Environmental Factors Shaping the Culturable Freshwater Fungi Diversity of Four Lakes in Yunnan Province, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Experimental Design
2.2. Sample Collection, Isolation, and Morphological Studies
2.3. DNA Extraction, PCR Amplification and Sequencing
2.4. Sequence Alignments and Phylogenetic Analyses
2.5. Measurement of Physical and Chemical Indicators of Water
2.6. Data Analysis
3. Results
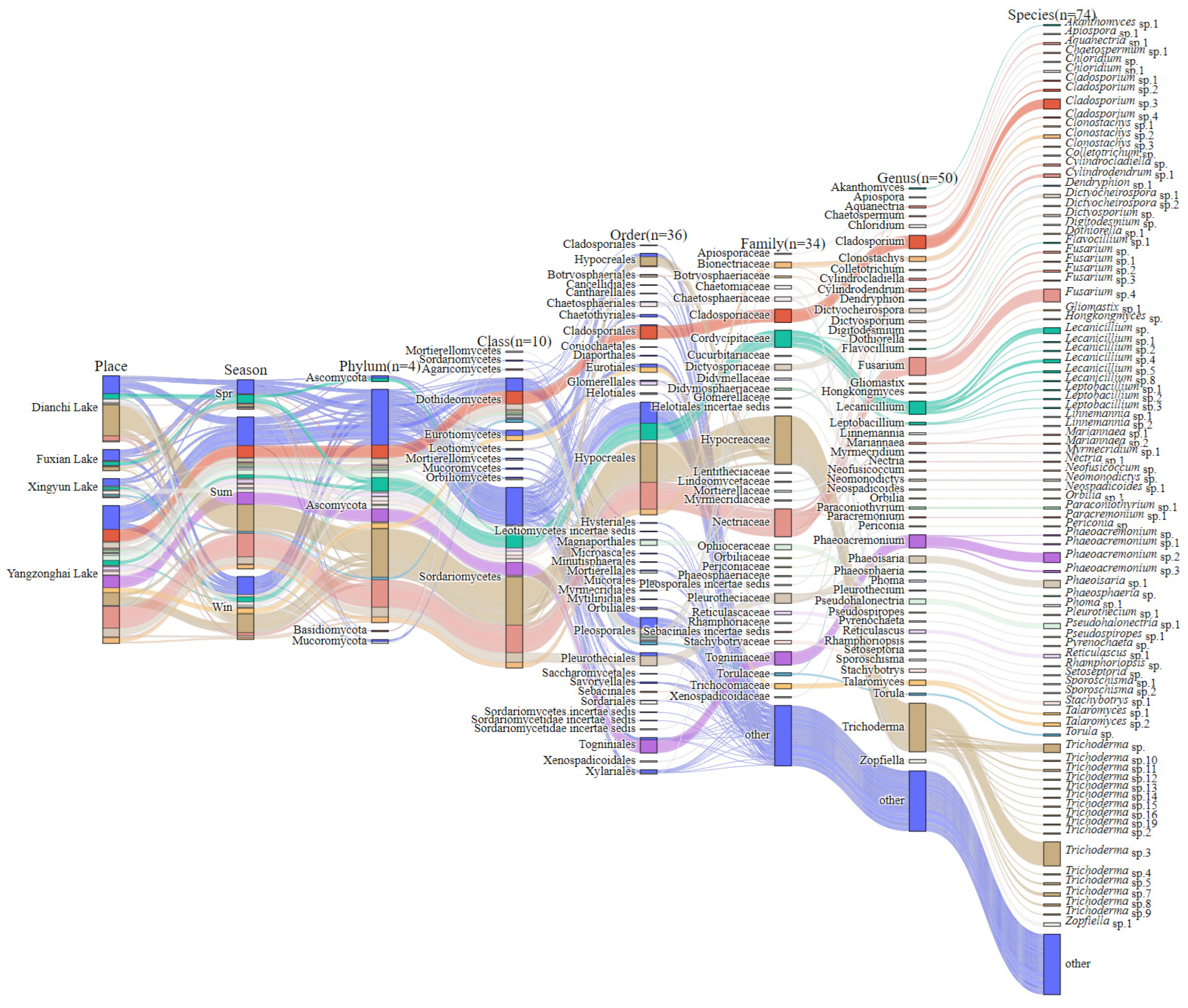
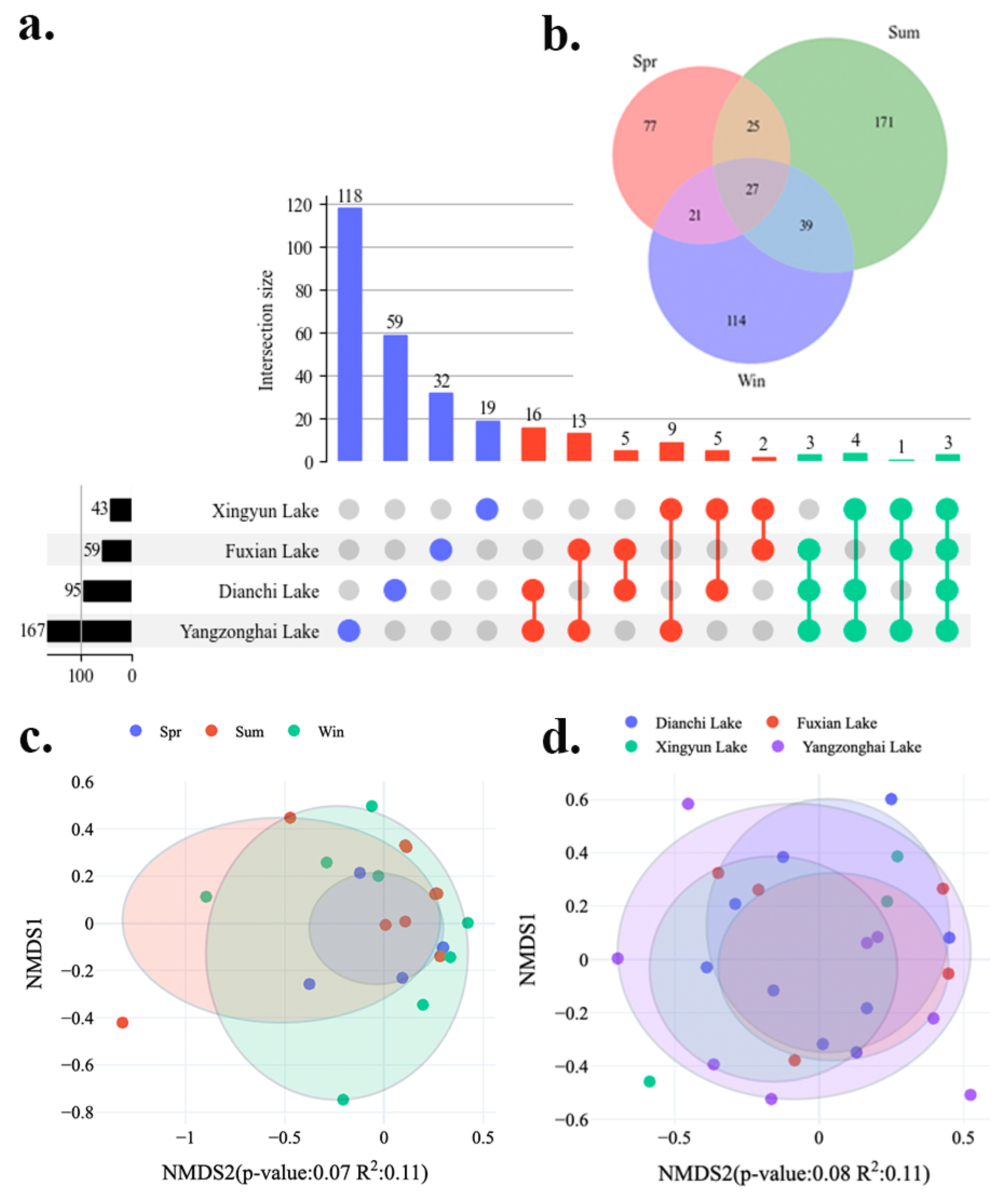
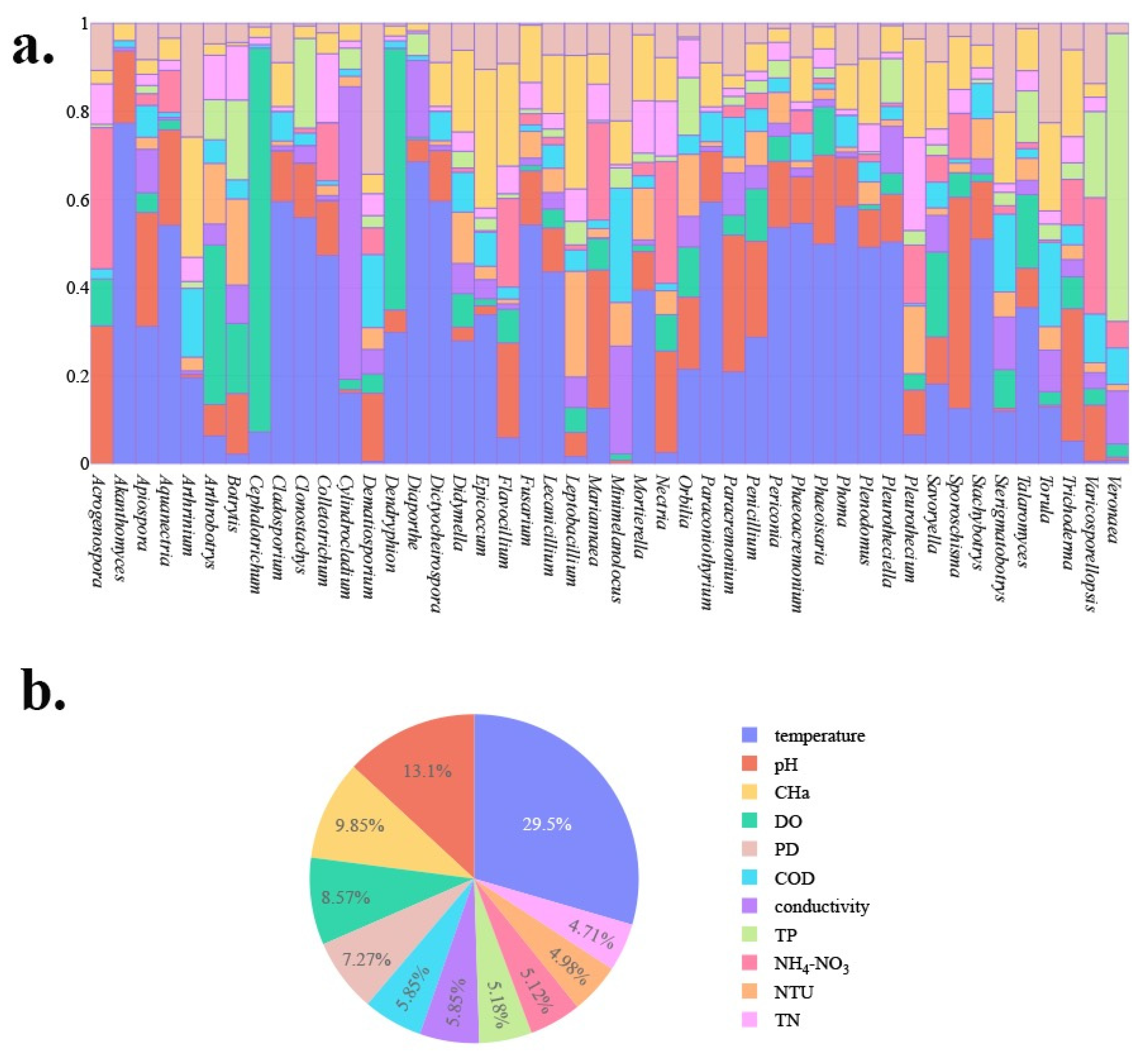
3.1. Culturable Freshwater Fungal Community Composition
Distribution of Culturable Freshwater Fungi Genera and Species in Four Lakes in Yunnan Province, China
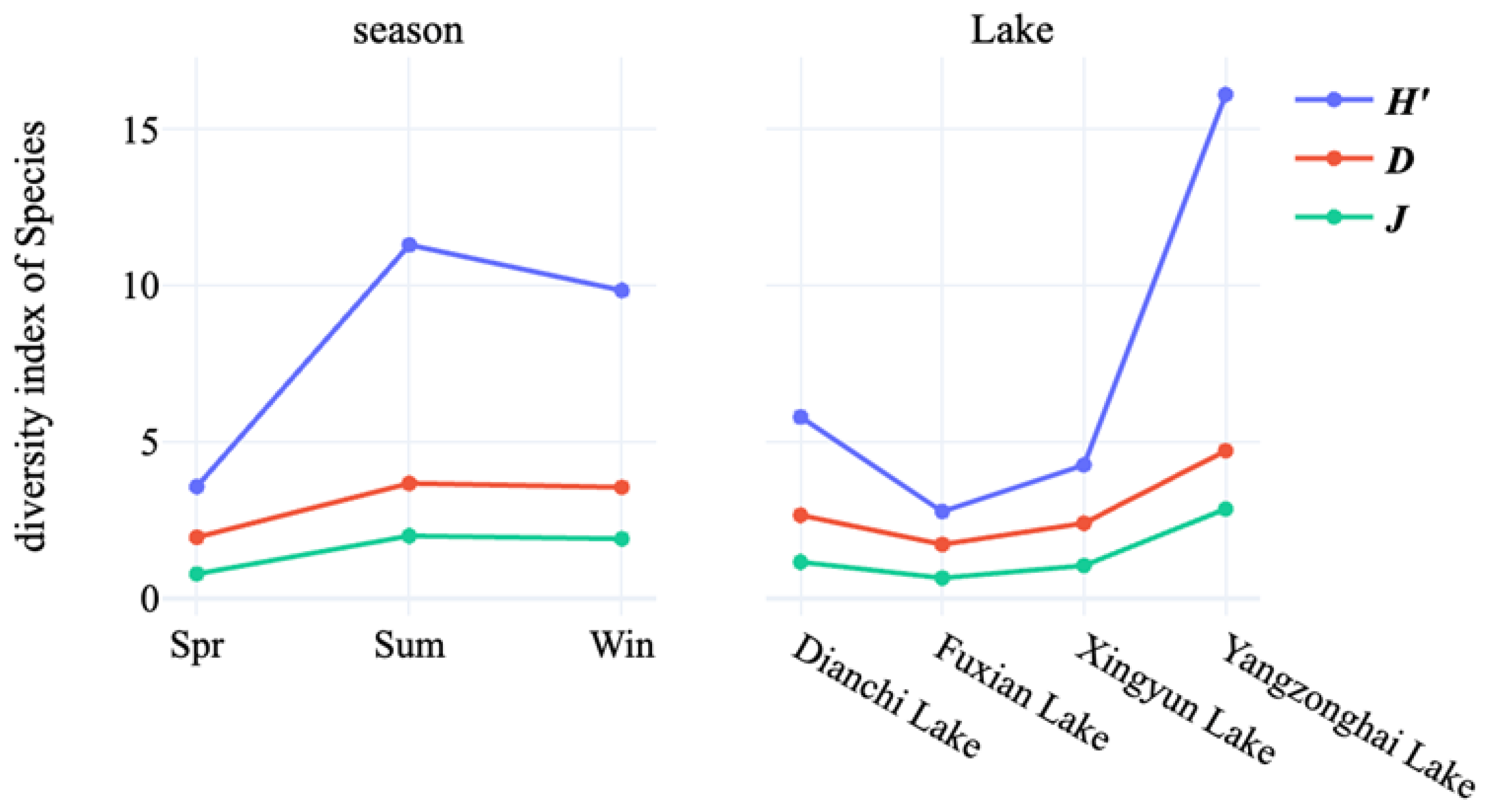
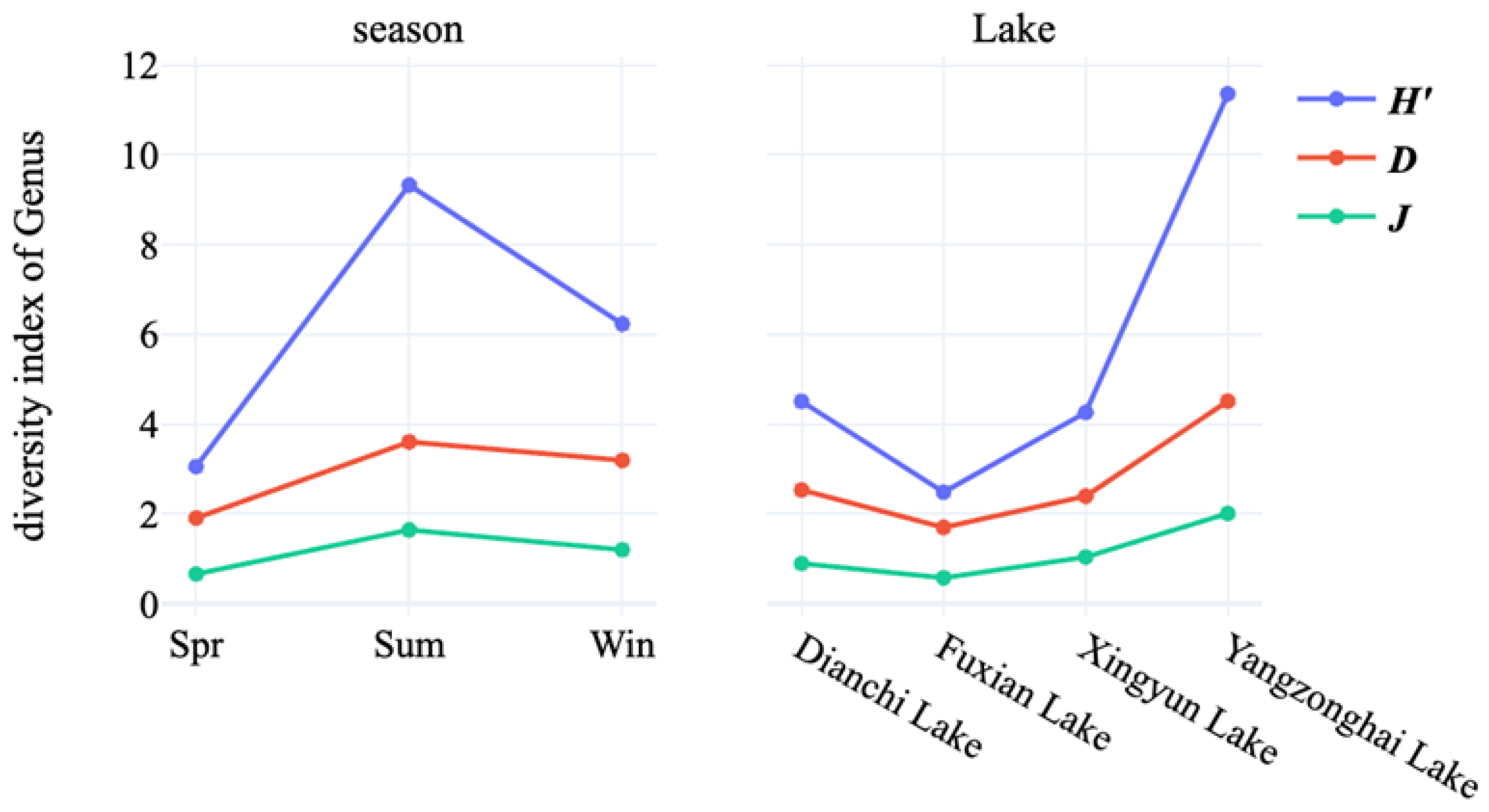
3.2. Diversity Analysis of Cultivable Freshwater Fungal Species in Four Lakes
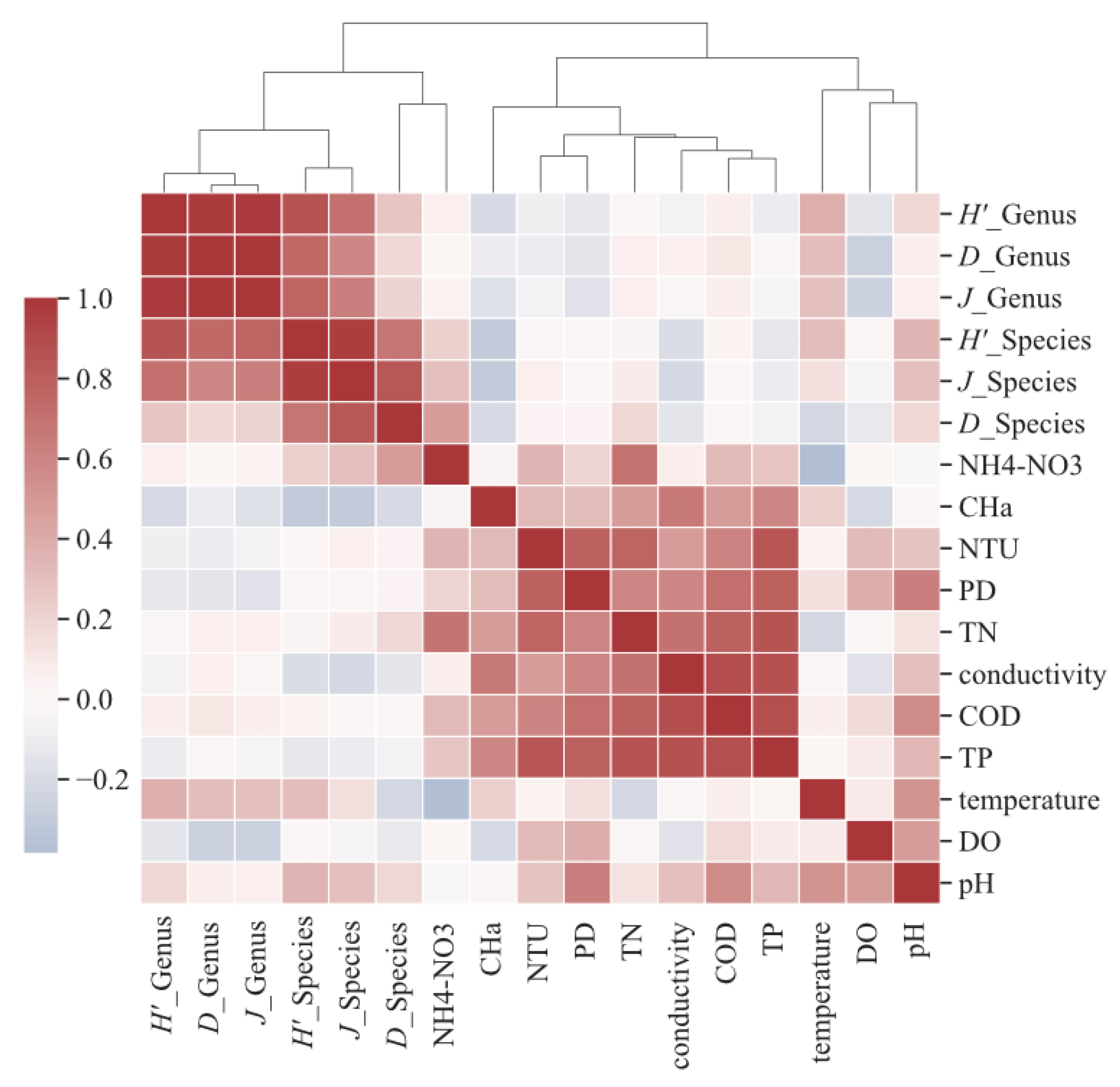
3.3. Correlation Analysis between Diversity Indices of Culturable Freshwater Fungi and Physical and Chemical Factors in Water Bodies
4. Discussion
4.1. Dominant Freshwater Fungal Communities in Four Lakes
4.2. Diversity of Freshwater Fungal Species in Four Lakes
4.3. Environmental Impact Factors of Freshwater Fungi in Four Lakes
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Season | Lake | CHa | COD | DO | NH3–NH4 | NTU | PD | TN | TP | Conductivity | pH | Temperature |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spring | Dianchi Lake | 0.007 | 5.76 | 7.22 | 0.27 | 30.3 | 9,713,549 | 1.98 | 0.074 | 475.9 | 8.38 | 16.8 |
| Spring | Fuxian Lake | 0.001 | 1.023333 | 7.9 | 0.025 | 1.833333 | 514,407.7 | 0.143333 | 0.010667 | 317.8333 | 8.533333 | 16.4 |
| Spring | Xingyun Lake | 0.003 | 7.076667 | 7.293333 | 0.035 | 22.13333 | 18,186,113 | 1.223333 | 0.083333 | 589.8667 | 8.766667 | 18.46667 |
| Spring | Yangzonghai Lake | 0.004 | 4.88 | 9.21 | 0.025 | 2.3 | 2,378,120 | 0.43 | 0.029 | 422.2 | 8.68 | 17.6 |
| Summer | Dianchi Lake | 0.004 | 5.49 | 9.68 | 0.025 | 40.7 | 38,632,520 | 0.89 | 0.075 | 404.7 | 9.13 | 25.5 |
| Summer | Fuxian Lake | 0.003667 | 1.166667 | 7.39 | 0.025 | 1.866667 | 710,880.7 | 0.153333 | 0.010667 | 310.1 | 8.593333 | 23.06667 |
| Summer | Xingyun Lake | 0.012 | 6.72 | 7.21 | 0.025 | 12.2 | 19,067,250 | 0.966667 | 0.074333 | 587.0667 | 8.853333 | 24.7 |
| Summer | Yangzonghai Lake | 0.002 | 4.9 | 7.6 | 0.025 | 2.6 | 4,274,750 | 0.47 | 0.026 | 407.2 | 8.92 | 26 |
| Winter | Dianchi Lake | 0.002 | 6.72 | 8.52 | 0.282 | 16.3 | 30,521,660 | 1.29 | 0.052 | 433 | 9.1 | 18.7 |
| Winter | Fuxian Lake | 0.00225 | 1.115 | 7.0075 | 0.025 | 1.4 | 604,847.8 | 0.1375 | 0.00925 | 314.475 | 8.3725 | 19.25 |
| Winter | Xingyun Lake | 0.00675 | 7.1175 | 7.5925 | 0.025 | 22.3 | 39,477,183 | 1.27 | 0.0825 | 600.125 | 8.89 | 18.7 |
