Abstract
The Indochinese thick-thumbed bat, Glischropus bucephalus, was described in 2011, but its molecular genetics (and the genetics of the whole genus Glischropus) are still poorly studied. We defined and annotated the complete mitogenome of Glischropus bucephalus (Chiroptera: Vespertilionidae) from Vietnam. The complete mitogenome is 17,023 bp in total length, which includes 13 complete protein-coding genes, 22 tRNA genes, 2 rRNA, and one non-coding region (the origin of replication). The nucleotide composition is 33.2% A, 29.7% T, 13.6% G, and 23.5% C. The mitochondrial protein-coding genes use the standard start codon (ATN), one complete stop codon (TAA), and two incomplete stop codons (TA- and T-). The phylogenetic analysis confirmed that the genus Glischropus belongs to the Pipistrellini tribe and revealed that Glischropus bucephalus is clustered with the “eastern” clade of Pipistrellus, supporting the paraphyletic nature of the latter genus.
1. Introduction
The name Glischropus was established by G. Dobson as a subgenus of Vesperugo for two species: African Vesperugo nanus (now—Afronycteris nanus) and Asian V. tylopus [1]. Later, it was raised to a full genus as its distribution is restricted to Southeast Asia (see Tate: [2]). Tate considered three species within the genus: the relatively widespread G. tylopus; G. batjanus Matschie, 1901; and G. javanus Chasen, 1929, with the two latter species known only from their type localities. Glischropus batjanus was then considered a partial synonym of G. tylopus, and a new species, G. bucephalus, was described as ranging from the mainland Asian north of the Isthmus of Kra [3]. Later, one extra species, G. aquillus, was described in Sumatra [4], and more recently, another new species, G. meghalayanus, was revealed in Meghalaya, India [5].
When separated as a genus, Glischropus was differentiated from Pipistrellus by its developed thumb pads (similar to that of Tylonycteris) and by the outer upper incisor, which is outwardly displaced from the row of teeth; otherwise, it was considered quite similar to Pipistrellus per se [2,6]. Chromosomal and genetic data support a close relationship between Glischropus and Pipistrellus [7,8], and it was placed within the tribe Pipistrellini [9]. Recent genetic data suggest that Glischropus represents one of the taxa that make Pipistrellus paraphyletic ([10]). However, whole mitogenomes, which seem to be useful tools for phylogenetic research (e.g., ref. [11]), were not yet studied for this genus.
Glischropus bucephalus is a small bat species (forearm length 31.7–35.7 mm) of pipistrelle-like appearance. Characteristic thickened pads on their thumb bases and plantar surface distinguish it from the similarly sized Pipistrellus. This feature is considered to have evolved as an adaptation for roosting in shelters with smooth surfaces like bamboo stems; however, this is still unconfirmed for this particular species [3]. This bat is sporadically found in forested areas at low and middle elevations across the mainland of Southeast Asia, from the north to the Isthmus of Kra. Its flight pattern and echolocation signals resemble those of Pipistrellus [12].
Although not ideal, the full mitochondrial DNA sequence is a functional tool for taxonomic studies at different levels, e.g., refs. [13,14], giving more reliable results than single gene sequences. Despite the fact that Glischropus bucephalus was described more than ten years ago, it was apparently involved in molecular genetic studies only twice [4,10], and its mitochondrion was not described. Moreover, not a single published mitochondrion of representatives of the genus Glischropus was known (although during the course of our research it was found that this was not entirely true). In order to fill the gap and provide additional clarity on the taxonomic position of Glischropus, we have isolated the complete mitochondrial DNA sequence of G. bucephalus.
2. Materials and Methods
2.1. DNA Extraction and Amplification
The DNA for this study was extracted from the alcohol-preserved tissue (muscle) of the museum specimen ZMMU S-195417, sourced in the vicinity of Ro Koi, Sa Thay District, Kon Tum Province, Vietnam (14.50° N, 107.72° E). Total genomic DNA was extracted using the standard phenol–chloroform deproteinization method [15]. A PCR was performed using an Encyclo Plus PCR kit (Evrogen, Moscow, Russia), following the manufacturer’s protocols. For PCR amplification of the mitogenome of G. bucephalus, we used 4 newly designed primer sets from multiple alignments of the complete mitogenomes of P. coromandra (GenBank: NC_029191.1, [16]) and P. abramus (KX355640.1, unpublished), which are available from GenBank. We used the following primers:
4 forward Pip0-4K_L 5′-CCCGTCACCCTCCTCAAG-3′;
Pip4-8K_L 5′-GGGCCCATACCCCGAAAA-3′;
Pip8-12K_Led 5′-ATGCCACARCTCAACACA-3′;
Glisch12-16K_L 5′-TCACCTGGTCAATCATAGAAT-3′;
and 4 reverse
Pip0-4K_R 5′-AATCCTGCCCCGGCTTC-3′;
Pip4-8K_R 5′-TGTAGGGGTAATGAAAGAGGCA-3′;
Pip8-12K_R 5′-CTATGGCAGAGGGGAGTCA-3′;
Glisch12-16K_R 5′-GGGCTGGTAGATCAATAAATG-3′.
