Abstract
This study is devoted to the investigation of Azygia (Digenea: Azygiidae) species diversity using classical morphological, recent molecular tools (28S rRNA and cox1 mtDNA for genetic-based inference) and robust statistical techniques (Principal component analysis, PCA). The analysis revealed that the genus Azygia included four valid species: A. lucii, A. longa, A. hwangtsiyui, and A. sibirica n. sp. The distribution of the type species A. lucii was confirmed in the largest Russian rivers: the Volga and the Ob. The worms isolated from Perccottus glenii were determined as the Chinese species A. hwangtsiyui, according to the genetic data for the cox1 mtDNA gene, at 1.32–1.56%. The new species, Azygia sibirica n. sp, was described from Esox lucius in the Ob River and differentiated from the type species A. lucii by the smaller ovary, testes and prostatic sac, wider body, very narrow pharyngeal lumen and form of anterior margin of ovary. In addition, multivariate analysis and three methods for species delimitation (ABGD, GMYC, bPTP) showed the subdivision of A. lucii and A. sibirica n. sp. into two separate groups, one from the Volga River and another from the Ob River, respectively. To conclude, A. lucii infects Esox lucius in the western (European part of Russia, the Volga River basin), and northern (Western Siberia, the Ob River basin) parts of Russia; A. sibirica n. sp. has also been found to infect Esox lucius in the Ob River, while A. hwangtsiyui infects Perccottus glenii in the South of the Russian Far East (the Artymovka River basin).
1. Introduction
The genus Azygia (Looss, 1899) comprises endoparasitic helminth found in the stomach and intestine of freshwater carnivorous fish from the orders Esociformes, Salmoniformes and Perciformes. Fish of the orders Cypriniformes, Siluriformes, Perciformes, Acipenseriformes, Scorpaeniformes, and Gadiformes are used as paratenic hosts. As a rule, azygiids are large worms up to 3–5 cm long. The intensity of infection often reaches several dozens or hundreds of individuals per fish. Some species are highly pathogenic to hosts. For instance, Azygia anguisticauda (Stafford, 1904) causes the pathologies of all intestine tissue layers (degeneration of villi, ruptured serosa, swelling, vacuolization in tunica muscularis and lamina propria) of the Dwarf snakehead Channa gachua (Hamilton, 1822) [1]. Azygia spp. are widely distributed in Europe, Asia, North America, and the European and Asian parts of Russia.
Based on the systematics suggested by Skrjabin and Guschanskaja [2], the genus Azygia includes 12 species and belongs to the family Azygiidae of the superfamily Azygioidea [3]. To date, the WORMS database includes 24 nominal species of Azygia, but validity has been confirmed only for Azygia sinipercae Wang and Pang, 1973 [4] and A. longa (Leidy, 1851), based on 28S rRNA gene [5], and for A. hwangtsiyui (Tsin, 1933), A. longa, and A. angusticauda, based on the cox1 mtDNA gene [6,7,8]. Seven species of Azygia were noted in the European, Siberian, and Far Eastern parts of Russia: A. amuriensis (Zmeev, 1936), A. hwangtsiyui, A. lucii (Müller, 1776), A. mirabilis (Braun, 1891), A. perryi (Fujita, 1918), A. robusta (Odhner, 1911), and A. volgensis (Linstow, 1907) [9,10,11,12,13,14,15].
Azygia lucii was described as from the Northern pike Esox lucius Linnaeus, 1758 by Müller [16], in Denmark (type locality). Currently, genetic data for Azygia spp. from Russia are completely absent, as well as validation of the type species A. lucii, registered there by many parasitologists [11,17,18,19,20]. However, limited morphological analyses have been performed for this species. Therefore, a detailed redescription, confirming that A. lucii exactly inhabits the rivers of the European and Siberian parts of Russia, is required. In addition, prior to this study, the Chinese species A. hwangtsiyui have already been observed in the rivers of the south of the Russian Far East [14,15,21].
