Abstract
The pygmy whitefish Prosopium coulterii (C. H. Eigenmann & R. S. Eigenmann, 1892) is a freshwater fish with a highly disjunct distribution ranging from the middle part of North America to Chukotka. There is still no consensus regarding its phylogeny and dispersal history due to limited information from the Chukotkan part of the range. We investigated 22 lakes over Chukotka and found a much broader distribution than it was previously thought. Pygmy whitefish was found to be a common species in the lakes that belong to rivers draining into the Arctic. Cytochrome B, cytochrome oxidase subunit 1, and ATP synthase F0 subunit 6 mitochondrial sites were analyzed from 25 samples to reconstruct the phylogenetic history of pygmy whitefish. Two haplogroups belonging to the east and west Chukotkan ranges were identified; both groups are closely related to Alaskan pigmy whitefish and distant from the Cascadia-Mackenzie (Peace) populations. Combining the distribution patterns, phylogenetic network topology, and the contemporary knowledge on the glaciation history of the region, we suggest a possible colonization pathway over Beringia region and beyond it. The basic biological characteristics (fork length, number of gill rakers, and pyloric caeca, age structure, and feeding) are also presented to characterize the populations over the investigated range.
1. Introduction
The diversity and distribution of the fish fauna inhabiting high latitude freshwaters is globally shaped by quaternary climatic cycles. The advancing of multiple glacial shields caused a dramatic reduction and fragmentation of ranges, while the glacial melting and warming led to global hydrological transformations, giving rise to broad opportunities of ecosystem colonization far beyond the boundaries of refugia [1,2,3,4]. The Beringian refugium, considered as a vital region for the origin of contemporary fish fauna in the northern North America and easternmost Asia, is one of the least studied sites even in the zoogeographical aspects. Several strictly freshwater species, particularly Dallia pectoralis Bean, 1880, Catostomus catostomus Forster, 1773, Coregonus pidschian Gmelin, 1789, Cottus cognatus Richardson, 1836, Thymallus arcticus Pallas, 1776, and Prosopium coulterii (Eigenmann et Eigenmann 1892), have been known to have trans-Beringian distribution, which is evidently defined by the water net transformations and a land bridge between North America and Asia during the cold ages. Several detailed phylogeographic reconstructions denote a specific colonization history with different resettlement pathways during different times for each of the abovementioned species [5,6,7,8,9,10]. However, the over-arching concept of the freshwater fauna formation and timing of resettlement is still lacking for the Beringian area. This makes each new historical reconstruction vital for a more profound understanding of the region’s history.
The pygmy whitefish Prosopium coulterii is a small (≤27 cm), silver-colored fish [11] widely distributed in North America [12,13] and also known to inhabit several lakes of Chukotka [14]. This species is of particular interest for phylogeographic studies, as its range is greatly disjunct [15] due to a long and complicated glacial history [16] as well as its inability to migrate even through the brackish waters with a salinity higher than 10 ppt [17]. The existing North American populations have been considered to have originated from the continental refugium in the Columbia River valley and the Beringian refugium. Temporary spillways between the upper course of the rivers of Pacific, Arctic, and Atlantic catchments contributed to the settlement of the North American and Alaskan waterbodies [17,18,19].
The northeastern border of Asia is known as one of the several hardest-to-reach places in the world, resulting in a limited exploration of the region. Until recently, the pygmy whitefish has been known to inhabit three lakes of the Amguema River drainage, which flows from the central part of Chukotka down to the Chukchi Sea from the central part of Chukotka [14,20,21]. No Asian populations have been previously found eastward or westward from this basin, making this habitat truly unique. Moreover, the phylogeny of the pygmy whitefish of Chukotkan lakes as well as the relation of Asian and North American populations have not been investigated yet. Since the pygmy whitefish is considered as species that dwelled northern North America throughout Pleistocene [17], the reconstruction of its lineage divergence may be regarded as a key mainstay for the zoogeographical reconstruction of fish resettlement in Beringia in general.
The aim of our study, therefore, was to reconstruct the distribution and phylogeography of the pygmy whitefish in the Chukotka region using the sequences of three coding regions of mitochondrial genome (COI, CytB, and ATP6). Additionally, some effort was made to characterize the fish over the range in general terms. We put forward several hypotheses. Firstly, the pygmy whitefish could be distributed much broader than it was considered before. Secondly, the pygmy whitefish from the Chukotkan part of the range could contain the haplogroup that originated from the Beringia refugium during the last glaciation. Thirdly, we suppose that the populations of the remote river basins would be isolated from each other since the last glaciation owing to the impossibility of migrations among the basins over the Arctic Ocean in current salinity conditions. Here we report the analysis the fish fauna of 22 lakes to shed light on these issues and to reconstruct the phylogeny of pygmy whitefish both in the region and worldwide.
2. Material and Methods
The investigation took place in summer 2019 and 2020 all over Chukotka. Twenty-two lakes of Arctic and Pacific draining were sampled (Figure 1, Table S1) using multipanel gillnets (15–45 mm mesh size) at the littoral (5–10 m), slope (15–30 m), and profundal (30+ m) zones. In total, twenty-five pygmy whitefish were collected from five lakes: three from Lake Kokanlegytgyn, eight from Vykvyrkapgytgyn, four from Ravkergytgyn, eight from Gytgypylgyn, and two from Gytgyl. Fin clips were sampled from each individual and fixed in the 95% alcohol. The following parameters were additionally determined: fork length, sex, and the numbers of pyloric caeca and gill rakers on the first left arch. To define the life expectancy, the scales were collected from each fish above the lateral line behind the dorsal fin. The food objects from stomachs were qualified by groups (planktonic organisms jointly, different benthic organisms by order).