Furthermore, additional primers were used to increase the overall base pair outcome for cytb:
L14108_pip 5′-AACCCACGACTAGTGACACGAAA-3′;
H15303_plec 5′-ACAAGAYCAGTGTAATTAATATACTACATAGAC-3′.
PCR amplification was performed in a final reaction volume of 20 μL, which contained 2 μL 10X Encyclo buffer, 0.4 μL 50X dNTP, 0.5 μL of each primer (C = 16 pmol/µL), 0.4 μL 50X Encyclo polymerase mix, and 1 μL of DNA sample. The conditions were as follows: 94 °C for 3 min (initial denaturation); then 95 °C for 10 s (denaturation); 55–65 °C for 30 s (annealing); and 68 °C for 8 min (extension) for 34 cycles with a final extension at 72 °C for 5 min. The PCR products were resolved by electrophoresis in 1.0% agarose gel and extracted using a DNA gel extraction kit (Qiagen, Valencia, CA, USA).
The four DNA fragments (~1800–2500 bp) that were the result of the amplification of the whole mitochondrial genomes from G. bucephalus were used for library preparation with an SQK-LSK110 ligation sequencing kit (1D) and a Native Barcoding Sequencing Kit, EXP-NBD (Oxford Nanopore Technologies, Oxford, UK). The sequencing library preparation procedure was performed as follows: the concentration of amplicons was measured by fluorometry using the Qubit 4.0 dsDNA BR assay, and a ~50–60 fmol mixture containing each of the four amplicons was prepared (in sum, 200–240 fmol). End repair and dA-tailing were performed on extracted DNA using the NEBNext Ultra II End Repair/dA-Tailing Module (NEB, E7546) and NEBNext FFPE Repair Mix (NEB, M6630). Next, the sample was purified at a ratio of 1:1 using MGIEasy DNA Clean Beads (MGI, 1000005278). Barcode ligation was performed using NEB Blunt/TA Ligase Master Mix (NEB, M0367) and a Native Barcoding Sequencing Kit, EXP-NBD104 (Oxford Nanopore Technologies). The barcode-ligated DNA was cleaned up by adding 1× volume of (MGI, 1000005278). Adapter ligation was performed using the Adapter Mix II from SQK-LSK110 (1D) and the NEBNext Quick Ligation Module (NEB, E6056). The adaptor-ligated DNA was cleaned up by adding 1× the volume (MGI, 1000005278) and sequenced using the workflow recommended by the manufacturer. The PromethION device (Oxford Nanopore Technologies, UK) was used for sequencing with flow cells FLO-PRO002 (R9.4.1 chemistry). Sequencing runs were controlled using MinKNOW software (version 22.03.4). The high-accuracy basecalling program integrated into the MinKNOW software was used for basecalling the raw signal, with a minimum Q-score of 9.
2.2. Mitogenome Assembling and Annotation
The raw data obtained in this work was used to assemble mitochondrial genomes. The draft assembly of the mtDNA reads was performed using CLC Genomic Workbench 8.5 [QIAGEN Aarhus A/S, US]. Reads that were presumably assigned to reference mtDNA were selected and used for assembly (10046 raw reads). The mitogenome attributed to Pipistrellus coromandra (GenBank:NC_0291919.1) was used as a reference sequence for the search. 10,046 raw reads were used as references for assembling the consensus sequences. The method of consensus polishing by raw ONT reads was used to improve and update the information obtained during the primary assembly of the mitochondrial genome.