The aim of the present study was to characterize the species diversity of azygiids from the largest Russian rivers that flow through the East European Plain, Western Siberia, and South of the Russian Far East; this was performed by clarifying the phylogenetic relationships of the worms, using both classical morphological and molecular genetic methods.
2. Materials and Methods
2.1. Sampling
During the field studies in the period of October 2018–June 2019, 120 individuals of Azygia hwangtsiyui were isolated from the stomach and anterior part of the intestine of Amur sleeper Perccottus glenii Dybowski, 1877, obtained from the Bolotnaya and Ivnyanka Rivers (the Artyomovka River basin) in the South of the Russian Far East [15]. The specimens were identified following the morphological characteristics of Azygia hwangtsiyui from P. glenii, caught by Besprozvannykh [14] in the Arsenyevka River (the Ussuri River basin).
Esox lucius were collected from the western and northern parts of Russia: West—the Rybinsk Reservoir (the Volga River) near Borok (58°06′ N, 38°23′ E) between September 2019 and August 2022; North—near four sites located along the Ob River: Nizhnevartovsk (60°93′ N, 76°55′ E), Surgut (61°24′ N, 73°39′ E), Khanty-Mansiysk (61°00′ N, 69°01′ E), and Priobye (62°54′ N, 65°64′ E), during July 2021. All fish were caught by gill nets or purchased from fishermen. Adult trematodes, belonging to the genus Azygia, were collected alive from freshly killed pikes.
In total, 40 specimens from E. lucius and 20 specimens from P. glenii, were selected for DNA extraction and preserved in 96% ethanol. The species names, definitive hosts and geographic location information about the material collected are given in Table 1.

Table 1.
Information on the azygiid species names, hosts, and accession numbers for the partial sequences of the 28S rRNA and cox1 mtDNA genes, downloaded from GenBank for phylogenetic analysis.
2.2. Morphological Examination
In total, 26 voucher specimens, used for morphological examination, were killed in warm (30–35 °C) 70% ethanol without crushing, stained with Mayer’s haematoxylin or alum carmine, dehydrated through a graded ethanol series, cleared in dimethyl phthalate and mounted in Canada balsam.
Mounted specimens were examined under a light microscope Olympus BX53F2 (Japan). Specimens were photographed with aid of a digital camera, and drawings were made using a drawing tube attached to the microscope. Measurements were taken using microscope software and a digital image analysis system. Voucher specimens were deposited in the helminthological collection of the I.D. Papanin Institute for Biology of Inland Waters RAS, Russia.
The following morphological characters were measured: 1. Body length; 2. Maximum body width 3. Body width/length ratio; 4. Forebody length; 5. Forebody width; 6. Forebody length as % of body length; 7. Forebody width/maximum body width; 8. Oral sucker length; 9. Oral sucker width; 10. Ventral sucker length; 11. Ventral sucker width; 12. Distance between suckers; 13. Pharynx length; 14. Pharynx width; 15. Pharynx width/length ratio; 16. Pharyngeal lumen; 17. Pharyngeal lumen/pharynx width ratio; 18. Reproductive zone length (from anterior margin of ovary to posterior margin of posterior testis); 19. Reproductive zone length as % of body length; 20. Ovary length; 21. Ovary width; 22. Ovary length/width ratio; 23. Anterior testes length; 24. Anterior testes width; 25. Distance of ovary from anterior testis; 26. Posterior testes length; 27. Posterior testes width; 28. Post-testicular space; 29. Post-testicular space as % of body length; 30. Vitellarium length (mean of two); 31. Distance of vitellarium from posterior end; 32. Prostatic sac length; 33. Prostatic sac width; 34. Vitelline follicles length and width (mean of 15 on each side of body); 35. Vitellarium length as % of body length; 36. Egg length; and 37. Egg width.