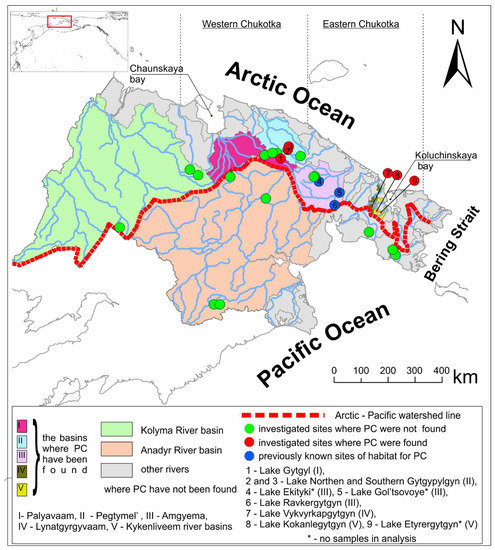
Figure 1.
Distribution of Prospium coulterii (PC) across the investigated sites.
Total DNA was extracted from samples using Chelex 100 resin (BioRad, Hercules, CA, USA). Three mitochondrial DNA (mtDNA) fragments were amplified: a 446 base pair (bp) fragment of 95 cytochrome oxidase subunit 1 (=COI, primers ProsCOIF 5′-CAACCACAAAGACATTGGCAC-3′ and ProsCOIR 5′-CCCGATTGAGGAGACAGTGTT-3′) [designed by the authors]; 1113 bp of cytochrome B (=CytB, ProsCytB-F 5′-GTCATAATTCCTGCTCGGAC-3′ and ProsCytB-R 5′-CCGACTTCCGGATTACAAGA-3′) [designed by the authors]; and 614 bp of ATP synthase F0 subunit 6 (=ATP6, H9208 5′-TATGCGTGTGCTTGGTGTGCCA-3′ and L8558 5′-AGCTTCTTCGACCAATTTATGAG-3′) [17]. Products from PCR were isolated using Exonuclease-I and Shrimp Alkaline Phosphatase protocol (Evrogen, Russia). For information about amplification mixture and PCR conditions, see Table S2. Purified DNA fragments were sequenced using AbiPrism 3730 (Applied Biosystems, Waltham, MA, USA), and the sequences were acquired in Chromas v.2.5 (Technelysium Pty, South Brisbane, Australia). The same primers as were applied for amplification were further used as sequencing primers.
Haplotype occurrence was analyzed considering the geographical subdivision of pygmy whitefish populations. We joined the following sample sets: western Chukotka (=WC, the Palavaam and Pegtymel river drainages); eastern Chukotka (=EC, the Amguema River drainage and Koluchninskaya Bay catchment); southern Alaska (=SA, waterbodies of the Alaska Range); and the continental part of North America (=NA, waterbodies of the Mackenzie, Yukon, and Lake Superior drainages). Haplotypes from SA and NA regions were taken from GeneBank (IDs in Supplementary Table S3). All haplotype sequences obtained during the study were submitted to GeneBank NCBI under OQ379951-OQ379954 (for COI haplotypes) and OQ405329-OQ405348 (for ATP and CytB haplotypes) accession numbers (IDs in Supplementary Table S4)
All sequences were aligned with ClustalW multiple alignment in BioEdit v.7.1.9 [22,23] and trimmed to the lengths of the Chukotkan variants; alignment errors were corrected manually. Haplotype networks were obtained using the TCS (statistical parsimony) method in PopArt v.1.7 [24]. The ATP gene was further studied and the GeneBank collection displayed much greater geographical coverage compared with the CytB and COI data. To provide an equivalent comparison of haplotype networks for different mtDNA fragments, the ATP6 haplotype diversity of SA and NA was reduced to the most common haplotypes, which also shared the collection sites with CytB and COI. Genetic divergence degree (d) was estimated in MEGA X [25] using the Maximum Composite Likelihood method [26]. Nucleotide (π) and haplotype (Hd) diversity indices were obtained from the polymorphism data block in DnaSP v.6 [27].
To provide an equivalent comparison of haplotype networks for different mtDNA fragments, the ATP6 haplotype diversity of SA and NA was reduced to the most frequent haplotypes. Each of these ATP6 haplotypes were associated with the closest collection sites for the (CytB + COI) combination. The total haplotype diversity of the ATP6 gene was presented in Supplementary Materials (Figure S2, Table S6). All of the obtained unique haplotypes of combined COI-CytB sequences for the whole species range, as well as COI-CytB-ATP6 sequences for the Chukotkan populations, were used for the subsequent phylogenetic analysis. Haplotype relationships were inferred by using the Maximum Likelihood method and the Hasegawa-Kishino-Yano model in MEGA X, implementing 1000 bootstrap replications. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary-rate differences among sites (4 categories (+G)). Codon positions included were 1st + 2nd + 3rd + Noncoding. All positions containing gaps and missing data were eliminated with a complete-deletion option. There were a total of 1476 positions in the final dataset of the COI-CytB sequences and 2080 positions in the COI-CytB-ATP6 dataset. The haplotype of Prosopuim cylindraceum obtained from full mitochondrion genome MF621767 was used as the outgroup. Additionally, TCS-networks of the unique COI-CytB and COI-CytB-ATP6 sequences were obtained in PopArt v.1.7.
3. Results
3.1. Distribution Patterns
The new populations of pygmy whitefish (Figure 1) were found in 5 out of 22 investigated lakes belonging to 4 different river basins. Firstly, the species’ presence was confirmed for the Amguema River drainage (Figure 1; one reinvestigated population and three previously known ones). To the west, pygmy whitefish was found in the Pegtymel and Palavaam river drainages. To the east, it inhabited the lakes of the Koluchinskaya bay catchment. No pygmy whitefish populations were observed southward in the rivers flowing into Pacific Ocean, as well as in the Kolyma River drainage.