| Winter | Yangzonghai Lake | 0.003 | 4.4 | 7.44 | 0.025 | 2 | 4,176,910 | 0.42 | 0.025 | 410.5 | 9.05 | 21.7 |
References
- Li, H. A study on the lake vegetation of Yunnan Plateau. Acta Bot. Yunnan. 1980, 2, 113–141, (In Chinese with English Abstract). [Google Scholar]
- Dong, Y.X.; Zhao, L.; Chen, Y.H. Succession of nine plateau lakes and regulation of ecological safety in Yunnan Province. Ecol. Econ. 2015, 31, 185–191. (In Chinese) [Google Scholar]
- Shen, H.W.; Bao, D.F.; Bhat, D.J.; Su, H.Y.; Luo, Z.L. Lignicolous freshwater fungi in Yunnan Province, China: An overview. Mycology 2022, 13, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.J.; Li, Y.L.; Shen, J.; Xie, P. Diatom community succession in the recent history of a eutrophic Yunnan Plateau Lake, Lake Dianchi, in subtropical China. Limnology 2009, 10, 247–253. [Google Scholar] [CrossRef]
- Luo, Z.L.; Hyde, K.D.; Liu, J.K.; Maharachchikumbura, S.S.N.; Jeewon, R.; Bao, D.F.; Bhat, D.J.; Lin, C.G.; Li, W.L.; Yang, J.; et al. Freshwater Sordariomycetes. Fungal Divers. 2019, 99, 451–660. [Google Scholar] [CrossRef]
- Dong, W.; Wang, B.; Hyde, K.D.; McKenzie, E.H.C.; Raja, H.A.; Tanaka, K.; Abdel-Wahab, M.A.; Abdel-Aziz, F.A.; Doilom, M.; Phookamsak, R.; et al. Freshwater Dothideomycetes. Fungal Divers. 2020, 105, 319–575. [Google Scholar] [CrossRef]
- Su, H.Y.; Hyde, K.D.; Maharachchikumbura, S.S.N.; Ariyawansa, H.A.; Luo, Z.L.; Promputtha, I.; Tian, Q.; Lin, C.G.; Shang, Q.J.; Zhao, Y.C.; et al. The families Distoseptisporaceae fam. nov., Kirschsteiniotheliaceae, Sporormiaceae and Torulaceae, with new species from freshwater in Yunnan Province, China. Fungal Divers. 2016, 80, 375–409. [Google Scholar] [CrossRef]
- Wang, R.X.; Luo, Z.L.; Hyde, K.D.; Bhat, D.J.; Su, X.J.; Su, H.Y. New species and records of Dictyocheirospora from submerged wood in north-western Yunnan, China. Mycosphere 2016, 7, 1357–1367. [Google Scholar] [CrossRef]
- Li, W.L.; Luo, Z.L.; Liu, J.K.; Bhat, D.J.; Bao, D.F.; Su, H.Y.; Hyde, K.D. Lignicolous freshwater fungi from China I: Aquadictyospora lignicola gen. et sp. nov. and new record of Pseudodictyosporium wauense from northwestern Yunnan Province. Mycosphere 2017, 8, 1587–1597. [Google Scholar] [CrossRef]
- Li, W.L.; Bao, D.F.; Bhat, D.J.; Su, H.Y. Tetraploa aquatica (Tetraplosphaeriaceae), a new freshwater fungal species from Yunnan Province, China. Phytotaxa 2020, 459, 181–189. [Google Scholar] [CrossRef]
- Luo, Z.L.; Hyde, K.D.; Bhat, D.J.; Jeewon, R.; Maharachchikumbura, S.S.N.; Bao, D.F.; Li, W.L.; Su, X.J.; Yang, X.Y.; Su, H.Y. Morphological and molecular taxonomy of novel species Pleurotheciaceae from freshwater habitats in Yunnan, China. Mycol. Prog. 2018, 17, 511–530. [Google Scholar] [CrossRef]
