc. B Biol. Sci. 2010, 277, 661–671. [Google Scholar] [CrossRef]
- Schmitt, T.; Varga, Z. Extra-Mediterranean refugia–the rule and not the exception? Front. Zool. 2012, 9, 22. [Google Scholar] [CrossRef]
- Morales-Barbero, J.; Martinez, P.A.; Ferrer-Castán, D.; Olalla-Tárraga, M.Á. Quaternary refugia are associated with higher speciation rates in mammalian faunas of the western Palaearctic. Ecography 2018, 41, 607–621. [Google Scholar] [CrossRef]
- Provan, J.; Bennett, K.D. Phylogeographic insights into cryptic glacial refugia. Trends Ecol. Evol. 2008, 23, 564–571. [Google Scholar] [CrossRef]
- Hofreiter, M.; Barnes, I. Diversity lost: Are all Holarctic large mammal species just relict populations? BMC Biol. 2010, 8, 46. [Google Scholar] [CrossRef]
- Hewitt, G.M. Post-glacial re-colonization of European biota. Biol. J. Linn. Soc. 1999, 68, 87–112. [Google Scholar] [CrossRef]
- Hewitt, G.M. Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. B Biol. Sci. 2004, 359, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Seddon, J.M.; Santucci, F.; Reeve, N.J.; Hewitt, G.M. DNA footprints of European hedgehogs, Erinaceus europaeus and E. concolor: Pleistocene refugia, postglacial expansion and colonization routes. Mol. Ecol. 2001, 10, 2187–2198. [Google Scholar] [PubMed]
- Bilton, D.T.; Mirol, P.M.; Mascheretti, S.; Fredga, K.; Zima, J.; Searle, J.B. Mediterranean Europe as an area of endemism for small mammals rather than a source for northwards postglacial colonization. Proc. R. Soc. B Biol. Sci. 1998, 265, 1219–1226. [Google Scholar] [CrossRef] [PubMed]
- Kotlík, P.; Deffontaine, V.; Mascheretti, S.; Zima, J.; Michaux, J.R.; Searle, J.B. A northern glacial refugium for bank voles (Clethrionomys glareolus). Proc. Natl. Acad. Sci. USA 2006, 103, 14860–14864. [Google Scholar] [CrossRef]
- Valdiosera, C.E.; García, N.; Anderung, C.; Dalén, L.; Crégut-Bonnoure, E.; Kahlke, R.; Stiller, M.; Brandström, M.; Thomas, M.G.; Arsuaga, J.L.; et al. Staying out in the cold: Glacial refugia and mitochondrial DNA phylogeography in ancient European brown bears. Mol. Ecol. 2007, 16, 5140–5148. [Google Scholar] [CrossRef]
- Bhagwat, S.A.; Willis, K.J. Species persistence in northerly glacial refugia of Europe: A matter of chance or biogeographical traits? J. Biogeogr. 2008, 35, 464–482. [Google Scholar] [CrossRef]
- Pedreschi, D.; García-Rodríguez, O.; Yannic, G.; Cantarello, E.; Diaz, A.; Golicher, D.; Korstjens, A.H.; Heckel, G.; Searle, J.B.; Gillingham, P.; et al. Challenging the European southern refugium hypothesis: Species-specific structures versus general patterns of genetic diversity and differentiation among small mammals. Glob. Ecol. Biogeogr. 2019, 28, 262–274. [Google Scholar] [CrossRef]
- Stewart, J.R.; Lister, A.M. Cryptic northern refugia and the origins of the modern biota. Trends Ecol. Evol. 2001, 16, 608–613. [Google Scholar] [CrossRef]
- Rull, V.; Schubert, C.; Aravena, R. Palynological studies in the Venezuelan Guayana Shield: Preliminary results. Curr. Res. Pleistocene 1988, 5, 54–56. [Google Scholar]
- Rull, V. Microrefugia. J. Biogeogr. 2009, 36, 481–484. [Google Scholar] [CrossRef]