Within the Asian range, the pygmy whitefish was found only in deep oligotrophic lakes without any brownification signatures, which occurs in the de-glaciated sites due to high organic inflow, resulting in tannins/peat accumulation in the lakes. The species commonly co-occurred with the lacustrine Salvelinus taranetzi Kaganowsky, 1955, Cottus cognatus, and Thymallus arcticus. No pygmy whitefish was found in the shallow-water and lowland lakes or in very headwaters of the basins, which were most likely covered with the perennial ice until the Holocene. According to the gillnet catches, pygmy whitefish commonly occurred on the lake slope deeper than 15–20 m.
3.2. Basic Biological Features
The mean fork length of mature fish was 10.9 ± 0.34 cm (from 8.9 to 13.8 cm); the maximum observed age was nine (females)—seven (males) years. The largest fish were sampled in Lake Vykvyrkapgytgyn, while the smallest and slow-growing ones—in the nearest Lake Kokanlegytgyn (both are situated in the Koluchinskaya bay catchment). The number of pyloric caeca was similar in all Chukotkan lakes, ranging from 15 to 21 (mean 18.7 ± 0.75). The gill rakers (from 16 to 21, 17.8 ± 0.49) were short and robust in all of the populations, and the mean raker number was the highest in Lake Kokanlegytgyn (19.3) and the lowest in Lake Gytgyl (16.0). The variations in gill raker number were ±1 across all the lakes except Lake Gytgypylgyn (Pegtymel River drainage), where they varied from 16 to 21. In all the lakes, the fish mainly fed on benthic chironomid, mayfly, and caddisfly larvae. In Lake Gytgypylgyn, single specimens were also observed to prey on zooplankton.
3.3. mtDNA Haplotype Diversity
All obtained Chukotkan sequences of the three analyzed mtDNA fragments did not correspond to those found earlier for the pygmy whitefish populations from the SA and NA regions. We found four new haplotypes of the COI fragment in Chukotka (Figure 2a): two in the lakes of EC and three in the lakes of WC. The CytB and ATP6 fragments displayed more pronounced differentiation among populations. Thirteen haplotypes of CytB and seven of ATP6 were found (Figure 2b,c): eight and four in EC and seven and six in WC, respectively. The distribution of haplotypes by populations is presented in Table 1. WC samples were defined by greater nucleotide (π 0.00149–0.00192 vs. 0.00190–0.00472) and haplotype (Hd 0.222–0.901 vs. 0.639–0.909) diversity comparing to EC (Table S5).
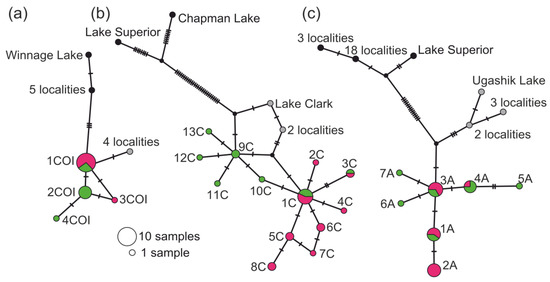
Figure 2.
Statistical parsimony networks for three mtDNA fragments of the Prosopium coulterii populations from four regions: COI (a), CytB (b) and ATP6 (c). Colours indicate different geographical clusters: green is WC, red is EC, grey is southern Alaska, and black is continental North America. The circle sizes correspond to the number of sequences only for Chukotkan haplotypes; for Alaskan and American ones, the number of localities where the haplotypes are found is indicated.

Table 1.
Occurrence of haplotypes of the three mtDNA fragments across Chukotkan populations of Prosopium coulterii. Number of individuals included into analysis was given after haplotype name.
The haplotype network demonstrated the subdivision of populations from four studied regions. For all three mtDNA fragments, shared as well as unique haplotypes were found in populations from EC and WC (Figure 2). All unique haplotypes differed by 2–4 substitutions from the shared basal haplotype. Moreover, the Chukotkan haplotypes of COI differed by at least 2 substitutions from SA and by 4 substitutions from NA haplotypes. For CytB, the Chukotkan haplotypes differed by 4–5 substitutions from SA and 41 substitutions from NA haplotypes; for ATP6, the difference was 6 and 19 substitutions, respectively. The pairwise evolutionary divergence between the groups under analysis from the WC and EC regions was two to four times less than between the Chukotkan and SA populations and four to nineteen times less than between the Chukotkan and NA populations (Table 2). The analysis of combined COI-CytB sequences involved twenty-two unique haplotypes (Figure 3); the groups from the Ravkergytgyn and Gytgyl lakes occupied the central part of the net, while the groups from Gytgypylgyn and Vykvyrkapgygtgyn lakes as well as the Alaskan/NA once were arranged on the net fringes. A similar picture of the Chukotkan populations differentiation was obtained via TCS analysis of COI-CytB-ATP6 sequences (Figure S1).

Table 2.
Statistically optimal pairwise estimates of genetic-divergence degree (d) based on COI; CytB; ATP6 fragment differences in the Prosopium coulterii populations from four regions.
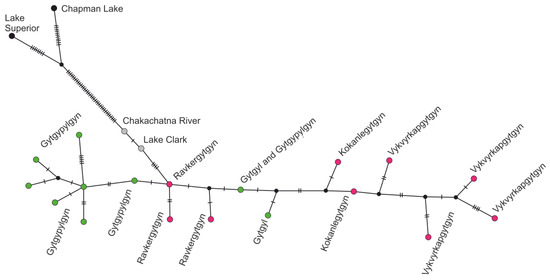
Figure 3.