- Luo, Z.L.; Hyde, K.D.; Liu, J.K.; Bhat, D.J.; Bao, D.F.; Li, W.L.; Su, H.Y. Lignicolous freshwater fungi from China II: Novel Distoseptispora (Distoseptisporaceae) species from northwestern Yunnan Province and a suggested unified method for studying lignicolous freshwater fungi. Mycosphere 2018, 9, 444–461. [Google Scholar] [CrossRef]
- Zhao, N.; Luo, Z.L.; Hyde, K.D.; Su, H.Y.; Bhat, D.J.; Liu, J.K.; Bao, D.F.; Hao, Y.E. Helminthosporium submersum sp. nov. (Massarinaceae) from submerged wood in north-western Yunnan Province, China. Phytotaxa 2018, 348, 269–278. [Google Scholar] [CrossRef]
- Bao, D.F.; McKenzie, E.H.C.; Bhat, D.J.; Hyde, K.D.; Luo, Z.L. Acrogenospora (Acrogenosporaceae, Minutisphaerales) appears to be a very diverse genus. Front. Microbiol. 2020, 11, 1606. [Google Scholar] [CrossRef]
- Wan, Y.L.; Bao, D.F.; Luo, Z.L.; Bhat, D.J.; Xu, Y.X.; Su, H.Y.; Hao, Y.E. Two new species of Minimelanolocus (Herpotrichiellaceae, Chaetothyriales) from submerged wood in Yunnan, China. Phytotaxa 2021, 480, 45–56. [Google Scholar] [CrossRef]
- Thomas, K. Australian freshwater. In Introductory Volume to the Fungi (Part2). Fungi of Australian; Grgurinovic, C.A., Ed.; Australian Biological Resources Study: Canberra, Australia, 1996; pp. 1–37. [Google Scholar]
- Calabon, M.S.; Hyde, K.D.; Jones, E.B.G.; Luo, Z.L.; Dong, W.; Hurdeal, V.G.; Gentekaki, E.; Rossi, W.; Leonardi, M.; Thiyagaraja, V.; et al. Freshwater Fungal Numbers. Fungal Divers. 2022, 114, 3–235. [Google Scholar] [CrossRef]
- Gleason, F.H.; Kagami, M.; Lefevre, E.; Sime-Ngando, T. The ecology of chytrids in aquatic ecosystems: Roles in food web dynamics. Fungal Biol. Reviews. 2008, 22, 17–25. [Google Scholar] [CrossRef]
- Wong, S.W.; Hyde, K.D.; Jones, E.B.G. Ultrastructure studies on freshwater ascomycetes, Fluminicolabipolaris gen. et sp. nov. Fungal Divers. 1999, 2, 189–197. [Google Scholar]
- Cai, L.; Zhang, K.Q.; Hyde, K.D. Freshwater ascomycetes. In Freshwater Mycology; Tsui, C.K.M., Hyde, K.D., Eds.; Fungal Diversity Press: Hong Kong, China, 2003; pp. 275–326. [Google Scholar]
- Cai, L.; Zhang, K.Q.; McKenzie, E.H.C.; Hyde, K.D. Freshwater fungi from bamboo and wood submergedin the Liput River in the Philippines. Fungal Divers. 2003, 13, 1–12. [Google Scholar]
- Jones, E.B.G.; Choeyklin, R. Ecology of marine and freshwater basidiomycetes. In Ecology of Saprotrophic Basidiomycetes; Boddy, L., Frankland, J.C., van West, P., Eds.; Academic Press: London, UK, 2008; pp. 301–324. [Google Scholar]
- Wong, M.K.; Goh, T.K.; Hodgkiss, I.J.; Hyde, K.D.; Ranghoo, V.M.; Tsui, C.K.; Ho, W.H.; Wong, W.S.; Yuen, T.K. Role of fungi in freshwater ecosystems. Biodivers. Conserv. 1998, 7, 1187–1206. [Google Scholar] [CrossRef]