- Gómez, A.; Lunt, D.H. Refugia within refugia: Patterns of phylogeographic concordance in the Iberian Peninsula. In Phylogeography of Southern European Refugia; Weiss, S., Ferrand, N., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 155–188. [Google Scholar]
- Randi, E. Phylogeography of South European mammals. In Phylogeography of Southern European Refugia; Weiss, S., Ferrand, N., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 101–126. [Google Scholar]
- Ersmark, E.; Klütsch, C.F.C.; Chan, Y.L.; Sinding, M.-H.S.; Fain, S.R.; Illarionova, N.A.; Oskarsson, M.; Uhlén, M.; Zhang, Y.-P.; Dalén, L.; et al. From the past to the present: Wolf phylogeography and demographic history based on the mitochondrial control region. Front. Ecol. Evol. 2016, 4, 134. [Google Scholar] [CrossRef]
- Dalén, L.; Nyström, V.; Valdiosera, C.; Germonpré, M.; Sablin, M.; Turner, E.; Angerbjörn, A.; Arsuaga, J.L.; Götherström, A. Ancient DNA reveals lack of postglacial habitat tracking in the arctic fox. Proc. Natl. Acad. Sci. USA 2007, 104, 6726–6729. [Google Scholar] [CrossRef] [PubMed]
- Lagerholm, V.K.; Sandoval-Castellanos, E.; Ehrich, D.; Abramson, N.I.; Nadachowski, A.; Kalthoff, D.C.; Germonpré, M.; Angerbjörn, A.; Stewart, J.R.; Dalén, L. On the origin of the Norwegian lemming. Mol. Ecol. 2014, 23, 2060–2071. [Google Scholar] [CrossRef]
- Palkopoulou, E.; Baca, M.; Abramson, N.I.; Sablin, M.; Socha, P.; Nadachowski, A.; Prost, S.; Germonpré, M.; Kosintsev, P.; Smirnov, N.G.; et al. Synchronous genetic turnovers across Western Eurasia in Late Pleistocene collared lemmings. Glob. Chang. Biol. 2016, 22, 1710–1721. [Google Scholar] [CrossRef]
- Leonard, J.A.; Vilà, C.; Fox-Dobbs, K.; Koch, P.L.; Wayne, R.K.; Van Valkenburgh, B. Megafaunal extinctions and the disappearance of a specialized wolf ecomorph. Curr. Biol. 2007, 17, 1146–1150. [Google Scholar] [CrossRef] [PubMed]
- Hofreiter, M. Pleistocene extinctions: Haunting the survivors. Curr. Biol. 2007, 17, R609–R611. [Google Scholar] [CrossRef]
- Lagerholm, V.K.; Sandoval-Castellanos, E.; Vaniscotte, A.; Potapova, O.R.; Tomek, T.; Bochenski, Z.M.; Shepherd, P.; Barton, N.; Van Dyck, M.-C.; Miller, R.; et al. Range shifts or extinction? Ancient DNA and distribution modelling reveal past and future responses to climate warming in cold-adapted birds. Glob. Chang. Biol. 2017, 23, 1425–1435. [Google Scholar]
- Maggs, C.A.; Castilho, R.; Foltz, D.; Henzler, C.; Jolly, M.T.; Kelly, J.; Olsen, J.; Perez, K.E.; Stam, W.; Väinölä, R.; et al. Evaluating signatures of glacial refugia for North Atlantic benthic marine taxa. Ecology 2008, 89, S108–S122. [Google Scholar] [CrossRef]
- Lenstra, J.A.; Ajmone-Marsan, P.; Beja-Pereira, A.; Bollongino, R.; Bradley, D.G.; Colli, L.; De Gaetano, A.; Edwards, C.J.; Felius, M.; Ferretti, L.; et al. Meta-analysis of mitochondrial DNA reveals several population bottlenecks during worldwide migrations of cattle. Diversity 2014, 6, 178–187. [Google Scholar] [CrossRef]
- Niedziałkowska, M. Phylogeography of European moose (Alces alces) based on contemporary mtDNA data and archaeological records. Mamm. Biol. 2017, 84, 35–43. [Google Scholar] [CrossRef]