2.3. Multivariate Analysis
A principal component analysis (PCA) and cluster analyses were performed for statistical data processing using the NTSYS 2.02k software package. In PCA, the eigenvectors were calculated using the correlation matrix; the length of the vector was set to 1. For cluster analysis, morphological distances between the studied individuals in the multidimensional space of characters were estimated using the Manhattan metric. Similarity matrices were analyzed by complete linkage clustering, illustrating the result by dendrogram. For multidimensional analysis, the absolute values of 19 body measurements were converted into relative indices by calculating their percentage ratio to body length. These 19 indices were as follows: maximum body width/body length, forebody length/body length, forebody width/body length, oral sucker length/body length, oral sucker width/body length, ventral sucker length/body length, ventral sucker width/body length, pharynx length/body length, pharynx width/body length, pharyngeal lumen/body length, ovary length/body length, ovary width/body length, anterior testes length/body length, anterior testes width/body length, distance of ovary from anterior testis/body length, posterior testes length/body length, posterior testes width/body length, post-testicular space/body length, and distance of vitellarium from posterior end/body length. In addition to the indices calculated as the ratio of a body measurement to the length of the body, four special indices were used: the width of the anterior part of the body/maximum width of the body, the ratio of the width and length of the pharynx, the ratio of the pharyngeal lumen to the width of the pharynx, and the ratio of the length of the ovary to its width. In total, the morphological relations of the studied individuals were estimated using the 23 body parameters.
2.4. DNA Extraction, Amplification, and Sequencing
Genomic DNA was extracted from 60 samples using the alkaline lysis method HotShot [24]: seven worms from the Volga River, 33 worms from the Ob River, and 20 worms from the Bolotnaya River. The 28S rRNA gene fragment was amplified using the forward primer U178 (5′-GCA CCC GCT GAA YTT AAG-3′) and the reverse primer L1642 (5′-CCA GCG CCA TCC ATT TTC A-3′) [25]. The cox1 mtDNA gene fragment was amplified using the forward primer JB3 (5′-TTT TTT GGG CAT CCT GAG GTT TAT-3′) [26] and the reverse primer CO1-R-trema (5′-CAA CAA ATC ATG ATG CAA AAG G-3′) [27]. The PCR mixture contained 2X DreamTaq Green PCR Master Mix (Thermo Scientific, USA), 0.5 μM forward and reverse primers and 4 μL templates in total volume of 20 μL. The amplification protocol was performed under the following conditions: for 28S, 2 min denaturation hold at 94 °C, 40 cycles of 30 s at 94 °C, 30 s at 52 °C, 2 min at 72 °C, and a 7 min extension hold at 72 °C; for cox1, 1 min denaturation hold at 94 °C, 30 cycles of 15 s at 94 °C, 30 s at 50 °C, 2 min at 72 °C, and a 7 min extension hold at 72 °C. Each PCR reaction included negative and positive controls, using both primers to detect possible contamination. PCR products were sequenced using the Big Dye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA) (as instructed by the manufacturer) and internal sequencing primers with 28S: 300F (5′-AGG GTT CGA TTC CGG AG-3′), 1200R (5′-GGG CAT CAC AGA CCT G-3′), 1200F (5′-CCC GAA AGA TGG TGA ACT ATG C-3′) [25]. The PCR products were read with a ABI 3130 genetic analyzer at the Federal Scientific Center of the East Asia Terrestrial Biodiversity FEB RAS.
In total, 81 newly sequences for three species were submitted to the GenBank database: 28S rRNA gene—21 sequences, 1708 bp (GenBank accession numbers OP794027-OP794047); and cox1 mtDNA—60 sequences, 852 bp (accession numbers OP804409-OP804468) (see Table 1).