- Gessner, M.O.; Gulis, V.; Kuehn, K.A.; Chauvet, E.; Suberkropp, K. Fungal decomposers of plant litter in aquatic ecosystems. In The Mycota. A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research. Environmental and Microbial Relationships; Kubicek, C.P., Druzhinina, I.S., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 4, pp. 301–324. [Google Scholar]
- Kuehn, K.A. The role of fungi in the decomposition of emergent wetland plants. Nov. Tech. Ideas Mycol. 2008, 20, 19–41. [Google Scholar]
- Krauss, G.J.; Sole, M.; Krauss, G.; Schlosser, D.; Wesenberg, D.; Baerlocher, F. Fungi in freshwaters: Ecology, physiology and biochemical potential. Fems Microbiol. Ecol. 2011, 35, 620–651. [Google Scholar] [CrossRef] [PubMed]
- Koske, R.E.; Duncan, I.W. Temperature effects on growth, sporulation, and germination of some “aquatic” hyphomycetes. Canad. J. Bot. 1973, 52, 1387–1391. [Google Scholar] [CrossRef]
- Zare-Maivan, H.; Shearer, C.A. Extracellular enzyme production and cell wall degradation by freshwater lignicolous fungi. Mycologia 1988, 80, 365–375. [Google Scholar] [CrossRef]
- Duarte, S.; Fernandes, I.; Nogueira, M.J.; Cássio, F.; Pascoal, C. Temperature alters interspecific relationships among aquatic fungi. Fungal Ecol. 2013, 6, 187–191. [Google Scholar] [CrossRef]
- Powers, S.M.; McCutchan, J.H.; Finlay, J.C. The Role of Freshwater Fungi in the Decomposition of Leaf Litter and Wood in Streams. J. Freshwater Ecol. 2021, 36, 105–122. [Google Scholar] [CrossRef]
- Gulis, V.; Suberkropp, K.; Dudgeon, D. Fungal Degradation of Wood in Stream Environments and Its Impact on Aquatic Invertebrates. Hydrobiologia 2022, 849, 639–652. [Google Scholar] [CrossRef]
- Kahl, J.S.; Lutz, C.; Miller, D. Effects of Freshwater Fungal Activity on Habitat Structure and Biodiversity in Stream Ecosystems. Aquat. Ecol. 2023, 57, 83–98. [Google Scholar] [CrossRef]
- Barton, D.C.; Gollnisch, R.E.; Liu, Z. Freshwater Fungi and Their Role in Water Quality and Nutrient Cycling. Water Res. 2023, 220, 1183–1194. [Google Scholar] [CrossRef]
- Eastwood, W.J.; Roberts, N.; Lamb, H.F.; Tibby, J.C. Holocene environmental change in southwest Turkey: A palaeoecological record of lake and catchment- related changes. Quat. Sci Rev. 1999, 18, 671–695. [Google Scholar] [CrossRef]
- Ye, X.C.; Zhang, Q.; Liu, J.; Li, X.H.; Xu, C.Y. Distinguishing the relative impacts of climate change and human activities on variation of streamflow in the Poyang Lake catchment, China. J. Hydrol. 2013, 494, 83–95. [Google Scholar] [CrossRef]
- Ho, J.C.; Michalak, A.M.; Pahlevan, N. Widespread global increase in intense lake phytoplankton blooms since the 1980s. Nature 2019, 574, 667–670. [Google Scholar] [CrossRef] [PubMed]