- Petit, R.J.; Aguinagalde, I.; de Beaulieu, J.L.; Bittkau, C.; Brewer, S.; Cheddadi, R.; Ennos, R.; Fineschi, S.; Grivet, D.; Lascoux, M.; et al. Glacial refugia: Hotspots but not melting pots of genetic diversity. Science 2003, 300, 1563–1565. [Google Scholar] [CrossRef] [PubMed]
- Kotlík, P.; Marková, S.; Horníková, M.; Escalante, M.A.; Searle, J.B. The bank vole (Clethrionomys glareolus) as a model system for adaptive phylogeography in the European theater. Front. Ecol. Evol. 2022, 10, 866605. [Google Scholar] [CrossRef]
- Lumibao, C.Y.; Hoban, S.M.; McLachlan, J. Ice ages leave genetic diversity ‘hotspots’ in Europe but not in Eastern North America. Ecol. Lett. 2017, 20, 1459–1468. [Google Scholar] [CrossRef]
- Gratton, P.; Marta, S.; Bocksberger, G.; Winter, M.; Trucchi, E.; Kühl, H. A world of sequences: Can we use georeferenced nucleotide databases for a robust automated phylogeography? J. Biogeogr. 2017, 44, 475–486. [Google Scholar] [CrossRef]
- Egeland, T.; Salas, A. A statistical framework for the interpretation of mtDNA mixtures: Forensic and medical applications. PLoS ONE 2011, 6, e26723. [Google Scholar] [CrossRef]
- Goodall-Copestake, W.P.; Tarling, G.A.; Murphy, E. On the comparison of population-level estimates of haplotype and nucleotide diversity: A case study using the gene Cox1 in animals. Heredity 2012, 109, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Pereira, L.; Cunha, C.; Amorim, A. Predicting sampling saturation of mtDNA haplotypes: An application to an enlarged Portuguese database. Int. J. Leg. Med. 2004, 118, 132–136. [Google Scholar] [CrossRef]
- Heck, J.R.; Kenneth, L.; van Belle, G.; Simberloff, D. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 1975, 56, 1459–1461. [Google Scholar] [CrossRef]
- Kalinowski, S.T. Counting alleles with rarefaction: Private alleles and hierarchical sampling designs. Conserv. Genet. 2004, 5, 539–543. [Google Scholar] [CrossRef]
- Petit, R.J.; Csaikl, U.M.; Bordács, S.; Burg, K.; Coart, E.; Cottrell, J.; van Dam, B.; Deans, J.D.; Dumolin-Lapègue, S.; Fineschi, S.; et al. Chloroplast DNA variation in European white oaks phylogeography and patterns of diversity based on data from over 2600 populations. For. Ecol. Manag. 2002, 156, 5–26. [Google Scholar] [CrossRef]
- Lucena-Perez, M.; Marmesat, E.; Kleinman-Ruiz, D.; Martínez-Cruz, B.; Węcek, K.; Saveljev, A.P.; Seryodkin, I.V.; Okhlopkov, I.; Dvornikov, M.G.; Ozolins, J.; et al. Genomic patterns in the widespread Eurasian lynx shaped by Late Quaternary climatic fluctuations and anthropogenic impacts. Mol. Ecol. 2020, 29, 812–828. [Google Scholar] [CrossRef] [PubMed]
- Sommer, R.; Benecke, N. Late- and Post-Glacial history of the Mustelidae in Europe. Mammal Rev. 2004, 34, 249–284. [Google Scholar] [CrossRef]
- Neumann, K.; Michaux, J.R.; Maak, S.; Jansman, H.A.H.; Kayser, A.; Mundt, G.; Gattermann, R. Genetic spatial structure of European common hamsters (Cricetus cricetus)–A result of repeated range expansion and demographic bottlenecks. Mol. Ecol. 2005, 14, 1473–1483. [Google Scholar] [CrossRef]