Statistical parsimony network for the combined COI-CytB sequences of the Prosopium coulterii populations from four regions. Colours indicate different geographical clusters: green is western Chukotka, red is eastern Chukotka, grey is southern Alaska, and black is continental North America. Haplotypes from Alaska and North America were combined from following Genebank sequences: Lake Superior (JX960823; JX960930); Lake Chapman (JX960824; JX960931); Chakachatna River (KT630748; KT630730 which is equal to KT630746; KT630728 from Kenai peninsula unnamed lake); and Lake Clark (KT630747; KT630729).
The ML-tree of COI-CytB sequences with the highest log likelihood (−3376.33) and gamma distribution = 0.1071 is shown in Figure 4 (the analysis involved 22 unique nucleotide sequences). The tree represented clasterization of the NA, SA, and Chukotkan groups with the bootstrap support ≥ 65%. The partition of Chukotkan fish to the EC and WC sub-clusters was not significant. Herewith, the unique haplotypes from Lake Gytgypylgyn were grouped on the tree in separate sub-cluster. Among the EC haplotypes, the Vykvyrkapgytgyn and Kokanlegytgyn haplogroups could be differentiated (81 and 68% of bootstrap, respectively), while the Ravkergytgyn haplogroup occupied the basal position. Using Chukotkan COI-CytB-ATP6 sequences in the ML-tree (this analysis involved 18 nucleotide sequences), the same-type clasterization was found with the 65% support of the separation of the unique Gytgypylgyn haplotypes (Figure S3).

Figure 4.
The ML-tree employed for the combined COI-CytB sequences of the Prosopium coulterii populations from four regions. The tree was drawn using Bootstrap consensus; the percentage of trees in which the associated haplotypes clustered together is shown next to the branches. Haplotypes from Alaska and North America (NA) were combined from following Genebank sequences of single vouchers: Lake Superior (JX960823; JX960930); Chapman Lake (JX960824; JX960931); Chakachatna River (KT630748; KT630730 which is equal to KT630746; KT630728 from Kenai peninsula unnamed lake); and Lake Clark (KT630747; KT630729).
4. Discussion
4.1. Distribution in Chukotka
The first catches of pygmy whitefish in Chukotka date back to the 90s in four lakes belonging to the Amguema River drainage [14,20,28]. Since then, no exploratory catches were undertaken in the region until 2019–2020. We checked twenty-two lakes over Chukotka and found that pygmy whitefish is much more spread than it was previously thought. The species was found in the lakes of five river basins draining the Arctic section of over 700 km length. It seems quite evident that not all populations dwelling in northern Chukotka have been found during the study.
Based on our and literature data, several criteria could be put forward to predict the occurrence of pygmy whitefish in the lakes over the studied region. Firstly, we found that all lakes populated by this species drain into the Arctic, and no populations were found in Pacific basins. This distribution pattern should be apparently determined by the colonization history of the region and, at first sight, looks quite puzzling since the populations of the Alaskan range are known in the Pacific, but not in the Arctic [17]. For the possible explanations, please see the Section 4.4. ‘Global phylogenetic history and colonization pathways’. Secondly, we found that pygmy whitefish inhabits only deep, clear lakes reflecting the low tolerance of this species to the near-bottom oxygen depletion due to the water brownification [29]. Thirdly, the absence of slope-dwelling predator is probably necessary for sustainable existence of the pygmy whitefish populations. No pygmy whitefish was found cohabiting with burbot Lota lota (Linnaeus, 1758), which also commonly occupies the slope zone. Similar patterns were previously demonstrated for Salvelinus alpinus (Linnaeus, 1758) that do not switch to benthic feeding in the presence of burbot [30]. Burbot was never recorded in transparent deep lakes in the highlands of Arctic Chukotka. At the same time, it was commonly observed for lowland Chukotkan lakes, the browninficated lakes, the lakes beyond the Arctic zone, and the Kolyma River drainage [21].
4.2. Basic Biological Characteristics
Variations in morphological or ecological features are challenging to estimate owing to the limited number of fish under analysis. Given the sample size, we would tentatively assume that pygmy whitefish is monotypic across the Chukotkan part of the range. It is a small-sized (up to 14 cm), benthivorous fish inhabiting the slopes of big mountain lakes. Some signs of intra-lake polymorphism could be noted for the Lake Gytgypylgyn population, where the highest variation in gill raker number and mtDNA haplotype diversity, as well as various types of feeding were observed. Gytgypylgyn is the largest and deepest waterbody in the region, and therefore, intra-lake trophic-based diversification is most probable for this particular object. Previously, three reproductively isolated morphs with different types of feeding and gill raker number were described for big Chignik Lake, Southern Alaska [16,31].