- Woolway, R.I.; Merchant, C.J. Worldwide alteration of lake mixing regimes in response to climate change. Nat. Geosci. 2019, 12, 271–276. [Google Scholar] [CrossRef]
- Chen, Y.X.; Lou, L.P.; Li, W.H. Effect of microorganisms on the biogenic matter circulation of lake. China water pollution control and ecological restoration technology advanced seminar, Hangzhou. 2004. Available online: https://ouci.dntb.gov.ua/en/works/7qbaJvB4/ (accessed on 2 June 2024)(In Chinese with English Abstract).
- Yang, J.X.; Wang, X.A.; Pan, X.F. An innovative restoration mode “macrophytes—fishes—benthons—birds” implemented in aesthetic plateau wetlands. Bull. Chin. Acad. Sci. 2023, 38, 1915–1923. (In Chinese) [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M., Gelfand, D., Shinsky, J., White, T., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Capella, G.S.; Silla, M.J.M.; Gabaldón, T. TrimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Miller, M.A.; Schwartz, T.; Pickett, B.E.; He, S.; Klem, E.B.; Scheuermann, R.H.; Passarotti, M.; Kaufman, S.; O’Leary, M.A.A. Restful API for access to phylogenetic tools via the CIPRES science gateway. Evol. Bioinform. 2015, 11, 43–48. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree. In Tree Figure Drawing Tool, Version 1.3.1; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2006. [Google Scholar]
- Zhang, Y.; Lu, J.; Wu, J.; Wang, J.; Lin, Y. Occurrence and distribution of antibiotic resistance genes in sediments in a semi-enclosed continental shelf sea. Sci. Total Environ. 2020, 720, 137712. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Z.; Xu, H.; Wang, L.; Liu, R.; Jia, X. Fate of antibiotic resistance genes and bacteria in a coupled water-processing system with wastewater treatment plants and constructed wetlands in coastal eco-industrial parks. Ecotoxicol. Environ. Saf. 2023, 252, 114606. [Google Scholar] [CrossRef] [PubMed]
- Sievert, C. Interactive Web-Based Data Visualization with R, Plotly, and Shiny; Chapman and Hall/CRC: Boca Raton, FL, USA, 2020; Available online: https://plotly-r.com (accessed on 2 June 2024).
- Lex, A.; Nils, G.; Hendrik, S.; Romain, V.; Hanspeter, P. UpSet: Visualization of Intersecting Sets. IEEE Trans. Vis. Comput. Graph. 2014, 20, 1983–1992. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Simpson, G.L.; Guillaume, F.; Roeland, B.; Pierre, K.; Peter, L.; Minchin, R.; O’Hara, R.B.; Solymos, P.; Stevens, M.; et al. Vegan: Community Ecology Package. Manual. 2022. Available online: https://CRAN.R-project.org/package=vegan (accessed on 2 June 2024).
- Virtanen, P.; Ralf, G.; Travis, E.; Matt, H.; Tyler, R.; David, C.; Evgeni, B.; Pearu, P.; Warren, W.; Jonathan, B. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 2 June 2024).