The G. bucephalus partial mitogenome was initially annotated using a P. coromandra (GenBank: NC_029191.1) mitogenome as a reference using Bowtie2 [17], then merged with Samtools [18]. The mean depth of coverage was calculated using bedtools (v.2.26.0). Next, a de novo assembly was performed. Annotations were characterized based on the P. coromandra, P. abramus, and P. kuhlii (GenBank: KU058655.1, [19]) mitogenomes. The 13 mitochondrial protein-coding genes (PCGs) sequences were translated into amino acid sequences in MEGA v.11.0.13 using the vertebrate mitogenome genetic code [20]. NCBI’s Conserved Domain Database was used for the annotation of the protein-coding genes [21]. The MITOS web server, the tRNA scan-SE search server, and the ARWEN web server (with default parameters) were used to identify the tRNA genes and potential stemloop secondary structures within these tRNA genes [22,23,24].
2.3. Phylogenetic Reconstruction
To elucidate the molecular phylogeny of the species under study, we reconstructed the phylogenetic trees under the maximum likelihood (ML) criterion based on concatenated sequences of all the mtDNA PCGs for 19 Vespertilioninae species, with 3 Myotis species used as outgroups. Sequences were aligned using the Clustal W algorithm, implemented in MEGA v.11.0.13 [20], and adjusted manually. The ML reconstructions were conducted in IQTree version 1.6 [25]. Each gene in the alignment was partitioned into three codon positions; the best-fitting substitution models were determined using the ModelFinder implementation [26] in IQTree (Table 1). Clade stability was inferred using Ultrafast Bootstrap [27] with 10,000 replicates. Phylogenetic trees for this dataset were also reconstructed with third codon positions omitted or with third codon positions and the Nd6 gene both omitted to avoid possible saturation bias.

Table 1.
Model types for protein-coding gene analysis configured by IQtree ModelFinder through ultrafast bootstrap (10,000 replicates) for the phylogenetic tree with 3 codons.
An additional ML phylogenetic reconstruction was performed based on the 54 cytb gene sequences of 26 Vespertilionid species using the same approach. The following substitution models were assigned for each codon position: TIM2e+G4 for the first, TIM3+F+I for the second, and TN+F+I+G4 for the third position.
GenBank accession numbers for specimens used in both analyses are provided in Table 2.

Table 2.
GenBank accession numbers for mitochondrion and cytb sequences used in analysis.
3. Results
The G. bucephalus complete mitogenome contains a total of 17,023 bp in length, which consists of 13 complete protein-coding genes (PCGs), 22 tRNA genes and 2 rRNA genes (Figure 1, Table 3). The mean depth of coverage is 9216. Eight tRNAs and Nd6 are transcribed from the light strand, while 11 PCGs, 14 tRNAs, and 2 rRNAs are located on the heavy strand. The H-strand base composition is 33.2% A, 29.7% T, 13.6% G, and 23.5% C, which differs only slightly from the Pipistrellus species. AT content is distinctly higher than GC content, which is common for Vespertilioninae. The gene order and orientation of the Glischropus mitochondrial genome were the same as those of “P. coromandra” (NC029191.1) and other Vespertilioninae.
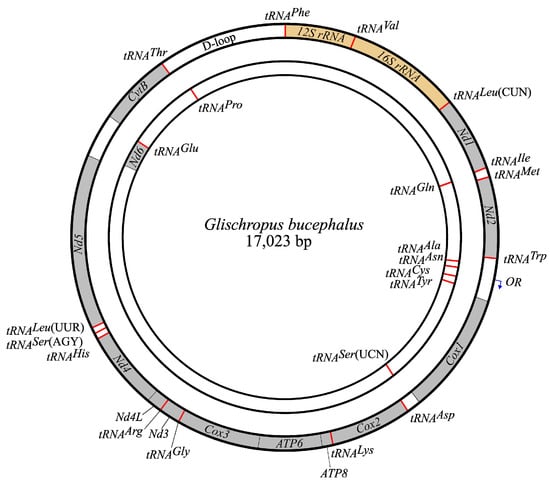
Figure 1.
Map of the G. bucephalus mitogenome. Gray color indicates the PCG regions; red color—tRNAs; yellow color—rRNAs. The heavy strand in the outer circle encodes 28 genes, whereas 9 genes are encoded in the light strand in the inner circle.

Table 3.
Gene organization and characterization of the G. bucephalus mitogenome.
The total length of the 13 mitochondrial PCGs in G. bucephalus is 11,379 bp, which could be translated into 3793 amino acids. ATG is the most common start codon and is used in 10 PCGs. However, the start codon ATT is used twice in both Nd2 and Nd3, and ATA is used only once in Nd5. TAA is the most common stop codon, which is used for the termination of 7 PCGs (Cox1, Cox2, ATP8, ATP6, Nd4L, Nd5, and Nd6). The incomplete stop codons are used for the termination of 5 PCGs (TA– for Nd1, Cox3, and Nd3; T– for Nd2 and Nd4). The replication origin (OR) is 35 bp in size and located between tRNAAsn and tRNACys within the WANCY tRNA cluster, as seen in most vertebrates [16].