4.3. Phylogeny within the Chukotkan Range
During the last ice age (MIS 2–4), the sea level dropped down for about 120 m, which resulted in the junction of numerous small rivers into a few large river basins, which continued on the exposed Arctic shelf [32]. Eastern rivers of Chukotka, including Amguema and Koluchinskaya bay tributaries, were combined into the Paleo-Amguema River, while in western Chukotka, the Palavaam and the Pegtymel formed the single Paleo-Palavaam drainage (Figure 5). Moreover, the headwaters of these huge river systems comprised numerous lakes, which edged the glacial shields, making it possible for fishes to survive during the cold periods and to distribute through the temporal pathways across the basins during glacial meltings. Analyzing the haplotype networks and the ML-tree for three coding mtDNA fragments, we found a partition of the pygmy whitefish populations over the Chukotkan range, wherein two groups of the area outskirts could be outlined by specific satellite haplotypes. The eastern Chukotkan group unifies fish from lakes belonging to the Koluchinskaya bay catchment, while the western group consists of fish from the Pegtymel’ drainage. Both groups formed its own unique haplotypes, differing in 2–4 substitutions from the shared presumably ancestral haplotypes. According to the haplotype net structure, the groups of the eastern part of Amguema, headwaters of Palavaam, and some specimens from Pegtymel’ rivers drainages could be considered as basal, which allows one to suppose that the headwaters of these rivers acted as a refugium during the ice age period. Moreover, the mutation rate for the CytB and ATP6 mitochondrial DNA fragments [33,34,35] approximately estimated for the fish can also point out the persistence of pygmy whitefish populations in that refugium throughout the last ice age. Since the major part of the Paleo-Palavaam River was partly covered with ice, while the west part of the Paleo-Amguema River drainage was free from glaciers [36], we considered that the invasion of pygmy whitefish into the majority of Pegtymel’s drainages and rivers of Koluchinskay bay occurred via temporary interconnections formed during the glacial melting after the last glaciation maximum. In respect of the genetic data, we can assume that the newly formed western and eastern populations have lost connection with the core group during the last ice age. Large water systems of Lake Gytgypylgyn and lakes of Koluchinskaya bay happened to be favorable for pygmy whitefish. The high nucleotide and haplotype diversity could signify the rapid population increase right after the invasion, it also could reflect the highly effective population size and potential concurrence of sympatric morphs in the ecosystem.
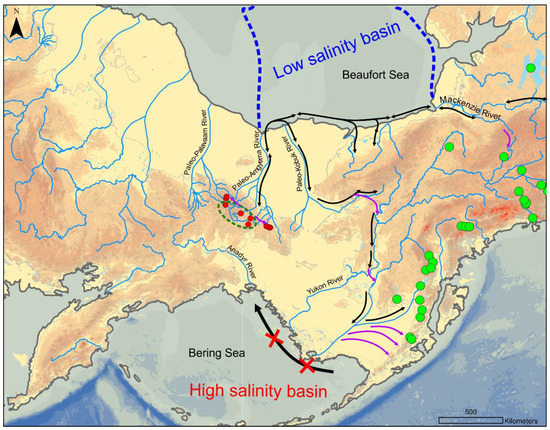
Figure 5.
Possible reconstruction of colonization pathways for pygmy whitefish in Beringia. Red dots mark known populations in the Chukotkan range, green dots mark some of North American populations; the dark-green dotted line shows the possible refugia site; black arrows demonstrate the migrations over the sea basin and rivers, while violet ones show the possible places where pygmy whitefish migrates from the temporary waterway connections.
The similar late colonization by pygmy whitefish was previously revealed for the Alsek and Copper River drainages, which both were populated from the headwaters of the Yukon River through the temporal water spillways [17,18]. We suggest that the Paleo-Palavaam could possibly be colonized by pygmy whitefish in the N68°23′11.3688″; E178°24′15.7397″; near Lake Eler’gytgyn (Figure S4). The local watershed is characterized by slight slopes, and former river valleys are still visible on the satellite images.
4.4. Global Phylogenetic History and Colonization Pathways
It is commonly accepted that pygmy whitefish is characterized by a greatly disjunct range shaped during Pleistocene glaciations, wherein the southernmost Alaskan sites and the continental part of Columbia River valley were the major refugia [17]. Given this fact, Cascadia-Mackenzie (Peace) populations are the mostly distinct on the ML-trees and, on all haplotype nets, they could be considered as ancestral for pygmy whitefish in general. In that scenario, the colonization of Beringia range took place from North America; furthermore, a high level of genetic subdivision between the Alaskan-Chukotkan and continental Cascadia-Mackenzie (Peace) populations [17] points to an early-middle Pleistocene disjunction of these groups. On the other hand, they could be also considered as the very first colonization wave from Beringia to the North America, as it was previously reported for Salvelinus charr [37,38]. That question could not have been ultimately considered without additional data from the Chukotkan-Alaskan part of the range. However, we suggest that the major refugia for the pygmy whitefish of the central part of North America were not in the Columbia River valley, but on the watershed of Peace and Yukon rivers instead of the ice shield that region was covered by huge pro-glacial lakes in the middle Pleistocene [39]. The identical regional distribution of pygmy whitefish, lake trout Salvelinus namaycush (Walbaum, 1792), and arctic charr S. alpinus [40,41,42] provides support for this refugium. Moreover, the eastern group of Coregonus clupeaformis (Mitchill, 1818) has been well separated from the other lineages of C. clupeaformis, which had probably survived during the Last Glaciation Maxima in the same refugium [43].
Our data also expand the understanding of the pygmy whitefish distribution and phylogeny in the western sector of the range. First of all, we propose that the western group of populations is highly likely to also be widespread in the Asian part of Arctic Beringia, and all Chukotkan populations jointly with Alaskan populations are genetically separated from the continental American group of populations. The structure of the haplotype networks suggests that the Pleistocene core of the western population system was associated with the Arctic draining in the center of Beringia, including the upper courses of Paleo-Amguema and Paleo-Palavaam rivers. That exact area was not glaciated during the late Pleistocene [36] and could have been covered by deep lakes suitable for pygmy whitefish.