- Cheng, X.; Li, W.; Zhang, T. A new species of Phaeoisaria from intertidal marine sediment collected in Weihai, China. Mycotaxon 2014, 127, 17–24. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Burgess, T.I.; Carnegie, A.J.; Hardy, G.E.; Smith, D.; Summerell, B.A.; Cano-Lira, J.F.; Guarro, J.; Houbraken, J.; et al. Fungal Planet description sheets: 625-715. Persoonia: Molecular Phylogeny and Evolution of Fungi. Persoonia 2017, 39, 270–467. [Google Scholar] [CrossRef]
- Hyde, K.D.; Tennakoon, D.S.; Jeewon, R.; Bhat, D.J.; Maharachchikumbura, S.S.N.; Rossi, W.; Leonardi, M.; Lee, H.B.; Mun, H.Y.; Houbraken, J.; et al. Fungal diversity notes 1036–1150: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2019, 96, 1–242. [Google Scholar] [CrossRef]
- Bucher, V.V.C.; Pointing, S.B.; Hyde, K.D.; Reddy, C.A. Production of wood decay enzymes, loss of mass, and lignin solubilization in wood by diverse tropical freshwater fungi. Microb. Ecol. 2004, 48, 331–337. [Google Scholar] [CrossRef]
- Vijaykrishna, D.; Jeewon, R.; Hyde, K.D. Fusoidispora aquatica: A new freshwater ascomycete from Hong Kong based on morphology and phylogeny inferred from rDNA gene sequences. Sydowia 2005, 57, 267–280. [Google Scholar]
- Hyde, K.D.; Fryar, S.; Tian, Q.; Bahkali, A.H.; Xu, J.C. Lignicolous freshwater fungi along a north-south latitudinal gradient in the Asian/Australian region; can we predict the impact of global warming on biodiversity and function? Fungal Ecol. 2016, 19, 190–200. [Google Scholar] [CrossRef]
- Guarro, J.; Vieira, L.A.; de Freitas, D.; Gené, J.; Zaror, L.; Hofling-Lima, A.L.; Fischman, O.; Zorat-Yu, C.; Figueras, M.J. Phaeoisaria clematidis as a cause of keratomycosis. J. Clin. Microbiol. 2000, 38, 2434–2437. [Google Scholar] [CrossRef] [PubMed]
- Chew, H.F.; Jungkind, D.L.; Mah, D.Y.; Raber, I.M.; Toll, A.D.; M, J.T.; Cohen, E.J. Posttraumatic fungal keratitis caused by Carpoligna sp. Cornea 2010, 29, 449–452. [Google Scholar] [CrossRef] [PubMed]
- Felse, P.A.; Panda, T. Production of xylanase by Trichoderma longibrachiatum on a mixture of wheat bran and wheat straw: Optimization of culture condition by Taguchi method. Enzyme Microb. Tech. 1999, 40, 801–805. [Google Scholar] [CrossRef]
- Azin, M.; Moravej, R.; Zareh, D. Self-directing optimization of parameters for extracellular chitinase production by Trichoderma harzianum in batch mode. Process Biochem. 2007, 34, 563–566. [Google Scholar] [CrossRef]
- O’Donnell, K.; Ward, T.J.; Robert, V.A.R.G.; Crous, P.W.; Geiser, D.M.; Kang, S. DNA Sequence-Based Identification of Fusarium: Current Status and Future Directions. Phytoparasitica 2015, 43, 583–595. [Google Scholar] [CrossRef]
- Gupta, V.K.; Varma, A. Fusarium: Pathogenicity and Biotechnology. In Fungi: Applications and Biotechnology, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2019; pp. 155–173. [Google Scholar]
- Pai, R.; Boloor, R.; Shreevidya, K.; Shenoy, D. Fusarium solani: An Emerging Fungus in Chronic Diabetic Ulcer. J. Lab. Physicians 2010, 2, 37–39. [Google Scholar] [CrossRef]
- Huang, Y.; Busk, P.K.; Lange, L. Cellulose and Hemicellulose-Degrading Enzymes in Fusarium commune Transcriptome and Functional Characterization of Three Identified Xylanases. Enzym. Microb. Technol. 2015, 73–74, 9–19. [Google Scholar] [CrossRef]
- El Hajj Assaf, C.; Zetina-Serrano, C.; Tahtah, N.; Khoury, A.E.; Atoui, A.; Oswald, I.P.; Puel, O.; Lorber, S. Regulation of Secondary Metabolism in the Penicillium Genus. Int. J. Mol. Sci. 2020, 21, 9462. [Google Scholar] [CrossRef]
- Cairns, T.C.; Nai, C.; Meyer, V. How a Fungus Shapes Biotechnology: 100 Years of Aspergillus niger Research. Fungal Biol. Biotechnol. 2018, 5, 13. [Google Scholar] [CrossRef]
- Li, Z.H.; Pubu, C.R.; Lyu, M.L.; Wang, M.; Liu, X.Y. Species diversity of zygomycotan fungi in the Tibet Autonomous Region. Microbiol. China 2018, 45, 1250–1261, (In Chinese with English Abstract). [Google Scholar]
- Liu, A.R.; Yang, T.; Xu, W.; Shangguan, Z.J.; Wang, J.Z.; Liu, H.Y.; Shi, Y.; Chu, H.Y.; He, J.S. Status, issues and prospects of belowground biodiversity on the Tibetan alpine grassland. Biodivers. Sci. 2018, 26, 972–987, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Liu, Y.X.; Cao, P.X.; Ma, H.M.; Liu, X. Research progress on soil microbial diversity and its influencing factors in Qinghai-Tibet Plateau. Environ. Ecol. 2019, 1, 1–7, (In Chinese with English Abstract). [Google Scholar]
- Kirchman, D.L. The ecology of Cytophaga-Flavobacteria in aquatic environments. Fems. Microbiol. Ecol. 2002, 39, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Zou, T.W.; Ren, L.W.; Zhang, Y.T.; Ding, R.; Zhang, H. The overview of the diversity of aquatic fungi. Heilongjiang Sci. 2015, 6, 25–29, (In Chinese with English Abstract). [Google Scholar]
- Tsui, C.K.M.; Hyde, K.D.; Hodgkiss, I.J. Colonization pattern of wood-inhabiting fungi on baits in Hong Kong rivers, with reference to the effects of organic pollution. Antonie Van Leeuwenhoek 2001, 79, 33–38. [Google Scholar] [CrossRef]
- Tsui, C.K.M.; Hyde, K.D.; Hodgkiss, I.J. Longitudinal and temporal distribution of freshwater ascomycetes and dematiaceous hyphomycetes on submerged wood in the Lam Tsuen River, Hong Kong. J. N. Am. Benthol. Soc. 2001, 20, 533–549. [Google Scholar] [CrossRef]
- Bärlocher, F. Aquatic hyphomycete spora in 10 streams of New Brunswick and Nova Scotia. Can. J. Bot. 2011, 65, 76–79. [Google Scholar] [CrossRef]
- Eaton, J.G.; Scheller, R.M. Effects of Climate Warming on Fish Thermal Habitat in Streams of the United States. Limnol. Oceanogr. 1996, 41, 1109–1115. [Google Scholar] [CrossRef]
- Chauvet, E.; Suberkropp, K. Temperature and sporulation of aquatic hyphomycetes. Appl. Environ. Microbiol. 1998, 64, 1522–1525. [Google Scholar] [CrossRef]
- Graca, M.A.S.; Bärlocher, F.; Gessner, M.O. Aquatic hyphomycetes: Ecology and biological activity. In Aquatic Mycology; Springer: Berlin/Heidelberg, Germany, 2015; pp. 69–99. [Google Scholar]
- Liu, X.; Xu, X.; Zhang, Y.; Wang, Y. Effects of dissolved oxygen on microbial diversity and enzyme activity in aquatic environments. Environ. Sci. Technol. 2013, 47, 12453–12460. [Google Scholar]
| Sites | Latitude | Longitude | Altitude (m) | Lake Area (km2) | Collection Time |
|---|---|---|---|---|---|
| Dianchi Lake | 24°22′–25°36′ | 102°22′–102°58′ | 1887.5 | 330 | 13 April 2022 |
| 26 July 2022 | |||||
| 13 October 2022 | |||||
| Fuxian Lake | 24°31′–24°51′ | 102°43′–102°59′ | 1720 | 212 | 21 April 2022 |