Both the tribe Pipistrellini (with the genera Pipistrellus, Nyctalus, and Glischropus) and its association with Vespertilionini have maximum support regardless of the data type. Similar to data published previously [10], Pipistrellini includes two well-supported clades, conditionally “western” and “eastern”, each of which includes species now assigned to Pipistrellus. Our G. bucephalus clearly belongs to the “eastern” clade, in which it occupies a sister position to NC029191.1 with maximum support (Figure 2).
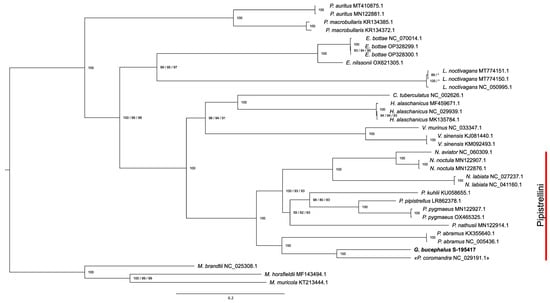
Figure 2.
The phylogenetic relationship between G. bucephalus and the other Vespertilioninae species is inferred by the maximum likelihood analysis based on the concatenated protein-coding gene sequences. The bootstrap values (indicated by the slashes on the branches) correspond to the trees constructed on full sequences (three codon positions), the first two codon positions (third positions omitted), and two positions with the exclusion of the Nd6 gene. The asterisks mark branches that in the second or third case have a different topology than shown. Myotis species are used as outgroups.
4. Discussion
In general, the obtained mitogenome of G. bucephalus in its length and structure is similar to the published mitogenomes of Pipistrellus spp. and Nyctalus spp., and especially to the mitogenome NC029191.1 attributed to P. coromandra. Moreover, the nucleotide composition has some differences: NC029191.1 has 1% more T and almost 1% less C (see [16]). Differences in the nucleotide composition with P. abramus (which is also very close on the phylogenetic tree) are less pronounced [28].
As expected, according to our data, the genus Glischropus undoubtedly belongs to the tribe Pipistrellini and is very closely related to the “eastern” clade of the genus Pipistrellus in its current recognition. In addition, both its position and the position of Nyctalus (forming a well-supported sister clade to the “western” Pipistrellus) obviously make Pipistrellus a paraphyletic taxon, which indicates its need for further revision. However, the position of G. bucephalus, sister to NC029191.1 and attributed to P. coromandra, is naturally surprising, because if this is correct, then Glischropus makes not only the whole Pipistrellus genus paraphyletic but also its “eastern” branch. This, on the one hand, could mean a special taxonomic status for P. abramus, but on the other hand, it contradicts all previously obtained results [10,29].
We included the cytb gene sequences taken from the mitochondrions of G. bucephalus and NC029191.1 in an expanded set of sequences of the same gene from different members of the Pipistrellini tribe (Figure 3). The tree, obtained as a result of the analysis, gives a somewhat different topology in which Glischropus stands in a sister position next to the other members of the tribe. However, the association of the “eastern” and “western” clades has very low support; hence, we could talk about trichotomy. These differences obviously indicate various resolution possibilities for an individual gene and genomic sequence for the phylogenetic constructions. Specimen NC029191.1 occupies a position within the G. tylopus clade, strongly suggesting its initial misattribution as P. coromandra. Thus, according to both the data of an individual gene and the mitochondrial genome, G. bucephalus forms a common clade with its congeneric, G. tylopus, while demonstrating undoubted differences in the species level from the latter.
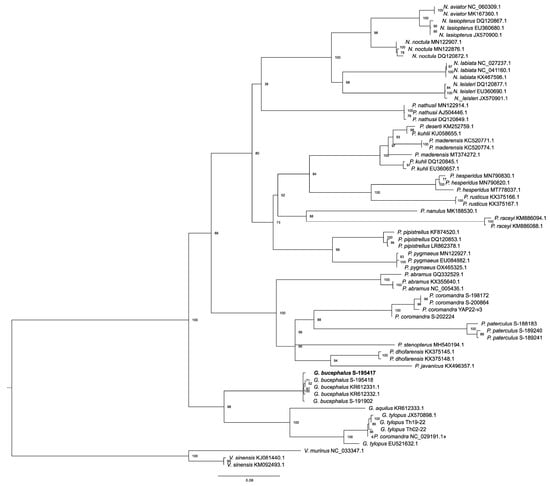
Figure 3.