According to the number of substitutions in mtDNA gene sequences, the contact between the Chukotkan and Alaskan populations was finally lost during the last Pleistocene ice age. We suggest that pygmy whitefish previously migrated from Beringia to the east directly via the Arctic Ocean rather than through the temporal spillways in river headwaters (Figure 5). It was recently revealed that the Arctic Ocean transformed into a few semi-isolated waterbodies with brackish salinity several times during the coldest periods of glaciation [41], which were possibly passable for freshwater fishes. The common sucker (Catostomus catostomus) likely colonized the rivers of Arctic draining into northeast Asia [7] using the same pathway. The colonization of the contemporary waterbodies in the Alaska Range became possible only after the local ice shield had melted; the Paleo-Kobuk River drainage was also partly frozen at the maximal glacial extent [44,45]. We suggest that the migration had possibly occurred through the Paleo-Kobuk River and several temporal spillways—(i) on the lowland waterbodies of Kobuk-Yukon rivers, (ii) Yukon- Kuskokwim rivers, and (iii) through the net of pro-glacial lakes along the glacial border. The later migration along the Bering Sea coastline back to the southern Chukotka was impossible because of the high salinity of that part of Pacific Ocean both during the cold and warm ages. That could apparently explain the observed distribution patterns of pygmy whitefish in Chukotkan range.
Finally, we suggest a sequence of events determining the contemporary distribution of pygmy whitefish over the range. The first-step separation happened long ago between the North America and Beringian populations. During the second step, the disjunction took place at the end of the ice age between the Alaskan and Chukotkan groups. The ocean level rose, which triggered the opening of the Bering Strait and paleo-rivers basin fragmentation, which could be considered as one of the most likely scenarios for the separation event. During the third step, the local groups within each of the Chukotkan and Alaskan ranges distributed from refugia over the postglacial ecosystems following the temporary water pathways formed on the borders of the retreated glacier. Therefore, the final step of the groups’ separation happened right after the end of the last glacial maxima when the modern water net had formed.
We also conclude that the newly analyzed data largely improve our knowledge of the evolution and settlement history for one more fish species dwelling in the Holarctic freshwaters. We expected that some populations of pygmy whitefish belonging to the Beringian group have not yet been discovered in the mountain lakes of the Kobuk River drainage and in the rivers draining the north slope of the Brooks Range. We also suppose that the pygmy whitefish is a species that possibly originated in Beringia and lately inhabited North America through Mackenzie River drainage. However, additional data from both Alaskan and Chukotkan sites are necessary to establish the overall phylogeny for this fish group.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d15040547/s1, Figure S1: Statistical parsimony network for the combined COI-CytB-ATP6 sequences of the Prosopium coulterii populations from Chukotka. Colours indicate different geographical clusters: green is western Chukotka, red is eastern Chukotka; Figure S2: Statistical parsimony network for 500 bp mtDNA ATP6gene fragment of the Prosopium coulterii populations from four regions. Colors indicate different geographical clusters western Chukotka (WC), red is eastern Chukotka (EC), grey is southern Alaska (SA) and black is continental North America (NA). The circle sizes correspond to the number of sequences. Haplotype names from SA and NA regions were taken from the original papers (H1-H26 from Whitt et al. (2011) [17]; H27–H31 from Gowell et al. (2012) [31]). All sequences were trimmed and aligned based on shortest sequences (H27–H31). Because of trimming to 500 bp sequence length some of known 650 bp-haplotypes from our data and Genebank stacked together, their names given trough «+» symbol; Figure S3: The ML-tree with the highest log likelihood (−4249,49) and Gamma distribution = 0.2366 employed for the combined COI-CytB-ATP6 sequences of the Prosopium coulterii populations from two Chukotkan regions. The tree is drawn using Bootstrap consensus; the percentage of trees in which the associated haplotypes clustered together is shown next to the branches; Figure S4: Potential temporary waterway connections at N68,38649200°; E178,40437189 near Lake Eler’gytgyn; Table S1: Locations of sampling sites over the eastern (EC) and western (WC) parts of Chukotka; Table S2: The amplification mixture and PCR conditions for each gene; Table S3: List of haplotypes which were taken from GeneBank. South Alaska (SA); central part of North America (NA). The bold highlighted haplotypes were used for network; all haplotypes were used to estimate the degrees of genetic divergence between groups; Table S4: List of accession numbers of the three mtDNA fragments across Chukotkan populations of Prosopium coulterii deposited to GenBank; Table S5: Nucleotide and haplotype diversity indices of COI-CytB-ATP6 sequences of the Prosopium coulterii populations from Western and Eastern Chukotka; Table S6: List of ATP synthase F0 subunit 6 haplotypes which were taken from GeneBank. Southern Alaska (SA)-clade 1; central part of North America (NA)-clade 2 according to Witt et al. (2011) [17].
Author Contributions
Conceptualization, G.N.M. and E.V.E.; methodology, G.N.M., E.V.E. and M.M.S.; software, N.O.M., G.V.I., E.N.K. and P.G.V.; formal analysis, N.O.M., G.V.I., P.G.V. and N.A.B.; investigation, G.N.M., E.V.E. (Chukotka region), N.A.B. and D.V.P. (Amguema region); resources, writing—original draft preparation, G.N.M. and E.V.E.; writing—review and editing, G.N.M., E.V.E. and N.O.M.; supervision, G.N.M. and E.V.E.; project administration, G.N.M., E.V.E. and M.M.S.; funding acquisition, G.N.M., E.V.E. and M.M.S. All authors have read and agreed to the published version of the manuscript.
Funding
CytB and COI sequencing were supported by the Russian Science Foundation (project #19-74-10054Π) while ATP6 sequencing and genetic analyses were supported by Russian international scientific collaboration program Mega-grant (mega-grant # 075-15-2022-1134).
Institutional Review Board Statement
The present research has met the requirements guided by the order of the High and Middle Education Ministry (care for vertebrate animal included in scientific experiments, #742 from 13 November 1984) and additionally by the Federal Law of the Russian Federation #498 FL (from 19 December 2018) with regard to the humane treatment of animals. The present study based on field research and involves the collection of fishes from a natural water bodies. Adult fish used in the current study were collected based on permeation number (#412020032162) obtained from Russian Federal Fisheries Agency (https://fish.gov.ru/) for Chukotkan region. The permit allowed us to collect fish in the specified water bodies. No other permits are required to carry out the present study in designated areas under Russian Federation laws.