- Baca, M.; Popović, D.; Baca, K.; Lemanik, A.; Doan, K.; Horáček, I.; López-García, J.M.; Bañuls-Cardona, S.; Pazonyi, P.; Desclaux, E.; et al. Diverse responses of common vole (Microtus arvalis) populations to Late Glacial and Early Holocene climate changes–Evidence from ancient DNA. Quat. Sci. Rev. 2020, 233, 106239. [Google Scholar] [CrossRef]
- Lorenzen, E.D.; Nogués-Bravo, D.; Orlando, L.; Weinstock, J.; Binladen, J.; Marske, K.A.; Ugan, A.; Borregaard, M.K.; Gilbert, M.T.P.; Nielsen, R.; et al. Species-specific responses of Late Quaternary megafauna to climate and humans. Nature 2011, 479, 359–364. [Google Scholar] [CrossRef]
- Marr, M.M.; Brace, S.; Schreve, D.C.; Barnes, I. Identifying source populations for the reintroduction of the Eurasian beaver, Castor fiber L. 1758, into Britain: Evidence from ancient DNA. Sci. Rep. 2018, 8, 2708. [Google Scholar]
- Lebarbenchon, C.; Poitevin, F.; Arnal, V.; Montgelard, C. Phylogeography of the weasel (Mustela nivalis) in the western-Palaearctic region: Combined effects of glacial events and human movements. Heredity 2010, 105, 449–462. [Google Scholar] [CrossRef]
- Stojak, J.; McDevitt, A.D.; Herman, J.S.; Kryštufek, B.; Uhlíková, J.; Purger, J.J.; Lavrenchenko, L.A.; Searle, J.B.; Wójcik, J.M. Between the Balkans and the Baltic: Phylogeography of a common vole mitochondrial DNA lineage limited to Central Europe. PLoS ONE 2016, 11, e0168621. [Google Scholar] [CrossRef]
- Grill, A.; Amori, G.; Aloise, G.; Lisi, I.; Tosi, G.; Wauters, L.A.; Randi, E. Molecular phylogeography of European Sciurus vulgaris: Refuge within refugia? Mol. Ecol. 2009, 18, 2687–2699. [Google Scholar] [CrossRef] [PubMed]
- McDevitt, A.D.; Yannic, G.; Rambau, R.V.; Hayden, T.J.; Searle, J.B. Postglacial recolonization of continental Europe by the pygmy shrew (Sorex minutus) inferred from mitochondrial and Y chromosomal DNA sequences. In Relict Species: Phylogeography and Conservation Biology; Springer: Berlin/Heidelberg, Germany, 2010; pp. 217–236. [Google Scholar]
- Vega, R.; Amori, G.; Aloise, G.; Cellini, S.; Loy, A.; Searle, J.B. Genetic and morphological variation in a Mediterranean glacial refugium: Evidence from Italian pygmy shrews, Sorex minutus (Mammalia: Soricomorpha). Biol. J. Linn. Soc. 2010, 100, 774–787. [Google Scholar] [CrossRef]
- Jaarola, M.; Searle, J.B. Phylogeography of field voles (Microtus agrestis) in Eurasia inferred from mitochondrial DNA sequences. Mol. Ecol. 2002, 11, 2613–2621. [Google Scholar] [CrossRef]
- Randi, E.; Alves, P.C.; Carranza, J.; Milošević-Zlatanović, S.; Sfougaris, A.; Mucci, N. Phylogeography of roe deer (Capreolus capreolus) populations: The effects of historical genetic subdivisions and recent nonequilibrium dynamics. Mol. Ecol. 2004, 13, 3071–3083. [Google Scholar] [CrossRef]
- Larson, G.; Albarella, U.; Dobney, K.; Rowley-Conwy, P.; Schibler, J.; Tresset, A.; Vigne, J.-D.; Edwards, C.J.; Schlumbaum, A.; Dinu, A.; et al. Ancient DNA, pig domestication, and the spread of the Neolithic into Europe. Proc. Natl. Acad. Sci. USA 2007, 104, 15276–15281. [Google Scholar] [CrossRef]