The phylogenetic relationship between G. bucephalus and the other Pipistrellini species is inferred by maximum likelihood analysis based on cytb sequences. The numbers in the branches show the bootstrap values. Vespertilio species are used as outgroups.
This example demonstrates the need to carefully handle the published data in addition to checking it when there is doubt. In general, our work confirms the assumption that there is a close relationship between Glischropus and Pipistrellus based on new data and again indicates a probable generic status for the “eastern” clade of the latter genus.
5. Conclusions
The mitochondrial genome of Glischropus bucephalus is 17,023 bp in length and includes 13 protein-coding genes. It is similar in structure, length, and composition to the previously published mitochondrial genomes of Pipistrellus abramus and “Pipistrellus coromandra”, but differs from both at a significant species level. Phylogenetic reconstruction using the newly obtained genome confirms the paraphyly of the genus Pipistrellus in its modern understanding and the need to revise its taxonomy. The somewhat paradoxical phylogenetic position of the G. bucephalus mitogenome prompted additional phylogenetic analysis of the individual cytb gene using sequences from the same specimens. The analysis showed that the mitogenome attributed to “Pipistrellus coromandra” actually belongs to Glischropus tylopus. This indicates the need for careful usage of sequences stored in GenBank.
Author Contributions
Conceptualization, S.V.K. and S.S.Z.; methodology, A.A.L. and A.S.S.; validation, A.S.S. and S.V.K.; formal analysis, S.S.Z., A.A.L. and A.S.S.; investigation, S.S.Z.; data curation, S.V.K.; writing—original draft preparation, S.V.K.; writing—review and editing, S.V.K., S.S.Z. and A.A.L.; visualization, S.S.Z.; supervision, S.V.K.; project administration, S.V.K.; funding acquisition, S.V.K. All authors have read and agreed to the published version of the manuscript.
Funding
The work was carried out with financial support from the Russian Science Foundation grant “Taxonomic and phylogenetic relationships of bats (Chiroptera, Mammalia) on the east of Asia” (RSF 22-24-00017).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Genetic data: data available in a publicly accessible repository in GenBank.
Acknowledgments
Our special thanks to V.S. Lebedev (Zoological Museum of Moscow University) for his valuable help with the data procession. The study of the collection materials was carried out in the Zoological Museum of Moscow University, using the collection facilities, with the support of its director, M.V. Kalyakin. The molecular genetic studies were performed at the facilities of the Vertebrate Zoology Department of Moscow University, with priceless support from A.A. Bannikova. Obtaining materials from Vietnam became possible through collaboration with the Joint Vietnamese-Russian Tropical Research and Technological Centre, with the support of Nguyen Dang Hoi and A.N. Kuznetsov. The work was performed at the facilities of the Vertebrate Zoology Department, Biological Faculty, and Zoological Museum of the Moscow State University, with the assistance of the administration of these departments and in line with the research theme of the Zoological Museum (no. 121032300105-0).
Conflicts of Interest
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of the paper. The funders had no role in the design of the study, in the collection, analysis, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
References
- Dobson, G.E. Descriptions of new or little-known species of bats of the genus Vesperugo. Proc. Zool. Soc. Lond. 1875, 470–474. [Google Scholar]
- Tate, G.H.H. Results of the Archbold Expeditions 47. Review of the vespertilionine bats, with special attention to genera and species of the Archbold Expeditions. Bull. Am. Mus. Nat. Hist. 1942, 80, 221–297. [Google Scholar]
- Csorba, G. A new species of Glischropus from the Indochinese Subregion (Mammalia: Chiroptera: Vespertilionidae). Zootaxa 2011, 2925, 41–48. [Google Scholar] [CrossRef]
- Csorba, G.; Görföl, T.; Wiantoro, S.; Kingston, T.; Bates, P.J.J.; Huang, J.C. Thumb-pads up—A new species of thickthumbed bat from Sumatra (Chiroptera: Vespertilionidae: Glischropus). Zootaxa 2015, 3980, 267–278. [Google Scholar] [CrossRef]
- Saikia, U.; Ruedi, M.; Csorba, G. Out of Southeast Asia: A new species of thick-thumbed bat (Chiroptera: Vespertilionidae: Glischropus) from Meghalaya, north-eastern India. Zootaxa 2022, 5154, 355–364. [Google Scholar] [CrossRef]
- Miller, G.S. The Families and Genera of Bats; Government Printing Office: Washington, DC, USA, 1907; pp. 1–282.