| 14 August 2022 | |||||
| 20 October 2022 | |||||
| 29 December 2022 | |||||
| 22 March 2023 | |||||
| 1 July 2023 | |||||
| Xingyun Lake | 24°33′ | 102°78′ | 1722 | 34.71 | 21 April 2022 |
| 13 August 2022 | |||||
| 20 October 2022 | |||||
| Yangzonghai Lake | 24°51′–24°58′ | 102°5′–103°02′ | 1770 | 31.1 | 10 March 2022 |
| 12 July 2022 | |||||
| 18 October 2022 |
| Genus | Dianchi Lake | Fuxian Lake | Xingyun Lake | Yangzonghai Lake | OF (%) | RA (%) |
|---|---|---|---|---|---|---|
| Acremonium | 1 | 0 | 0 | 0 | 25 | 0.18 |
| Acrogenospora | 1 | 0 | 0 | 0 | 25 | 0.18 |
| Akanthomyces | 0 | 0 | 0 | 3 | 25 | 0.53 |
| Alternaria | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Apiospora | 6 | 0 | 3 | 1 | 75 | 1.77 |
| Aquanectria | 1 | 0 | 0 | 3 | 50 | 0.71 |
| Arthrinium | 1 | 1 | 0 | 0 | 50 | 0.35 |
| Arthrobotrys | 1 | 1 | 0 | 1 | 75 | 0.53 |
| Aspergillus | 0 | 0 | 0 | 3 | 25 | 0.53 |
| Atractium | 0 | 0 | 0 | 2 | 25 | 0.35 |
| Bipolaris | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Botryosphaeria | 0 | 1 | 0 | 0 | 25 | 0.18 |
| Botrytis | 0 | 1 | 0 | 0 | 25 | 0.18 |
| Cancellidium | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Capronia | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Cephalosporium | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Cephalotrichum | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Chaetomium | 0 | 1 | 0 | 0 | 25 | 0.18 |
| Chaetosphaeria | 1 | 0 | 0 | 0 | 25 | 0.18 |
| Chloridium | 6 | 0 | 0 | 0 | 25 | 1.06 |
| Circinella | 3 | 0 | 0 | 0 | 25 | 0.53 |
| Cladosporium | 0 | 3 | 2 | 15 | 75 | 3.54 |
| Clohesyomyces | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Clonostachys | 1 | 0 | 1 | 14 | 75 | 2.83 |
| Colletotrichum | 2 | 0 | 0 | 2 | 50 | 0.71 |
| Coniochaeta | 0 | 1 | 0 | 0 | 25 | 0.18 |
| Cordyceps | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Cosmospora | 2 | 0 | 0 | 0 | 25 | 0.35 |
| Cylindrocladiella | 1 | 0 | 0 | 3 | 50 | 0.71 |
| Cylindrocladium | 0 | 1 | 0 | 0 | 25 | 0.18 |
| Cylindrodendrum | 0 | 0 | 0 | 4 | 25 | 0.71 |
| Dactylium | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Dactylonectria | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Dematiosporium | 2 | 0 | 1 | 0 | 50 | 0.53 |
| Dendryphion | 0 | 0 | 0 | 3 | 25 | 0.53 |
| Diaporthe | 0 | 1 | 0 | 1 | 50 | 0.35 |
| Dictyocheirospora | 1 | 1 | 1 | 7 | 100 | 1.77 |
| Dictyocheirospora | 0 | 1 | 0 | 0 | 25 | 0.18 |
| Dictyosporium | 0 | 0 | 0 | 5 | 25 | 0.88 |
| Didymella | 0 | 3 | 0 | 4 | 50 | 1.24 |
| Didymellaceae | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Digitodesmium | 0 | 0 | 2 | 0 | 25 | 0.35 |
| Distoseptispora | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Dothiorella | 0 | 0 | 2 | 1 | 50 | 0.53 |
| Entoleuca | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Epicoccum | 0 | 1 | 0 | 1 | 50 | 0.35 |
| Exophiala | 1 | 0 | 2 | 3 | 75 | 1.06 |
| Flavocillium | 3 | 1 | 0 | 1 | 75 | 0.88 |
| Fusarium | 10 | 3 | 4 | 22 | 100 | 6.9 |
| Fusicolla | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Galactomyces | 0 | 1 | 0 | 0 | 25 | 0.18 |
| Geosmithia | 0 | 1 | 0 | 0 | 25 | 0.18 |
| Gliomastix | 1 | 0 | 0 | 3 | 50 | 0.71 |
| Graphium | 0 | 1 | 0 | 0 | 25 | 0.18 |
| Halobyssothecium | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Helminthosporium | 1 | 0 | 0 | 0 | 25 | 0.18 |
| Hongkongmyces | 0 | 0 | 0 | 2 | 25 | 0.35 |
| Hyalorbilia | 1 | 0 | 0 | 0 | 25 | 0.18 |
| Hydropisphaera | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Idriella | 1 | 0 | 0 | 0 | 25 | 0.18 |
| Juxtiphoma | 1 | 0 | 0 | 0 | 25 | 0.18 |
| Lecanicillium | 9 | 7 | 5 | 10 | 100 | 5.49 |
| Lentithecium | 0 | 0 | 1 | 1 | 50 | 0.35 |
| Leptobacillium | 4 | 0 | 3 | 4 | 75 | 1.95 |
| Linnemannia | 3 | 0 | 0 | 2 | 50 | 0.88 |
| Lophiostoma | 0 | 1 | 0 | 0 | 25 | 0.18 |
| Mariannaea | 8 | 0 | 2 | 3 | 75 | 2.3 |
| Memnoniella | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Minimelanolocus | 0 | 0 | 1 | 0 | 25 | 0.18 |
| Montagnula | 0 | 1 | 0 | 0 | 25 | 0.18 |
| Mortierella | 1 | 0 | 0 | 2 | 50 | 0.53 |
| Mucor | 2 | 0 | 0 | 0 | 25 | 0.35 |
| Myrmecridium | 0 | 0 | 0 | 2 | 25 | 0.35 |
| Mytilinidion | 1 | 0 | 0 | 0 | 25 | 0.18 |
| Myxotrichum | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Nectria | 4 | 0 | 0 | 0 | 25 | 0.71 |
| Nectriopsis | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Neofusicoccum | 0 | 0 | 0 | 3 | 25 | 0.53 |
| Neomonodictys | 0 | 0 | 0 | 4 | 25 | 0.71 |
| Neomultiseptospora | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Neopestalotiopsis | 1 | 0 | 0 | 1 | 25 | 0.35 |
| Neopyrenochaeta | 1 | 1 | 1 | 1 | 100 | 0.71 |
| Neospadicoides | 2 | 0 | 0 | 0 | 25 | 0.35 |
| Nigrospora | 0 | 1 | 0 | 0 | 25 | 0.18 |
| Orbilia | 1 | 2 | 0 | 1 | 75 | 0.71 |
| Paracamarosporium | 1 | 0 | 0 | 1 | 50 | 0.35 |
| Paraconiothyrium | 0 | 1 | 0 | 4 | 50 | 0.88 |
| Paracremonium | 1 | 0 | 3 | 3 | 75 | 1.24 |
| Penicillium | 2 | 2 | 1 | 3 | 100 | 1.42 |
| Periconia | 1 | 1 | 1 | 3 | 100 | 1.06 |
| Pestalotiopsis | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Phaeoacremonium | 6 | 4 | 1 | 14 | 100 | 4.42 |
| Phaeoisaria | 2 | 1 | 0 | 9 | 75 | 2.12 |
| Phaeosphaeria | 0 | 0 | 0 | 2 | 25 | 0.35 |
| Phoma | 1 | 2 | 0 | 3 | 75 | 1.06 |
| Phomopsis | 1 | 0 | 0 | 0 | 25 | 0.18 |
| Plectosphaerella | 0 | 1 | 0 | 0 | 25 | 0.18 |
| Plenodomus | 1 | 1 | 0 | 0 | 25 | 0.35 |
| Pleurotheciella | 0 | 1 | 1 | 3 | 75 | 0.88 |
| Pleurothecium | 3 | 0 | 0 | 0 | 25 | 0.53 |
| Podila | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Pseudoastrosphaeriella | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Pseudohalonectria | 0 | 0 | 1 | 7 | 50 | 1.42 |
| Pseudorobillarda | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Pseudospiropes | 0 | 0 | 0 | 2 | 25 | 0.35 |
| Purpureocillium | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Pyrenochaeta | 0 | 0 | 0 | 2 | 25 | 0.35 |
| Pyrenochaetopsis | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Reticulascus | 0 | 0 | 0 | 4 | 25 | 0.71 |
| Rhamphoriopsis | 2 | 0 | 0 | 0 | 25 | 0.35 |
| Rhinocladiella | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Rhytidhysteron | 1 | 0 | 0 | 1 | 50 | 0.35 |
| Samsoniella | 0 | 1 | 0 | 0 | 25 | 0.18 |
| Savoryella | 0 | 1 | 1 | 1 | 75 | 0.53 |
| Setoseptoria | 0 | 0 | 0 | 2 | 25 | 0.35 |
| Simplicillium | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Sporoschisma | 3 | 0 | 0 | 2 | 50 | 0.88 |
| Stachybotrys | 0 | 3 | 0 | 5 | 50 | 1.42 |