- Olano-Marin, J.; Plis, K.; Sonnichsen, L.; Borowik, T.; Niedzialkowska, M.; Jedrzejewska, B. Weak population structure in European roe deer (Capreolus capreolus) and evidence of introgressive hybridization with Siberian roe deer (C. pygargus) in Northeastern Poland. PLoS ONE 2014, 9, e109147. [Google Scholar]
- Teacher, A.G.; Thomas, J.A.; Barnes, I. Modern and ancient red fox (Vulpes vulpes) in Europe show an unusual lack of geographical and temporal structuring, and differing responses within the carnivores to historical climatic change. BMC Evol. Biol. 2011, 13, 214. [Google Scholar] [CrossRef] [PubMed]
- McDevitt, A.D.; Coscia, I.; Browett, S.S.; Ruiz-González, A.; Statham, M.J.; Ruczyńska, I.; Roberts, L.; Stojak, J.; Frantz, A.C.; Norén, K.; et al. Next-generation phylogeography resolves post-glacial colonization patterns in a widespread carnivore, the red fox (Vulpes vulpes), in Europe. Mol. Ecol. 2022, 31, 993–1006. [Google Scholar] [CrossRef] [PubMed]
- Hofreiter, M.; Serre, D.; Rohland, N.; Rabeder, G.; Nagel, D.; Conard, N.; Münzel, S.; Pääbo, S. Lack of phylogeography in European mammals before the last glaciation. Proc. Natl. Acad. Sci. USA 2004, 101, 12963–12968. [Google Scholar] [CrossRef]
- Vilà, C.; Amorim, I.R.; Leonard, J.A.; Posada, D.; Castroviejo, J.; Petrucci-Fonseca, F.; Crandall, K.A.; Ellegren, H.; Wayne, R.K. Mitochondrial DNA phylogeography and population history of the grey wolf Canis lupus. Mol. Ecol. 1999, 8, 2089–2103. [Google Scholar] [CrossRef]
- Ersmark, E.; Baryshnikov, G.; Higham, T.; Argant, A.; Castaños, P.; Döppes, D.; Gasparik, M.; Germonpré, M.; Lidén, K.; Lipecki, G.; et al. Genetic turnovers and northern survival during the last glacial maximum in European brown bears. Ecol. Evol. 2019, 9, 5891–5905. [Google Scholar] [CrossRef] [PubMed]
- Meiri, M.; Lister, A.M.; Higham, T.F.; Stewart, J.R.; Straus, L.G.; Obermaier, H.; Gonzalez Morales, M.R.; Marín-Arroyo, A.B.; Barnes, I. Late-glacial recolonization and phylogeography of European red deer (Cervus elaphus L.). Mol. Ecol. 2013, 22, 4711–4722. [Google Scholar] [CrossRef]
- Brace, S.; Ruddy, M.; Miller, R.; Schreve, D.C.; Stewart, J.R.; Barnes, I. The colonization history of British water vole (Arvicola amphibius (Linnaeus, 1758)): Origins and development of the Celtic fringe. Proc. R Soc. B 2016, 283, 20160130. [Google Scholar] [CrossRef] [PubMed]
- Queiros, J.; Acevedo, P.; Santos, J.P.; Barasona, J.; Beltran-Beck, B.; Gonzalez-Barrio, D.; Armenteros, J.A.; Diez-Delgado, I.; Boadella, M.; Fernandez de Mera, I.; et al. Cryptic refugia and their role in European postglacial colonization history: A case study in red deer (Cervus elaphus). PLoS ONE 2019, 14, e0210282. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.R.; Stringer, C.B. Human evolution out of Africa: The role of refugia and climate change. Science 2012, 335, 1317–1321. [Google Scholar] [CrossRef]
- Posth, C.; Wißing, C.; Kitagawa, K.; Pagani, L.; van Holstein, L.; Racimo, F.; Wehrberger, K.; Conard, N.J.; Kind, C.J.; Bocherens, H.; et al. Deeply divergent archaic mitochondrial genome provides lower time boundary for African gene flow into Neanderthals. Nat. Comm. 2017, 8, 16046. [Google Scholar] [CrossRef]