Data Availability Statement
The sequencing data in this study (Table 1) have been uploaded to Gen Bank (http://www.ncbi.nlm.nih.gov, accessed on 14 March 2023) under following numbers Table S4.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hewitt, G. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef]
- Hewitt, G.M. Quaternary phylogeography: The roots of hybrid zones. Genetica 2011, 139, 617–638. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.R.; Lister, A.M. Cryptic northern refugia and the origins of the modern biota. Trends Ecol. Evol. 2001, 16, 608–613. [Google Scholar] [CrossRef]
- Shafer, A.B.; Cullingham, C.I.; Cote, S.D.; Coltman, D.W. Of glaciers and refugia: A decade of study sheds new light on the phylogeography of northwestern North America. Mol. Ecol. 2010, 19, 4589–4621. [Google Scholar] [CrossRef] [PubMed]
- Stamford, M.D.; Taylor, E.B. Phylogeographical lineages of Arctic grayling (Thymallus arcticus) in North America: Divergence, origins and affinities with Eurasian Thymallus. Mol. Ecol. 2004, 13, 1533–1549. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.N.; Taylor, E.B. Pleistocene glaciations and contemporary genetic diversity in a Beringian fish, the broad whitefish, Coregonus nasus (Pallas): Inferences from microsatellite DNA variation. J. Evol. Biol. 2010, 23, 72–86. [Google Scholar] [CrossRef]
- Bachevskaya, L.T.; Pereverzeva, V.V.; Ivanova, G.D.; Agapova, G.A.; Primak, A.A. Genetic structure of the Siberian Sucker (Catostomus catostomus rostratus) according to data on sequence variation of the mtDNA cytochrome b gene. Biol. Bull. 2014, 41, 306–311. [Google Scholar] [CrossRef]
- Bachevskaya, L.T.; Pereverzeva, V.V.; Primak, A.A.; Agapova, G.A. Genetic Variability of the Siberian Sucker Catostomus catostomus rostratus (Teleostei: Catastomidae) from Water Bodies of the Northeast of Russia. Russ. J. Genet. 2022, 58, 428–435. [Google Scholar] [CrossRef]
- Campbell, M.A.; Lopéz, J.A. Mitochondrial phylogeography of a Beringian relict: The endemic freshwater genus of blackfish Dallia (Esociformes). J. Fish Biol. 2014, 84, 523–538. [Google Scholar] [CrossRef]
- Campbell, M.A.; Takebayashi, N.; López, J.A. Beringian sub-refugia revealed in blackfish (Dallia): Implications for understanding the effects of Pleistocene glaciations on Beringian taxa and other Arctic aquatic fauna. BMC Evol. Biol. 2015, 15, 144. [Google Scholar] [CrossRef]
- McCart, P.J. Growth and Morphometry of the Pygmy Whitefish (Prosopium coulteri) in British Columbia. Ph.D. Thesis, University of British Columbia, Vancouver, BC, Canada, 1963; pp. 1–97. [Google Scholar]
- Scott, W.B.; Crossman, E.J. Freshwater Fishes of Canada. Fish. Res. Board Can. Bull. 1973, 184, 966. [Google Scholar]
- Vecsei, P.; Panayi, D. Range extension for Pygmy Whitefish (Prosopium coulterii) in the Northwest Territories, Canada. Can. Field-Nat. 2015, 129, 70–75. [Google Scholar] [CrossRef]
- Chereshnev, I.A.; Skopets, M.B. A new record of the pygmy whitefish, Prosopium coulteri, from the Amguem River Basin, (Chukotski Peninsula). J. Ichthyol. 1992, 32, 46–55. [Google Scholar]
- Eschmeyer, P.H.; Bailey, R.M. The pygmy whitefish, Coregonus coulteri, in Lake Superior. Trans. Am. Fish. Soc. 1955, 84, 161–199. [Google Scholar] [CrossRef]
- Lindsey, C.C.; Franzin, W.G. New complexities in zoogeography and taxonomy of the pygmy whitefish (Prosopium coulteri). J. Fish. Board Can. 1972, 29, 1772–1775. [Google Scholar] [CrossRef]
- Witt, J.D.; Zemlak, R.J.; Taylor, E.B. Phylogeography and the origins of range disjunctions in a north temperate fish, the pygmy whitefish (Prosopium coulterii), inferred from mitochondrial and nuclear DNA sequence analysis. J. Biogeogr. 2011, 38, 1557–1569. [Google Scholar] [CrossRef]
- Wiedmer, M.; Montgomery, D.R.; Gillespie, A.R.; Greenberg, H. Late Quaternary megafloods from Glacial Lake Atna, Southcentral Alaska, USA. Quat. Res. 2010, 73, 413–424. [Google Scholar] [CrossRef]
- Blanchfield, P.J.; Taylor, E.B.; Watkinson, D.A. Morphological and genetic analyses identify a new record of a glacial relict: Pygmy Whitefish (Prosopium coulterii) from Northwestern Ontario. Can. J. Zool. 2014, 92, 267–271. [Google Scholar] [CrossRef]
- Chereshnev, I.A.; Volobuev, V.V.; Shestakov, A.V.; Frolov, S.V. Salmonoid Fishes in Russian North-East. [Lososevidnye ryby Severo-Vostoka Rossii]; Dal’nauka: Vladivostok, Russia, 2002; p. 496. (In Russian) [Google Scholar]