| Stephanonectria | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Sterigmatobotrys | 1 | 1 | 1 | 0 | 75 | 0.53 |
| Striatibotrys | 1 | 0 | 0 | 0 | 25 | 0.18 |
| Talaromyces | 0 | 1 | 1 | 10 | 75 | 2.12 |
| Thelonectria | 1 | 0 | 0 | 0 | 25 | 0.18 |
| Thyridium | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Torula | 0 | 2 | 4 | 0 | 50 | 1.06 |
| Trematophoma | 0 | 0 | 0 | 1 | 25 | 0.18 |
| Trichoderma | 33 | 13 | 7 | 29 | 100 | 14.51 |
| Varicosporellopsis | 1 | 0 | 1 | 0 | 50 | 0.35 |
| Veronaea | 1 | 0 | 1 | 0 | 50 | 0.35 |
| Verticillium’ clade | 0 | 1 | 0 | 1 | 50 | 0.35 |
| Zopfiella | 0 | 0 | 5 | 0 | 25 | 0.88 |
| unclassified Hyaloscyphaceae | 1 | 0 | 0 | 0 | 25 | 0.18 |
| Xenoacremonium | 1 | 0 | 0 | 0 | 25 | 0.18 |
| TA(cfu/L) | 151 | 74 | 60 | 280 |
| Culture Number | GenBank Number | Blast Result | |
|---|---|---|---|
| Species Name | Identity (%) | ||
| KUNCC23-13242 | PP620729 | Apiospora sp.1 | 99.09% |
| KUNCC23-13240 | PP620730 | Apiospora sp.2 | 100% |
| KUNCC23-13224 | PP620731 | Apiospora sp.3 | 100% |
| KUNCC22-12641 | PP620732 | Apiospora sp. | 100% |
| KUNCC23-13007 | PP620733 | Apiospora sp.4 | 97.21% |
| KUNCC23-13264 | PP620734 | Cladosporium sp.1 | 99.80% |
| KUNCC23-12949 | PP620735 | Cladosporium sp.2 | 100% |
| KUNCC22-12617 | PP620736 | Cladosporium sp.3 | 100% |
| KUNCC23-13256 | PP620737 | Cladosporium sp.4 | 100% |
| KUNCC23-13678 | PP620738 | Cladosporium sp.5 | 100% |
| KUNCC22-12559 | PP620739 | Cladosporium sp. | 100% |
| KUNCC23-13198 | PP620740 | Cladosporium sp.6 | 100% |
| KUNCC22-12584 | PP620741 | Cladosporium sp.7 | 100% |
| KUNCC22-12605 | PP620742 | Clonostachys sp.1 | 100% |
| KUNCC23-12936 | PP620743 | Clonostachys sp.2 | 100% |
| KUNCC23-12897 | PP620744 | Clonostachys sp. | 100% |
| KUNCC22-12575 | PP620745 | Dictyocheirospora sp.1 | 100% |
| KUNCC23-13639 | PP620746 | Dictyocheirospora sp.2 | 99.81% |
| KUNCC23-16675 | PP620747 | Dictyocheirospora sp.3 | 100% |
| KUNCC22-12506 | PP620748 | Fusarium sp.7 | 100% |
| KUNCC23-12721 | PP620749 | Fusarium sp.6 | 100% |
| KUNCC23-14455 | PP620750 | Fusarium sp.5 | 100% |
| KUNCC23-13216 | PP620751 | Fusarium sp.4 | 100% |
| KUNCC23-12923 | PP620752 | Fusarium sp.3 | 99.81% |
| KUNCC23-14545 | PP620753 | Fusarium sp.2 | 100% |
| KUNCC22-12579 | PP620754 | Fusarium sp.1 | 100% |
| KUNCC22-12583 | PP620755 | Fusarium sp. | 99.24% |
| KUNCC23-12960 | PP620756 | Lecanicillium sp.1 | 100% |
| KUNCC22-12552 | PP620757 | Lecanicillium sp.2 | 99.64% |
| KUNCC22-12486 | PP620758 | Lecanicillium sp.3 | 100% |
| KUNCC22-12491 | PP620759 | Lecanicillium sp. | 100% |
| KUNCC22-12478 | PP620760 | Leptobacillium sp.1 | 99% |
| KUNCC23-13273 | PP620761 | Leptobacillium sp.2 | 100% |
| KUNCC23-12959 | PP620762 | Mariannaea sp.1 | 100% |
| KUNCC22-12581 | PP620763 | Mariannaea sp.2 | 100% |
| KUNCC23-13280 | PP620764 | Mariannaea sp.3 | 99.81% |
| KUNCC22-12571 | PP620765 | Mariannaea sp.4 | 99.81% |
| KUNCC23-13186 | PP620766 | Phaeoacremonium sp.1 | 99.57% |
| KUNCC23-13636 | PP620767 | Phaeoacremonium sp.2 | 100% |
| KUNCC23-14496 | PP620768 | Phaeoacremonium sp.3 | 99.44% |
| KUNCC23-13666 | PP620769 | Phaeoacremonium sp.4 | 99.27% |
| KUNCC23-13208 | PP620770 | Phaeoacremonium sp.5 | 99.64% |
| KUNCC22-12502 | PP620771 | Phaeoacremonium sp.6 | 99.82% |
| KUNCC23-12925 | PP620772 | Phaeoacremonium sp. | 99.28% |
| KUNCC23-12900 | PP620773 | Phaeoisaria sp.1 | 97.58% |
| KUNCC22-12546 | PP620774 | Phaeoisaria sp.2 | 100% |
| KUNCC22-12501 | PP620775 | Talaromyces sp.1 | 100% |
| KUNCC23-14471 | PP620776 | Talaromyces sp.2 | 100% |
| KUNCC23-14506 | PP620777 | Talaromyces sp.3 | 100% |
| KUNCC23-14476 | PP620778 | Talaromyces sp. | 99.44% |
| KUNCC23-12883 | PP620779 | Talaromyces sp.4 | 100% |
| KUNCC23-14456 | PP620780 | Trichoderma sp.1 | 100% |
| KUNCC23-13215 | PP620781 | Trichoderma sp.2 | 100% |
| KUNCC23-13245 | PP620782 | Trichoderma sp.3 | 100% |
| KUNCC23-13344 | PP620783 | Trichoderma sp.4 | 99.82% |
| KUNCC23-13606 | PP620784 | Trichoderma sp.5 | 100% |
| KUNCC22-12488 | PP620785 | Trichoderma sp.6 | 100% |
| KUNCC23-13617 | PP620786 | Trichoderma sp.7 | 100% |
| KUNCC23-13608 | PP620787 | Trichoderma sp.9 | 100% |
| KUNCC23-13181 | PP620788 | Trichoderma sp.10 | 100% |
| KUNCC23-12888 | PP620789 | Trichoderma sp.11 | 100% |
| KUNCC23-13610 | PP620790 | Trichoderma sp.12 | 99.81% |
| KUNCC22-12614 | PP620791 | Trichoderma sp. | 100% |
| KUNCC22-12555 | PP620792 | Trichoderma sp.13 | 100% |
| KUNCC23-12720 | PP620793 | Trichoderma sp.14 | 100% |
| KUNCC23-13283 | PP620794 | Trichoderma sp.15 | 100% |
| Season | Shannon Index | Pielou Index | Simpson Index |
| Spring | 1.95 | 3.57 | 0.77 |
| Summer | 3.67 | 11.3 | 1.99 |
| Winter | 3.55 | 9.84 | 1.9 |
| Lake | Shannon Index | Pielou Index | Simpson Index |
| Dianchi Lake | 2.65 | 5.79 | 1.15 |
| Fuxian Lake | 1.72 | 2.77 | 0.64 |
| Xingyun Lake | 2.4 | 4.26 | 1.04 |
| Yangzonghai Lake | 4.71 | 16.1 | 2.85 |
| Season | Shannon Index | Pielou Index | Simpson Index |
| Spring | 1.91 | 3.06 | 0.66 |
| Summer | 3.61 | 9.33 | 1.64 |
| Winter | 3.19 | 6.23 | 1.2 |
| Lake | Shannon Index | Pielou Index | Simpson Index |
| Dianchi Lake | 2.54 | 4.51 | 0.9 |
| Fuxian Lake | 1.7 | 2.49 | 0.58 |
| Xingyun Lake | 2.4 | 4.26 | 1.04 |
| Yangzonghai Lake | 4.51 | 11.36 | 2.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Zhao, Z.-X.; Gui, H.; Wang, X.-A.; Xing, P.; Karunarathna, S.C.; Cheewangkoon, R. Environmental Factors Shaping the Culturable Freshwater Fungi Diversity of Four Lakes in Yunnan Province, China. Diversity 2024, 16, 612. https://doi.org/10.3390/d16100612
Li L, Zhao Z-X, Gui H, Wang X-A, Xing P, Karunarathna SC, Cheewangkoon R. Environmental Factors Shaping the Culturable Freshwater Fungi Diversity of Four Lakes in Yunnan Province, China. Diversity. 2024; 16(10):612. https://doi.org/10.3390/d16100612
Chicago/Turabian StyleLi, Lu, Zhen-Xiong Zhao, Heng Gui, Xiao-Ai Wang, Peng Xing, Samantha C. Karunarathna, and Ratchadawan Cheewangkoon. 2024. "Environmental Factors Shaping the Culturable Freshwater Fungi Diversity of Four Lakes in Yunnan Province, China" Diversity 16, no. 10: 612. https://doi.org/10.3390/d16100612
APA StyleLi, L., Zhao, Z.-X., Gui, H., Wang, X.-A., Xing, P., Karunarathna, S. C., & Cheewangkoon, R. (2024). Environmental Factors Shaping the Culturable Freshwater Fungi Diversity of Four Lakes in Yunnan Province, China. Diversity, 16(10), 612. https://doi.org/10.3390/d16100612