- Vega, R.; McDevitt, A.D.; Stojak, J.; Mishta, A.; Wójcik, J.M.; Kryštufek, B.; Searle, J.B. Phylogeographical structure of the pygmy shrew: Revisiting the roles of southern and northern refugia in Europe. Biol. J. Linn. Soc. 2020, 129, 901–917. [Google Scholar] [CrossRef]
- Gamble, C.; Davies, W.; Pettitt, P.; Hazelwood, L.; Richards, M. The archaeological and genetic foundations of the European population during the Late Glacial: Implications for ‘agricultural thinking’. Camb. Archaeol. J. 2005, 15, 193–223. [Google Scholar] [CrossRef]
- Posth, C.; Renaud, G.; Mittnik, A.; Drucker, D.G.; Rougier, H.; Cupillard, C.; Valentin, F.; Thevenet, C.; Furtwängler, A.; Wißing, C.; et al. Pleistocene mitochondrial genomes suggest a single major dispersal of non-Africans and a Late Glacial population turnover in Europe. Curr. Biol. 2016, 26, 827–833. [Google Scholar] [CrossRef]
- Fu, Q.; Posth, C.; Hajdinjak, M.; Petr, M.; Mallick, S.; Fernandes, D.; Furtwängler, A.; Haak, W.; Meyer, M.; Mittnik, A.; et al. The genetic history of Ice Age Europe. Nature 2016, 534, 200–205. [Google Scholar] [CrossRef]
- Pereira, J.B.; Costa, M.D.; Vieira, D.; Pala, M.; Bamford, L.; Harich, N.; Cherni, L.; Alshamali, F.; Hatina, J.; Rychkov, S.; et al. Reconciling evidence from ancient and contemporary genomes: A major source for the European Neolithic within Mediterranean Europe. Proc. R. Soc. B 2017, 284, 20161976. [Google Scholar] [CrossRef] [PubMed]
- Posth, C.; Yu, H.; Ghalichi, A.; Rougier, H.; Crevecoeur, I.; Huang, Y.; Ringbauer, H.; Rohrlach, A.B.; Nägele, K.; Villalba-Mouco, V.; et al. Palaeogenomics of Upper Palaeolithic to Neolithic European hunter-gatherers. Nature 2023, 615, 117–126. [Google Scholar] [CrossRef]
- Villalba-Mouco, V.; van de Loosdrecht, M.S.; Rohrlach, A.B.; Fewlass, H.; Talamo, S.; Yu, H.; Aron, F.; Lalueza-Fox, C.; Cabello, L.; Duarte, P.C.; et al. A 23,000-year-old southern Iberian individual links human groups that lived in Western Europe before and after the Last Glacial Maximum. Nat. Ecol. Evol. 2023, 7, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.; Richards, M.; Goios, A.; Alonso, A.; Albarrán, C.; Garcia, O.; Behar, D.M.; Gölge, M.; Hatina, J.; Al-Gazali, L.; et al. High-resolution mtDNA evidence for the late-glacial resettlement of Europe from an Iberian refugium. Genome Res. 2005, 15, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Terberger, T.; Street, M. Hiatus or continuity? New results for the question of pleniglacial settlement in Central Europe. Antiquity 2002, 76, 691–698. [Google Scholar]
- Pârâu, L.G.; Wink, M. Common patterns in the molecular phylogeography of western Palearctic birds: A comprehensive review. J. Ornithol. 2021, 162, 937–959. [Google Scholar] [CrossRef]
- Stern, J.M.; Simes, R.J. Publication bias: Evidence of delayed publication in a cohort study of clinical research projects. BMJ 1997, 315, 640–645. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).