- Chereshnev, I.A. Pygmy Whitefish. [Karlikovyj valek]. In Red Data Book of the Chukchi Autonomous District Vol. 1 Animals; Dikii Sever Publ: Magadan, Russia, 2008; p. 235. (In Russian) [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 1, 2–3. [Google Scholar] [CrossRef]
- Hall, T. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Chereshnev, I.A. Biological Diversity of Freshwater Fish Fauna in the Russian North-East [Biologicheskoe Raznoobrazie Presnovodnoj Ihtiofauny Severo-Vostoka Rossii]; Dal’nauka: Vladivostok, Russia, 1996; p. 196. (In Russian) [Google Scholar]
- Knoll, L.B.; Williamson, C.E.; Pilla, R.M.; Leach, T.H.; Brentrup, J.A.; Fisher, T.J. Browning-related oxygen depletion in an oligotrophic lake. Inland Waters 2018, 8, 255–263. [Google Scholar] [CrossRef]
- Knudsen, R.; Amundsen, P.A.; Klemetsen, A. Arctic charr in sympatry with burbot: Ecological and evolutionary consequences. Hydrobiologia 2010, 650, 43–54. [Google Scholar] [CrossRef]
- Gowell, C.P.; Quinn, T.P.; Taylor, E.B. Coexistence and origin of trophic ecotypes of pygmy whitefish, Prosopium coulterii, in a south-western Alaskan lake. J. Evol. Biol. 2012, 25, 2432–2448. [Google Scholar] [CrossRef]
- Pavlidis, Y.A.; Ionin, A.S.; Medvedev, V.S. Paleogeography of the Late Wurm of the Beringian Shelf [Paleogeografiya pozdnego vyurma shel’fa Beringii]. In Geology and Geomorphology of Shelves and Continental Slopes; [Geologiya i Geomorfologiya Shel’fov i Materikovyh Sklonov]; Nauka: Moscow, Russia, 1985; pp. 65–76. (In Russian) [Google Scholar]
- Zardoya, R.; Doadrio, I. Molecular evidence on the evolutionary and biogeographical patterns of European cyprinids. J. Mol. Evol. 1999, 49, 227–237. [Google Scholar] [CrossRef]
- Oleinik, A.G. On the mutation rates of the mitochondrial and nuclear genomes of salmonid fishes. Russ. J. Mar. Biol. 2000, 26, 432–438. [Google Scholar] [CrossRef]
- Machordom, A.; Doadrio, I. Evidence of a Cenozoic Betic-Kabilian connection based on freshwater fish phylogeography (Luciobarbus, Cyprinidae). Mol. Phylogenet. Evol. 2001, 18, 252–263. [Google Scholar] [CrossRef]
- Glushkova, O.Y. Late Pleistocene glaciations in north-east Asia. Dev. Quat. Sci. 2011, 15, 865–875. [Google Scholar] [CrossRef]
- Oleinik, A.G.; Skurikhina, L.A.; Brykov, V.A. Phylogeny of charrs of the genus Salvelinus based on mitochondrial DNA data. Russ. J. Genet. 2015, 51, 55–68. [Google Scholar] [CrossRef]
- Lecaudey, L.A.; Schliewen, U.K.; Osinov, A.G.; Taylor, E.B.; Bernatchez, L.; Weiss, S.J. Inferring phylogenetic structure, hybridization and divergence times within Salmoninae (Teleostei: Salmonidae) using RAD-sequencing. Mol. Phylogenet. Evol. 2018, 124, 82–99. [Google Scholar] [CrossRef] [PubMed]
- Duk-Rodkin, A.; Barendregt, R.W. Stratigraphical record of glacials/interglacials in northwest Canada. Dev. Quat. Sci. 2011, 15, 661–698. [Google Scholar] [CrossRef]
- COSEWIC. COSEWIC Assessment and Status Report on the Pygmy Whitefish Prosopium Coulterii, Southwestern Yukon Beringian Populations, Yukon River Populations, Pacific Populations, Western Arctic Populations, Great Lakes—Upper St. Lawrence populations, Water; Committee on the Status of Endangered Wildlife in Canada: Ottawa, ON, Canada, 2016. [Google Scholar]
- Esin, E.V.; Markevich, G.N. Evolution of the charrs, genus Salvelinus (Salmonidae). 1. origins and expansion of the species. J. Ichthyol. 2018, 58, 187–203. [Google Scholar] [CrossRef]
- Budy, P.; Rogers, K.B.; Kanno, Y.; Penaluna, B.E.; Hitt, N.; Thiede, G.P.; Dunham, J.; Mellison, C.; Somer, W.L.; DeRito, J. Distribution and status of trout and char in North America. In Trout and Char of the World; Kershner, J.L., Williams, J.E., Gresswell, R.E., Lobón-Cerviá, J., Eds.; American Fisheries Society: Vancouver, BC, Canada, 2019; Chapter 7; pp. 1–58. [Google Scholar] [CrossRef]
- Bernatchez, L.; Dodson, J.J. Phylogeographic structure in mitochondrial DNA of the lake whitefish (Coregonus clupeaformis) and its relation to Pleistocene glaciations. Evolution 1991, 45, 1016–1035. [Google Scholar]
- Geibert, W.; Matthiessen, J.; Stimac, I.; Wollenburg, J.; Stein, R. Glacial episodes of a freshwater Arctic Ocean covered by a thick ice shelf. Nature 2021, 590, 97–102. [Google Scholar] [CrossRef]
- Kaufman, D.S.; Young, N.E.; Briner, J.P.; Manley, W.F. Alaska palaeo-glacier atlas (version 2). Dev. Quat. Sci. 2011, 15, 427–445. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).