- Volleth, M.; Bronner, G.; Göpfert, M.C.; Heller, K.-G.; von Helversen, O.; Yong, H.-S. Karyotype comparison and phylogenetic relationships of Pipistrellus-like bats (Vespertilionidae; Chiroptera; Mammalia). Chromosome Res. 2001, 9, 25–46. [Google Scholar] [CrossRef] [PubMed]
- Heaney, L.R.; Balete, D.S.; Alviola, P.; Rickart, E.A.; Ruedi, M. Nyctalus plancyi and Falsistrellus petersi (Chiroptera: Vespertilionidae) from northern Luzon, Philippines: Ecology, phylogeny, and biogeographic implications. Acta Chiropterol. 2012, 14, 265–278. [Google Scholar] [CrossRef]
- Simmons, N.B. Order Chiroptera. In Mammal Species of the World: A Taxonomic and Geographic Reference, 3rd ed.; Wilson, D.E., Reeder, D.M., Eds.; The Johns Hopkins Univ. Press: Baltimore, MD, USA, 2005; pp. 312–529. [Google Scholar]
- Zhukova, S.S.; Solovyeva, E.N.; Artyushin, I.V.; Kruskop, S.V. Paraphyly of the Pipistrelles (Pipistrellus; Vespertilionidae) is confirmed by the analysis of the nuclear gene markers. Doklady Biochem. Biophys. 2022, 507, 302–306. [Google Scholar] [CrossRef]
- Mackiewicz, P.; Matosiuk, M.; Świsłocka, M.; Zachos, F.E.; Hajji, G.M.; Saveljev, A.P.; Seryodkin, I.V.; Farahvash, T.; Rezaei, H.R.; Torshizi, R.V.; et al. Phylogeny and evolution of the genus Cervus (Cervidae, Mammalia) as revealed by complete mitochondrial genomes. Sci. Rep. 2022, 12, 16381. [Google Scholar] [CrossRef]
- Kruskop, S.V. Bats of Vietnam. Checklist and an Identification Manual, 2nd ed.; Revised and Supplemented; KMK Scientific Press: Moscow, Russia, 2013; pp. 1–300. [Google Scholar]
- Hassanin, A.; Hugot, J.-P.; van Vuuren, B.J. Comparison of mitochondrial genome sequences of pangolins (Mammalia, Pholidota). Comptes Rendus Biol. 2015, 338, 260–265. [Google Scholar] [CrossRef]
- Zhang, Y.-C.; Lwin, Y.H.; Li, R.; Maung, K.-W.; Li, G.-G.; Quan, R.-C. Molecular phylogeny of the genus Muntiacus with special emphasis on the phylogenetic position of Muntiacus gongshanensis. Zool. Res. 2019, 42, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbour Lab Press: New York, NY, USA, 1989; pp. 1–1659. [Google Scholar]
- Rahman, M.M.; Yoon, K.B.; Kim, J.Y.; Hussin, M.Z.; Park, Y.C. Complete mitochondrial genome sequence of the Indian pipistrelle Pipistrellus coromandra (Vespertilioninae). Animal Cells Sys. 2016, 20, 86–94. [Google Scholar] [CrossRef][Green Version]
- Langmead, B.; Wilks, C.; Antonescu, V.; Charles, R. Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics 2019, 35, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Locatelli, A.G.; Jebb, D.; Teeling, E.C. The complete mitochondrial genome of Kuhl’s pipistrelle, Pipistrellus kuhlii (Chiroptera: Vespertilionidae). Mitochondrial DNA Part B 2016, 1, 423–424. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2010, 39, D225–D229. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phyl. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Laslett, D.; Canbäck, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Kim, H.R.; Lim, S.J.; Kim, H.J.; Cho, J.Y.; Park, Y.C. Complete mitochondrial genome of the house bat Pipistrellus abramus (Mammalia: Chiroptera) from Korea. Mitochondrial DNA Part B 2017, 2, 540–541. [Google Scholar] [CrossRef]
- Kruskop, S.V.; Solovyeva, E.N.; Kaznadzey, A.D. Unusual Pipistrelle: Taxonomic position of the Malayan Noctule (Pipistrellus stenopterus; Vespertilionidae; Chiroptera). Zool. Stud. 2018, 57, 1–15. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).