2.5. Alignment and Phylogenetic Analyses
Nucleotide sequences were assembled and aligned using MEGA X [28]. The alignment of partial 28S rDNA sequences were trimmed to the length of the shortest sequence downloaded from GenBank (No. LN865022, see Table 1). Genetic divergence was estimated using genetic p-distance values, which were calculated by including all substitution types in MEGA X. Phylogenetic analyses (based on 26 partial sequences of the 28S rRNA gene (1235 bp length) and 16 partial sequences of the cox1 mtDNA gene (678 bp length)) were carried out using the Bayesian Inference (BI) algorithm [29] with the GTR+I+G model (chosen using the Bayesian information criteria (BIC)); this included gamma and invariant sites, selected in jModeltest v. 2.1.5, as the best [30]. The MCMC algorithm was performed using 300,000 generations and two independent runs (average standard deviation of split frequencies was less than 0.01); in total, 25% of generations were discarded as burn-in in MrBayes v. 3.1.2 software [29]. Additional phylogenetic trees for the same samplings were reconstructed using the maximum likelihood (ML) method, based on the same GTR+G model in the MEGA X software, with 1000 bootstrap replicates.
Outgroup taxa used for 28S tree reconstruction were presented with the species from the family Gonocercidae (Hemipera manteri (accession no. AY222196), Gonocerca crassa (KY197012), G. phycidis (KY197010), G. oshoro (KY197013), and G. muraenolepisi (LN865025), Gonocerca sp. (HF543941, LN650651)), and one species from Derogenidae—Thometrema lotzi (KC985236). The 28S tree was rooted with the diplostomid species Diplostomum phoxini (AY222173).
ABGD (Automatic Barcode Gap Discovery) [31], GMYC (Generalized Mixed Yule Coalescent) [32] and bPTP (an updated version of the original maximum likelihood PTP (Poisson Tree Processes), which adds Bayesian support (BS) values to delimited species on the input tree) [33], were used on the web platforms to reveal cryptic lineages. ABGD scanned a range of Prior Intraspecific divergence from Pmin = 0.001 to Pmax = 0.1, with P Steps = 10, using the Kimura (K80) model, TS/TV = 2. For GMYC input, we used a fully resolved and an ultrametric tree obtained in BEAST to detect the transition in the tree where the branching pattern switches from speciation to intra-species coalescent process. According to the best result of the GMYC species delimitation method, the following was obtained: single likelihood of null model—505.6114; maximum likelihood of GMYC model—519.5971; likelihood ratio—27.97143; result of LR test—8.434944e-07; number of ML clusters—3; confidence interval 3–13; number of ML entities—3; confidence interval 3–14; threshold time—0.008151962. bPTP used phylogenetic input tree: unrooted; number of MCMC generations—100000; thinning every 100; and burn-in 0.1. The most supported partition, found by a simple heuristic search in bPTP, was as follows: Species 1 (support = 0.544), Species 2 (support = 0.873), Species 3 (support = 0.900).
3. Results
3.1. Redescription of Azygia Lucii (Müller, 1776) (Figure 1)
Taxonomic Summary
Synonyms: Azygia tereticollis (Rudolphi, 1802); Azygia volgensis (Linstow, 1907); Azygia lucii johanseni (Pavlov, 1931).
Host: Esox lucius (Linnaeus, 1758).
Locality: West—the Rybinsk Reservoir (the Volga River basin) near Borok, Russia (58°06′ N, 38°23′ E).
Voucher material: No. 2/44 (1–6) are deposited in the Parasites Collection of the I. D. Papanin Institute for Biology of Inland Waters RAS, Russia.
Genetic data: The 28S rRNA gene—OP794033–OP794042; cox1 mtDNA gene—OP804437–OP804468.
Morphology. Based on 10 adult specimens. Measurements reported in millimeters. Body long, moderately or slightly flattened, with barely noticeable expansion behind ventral sucker, 9.37–18.9 (12.8, 10) long, with maximum width 1.08–1.95 (1.39, 10) at level of ovary or anterior testis. Posterior end bluntly rounded; body width-to-length ratio: 0.068–0.119 (0.095, 10). Forebody short, 1.62–4.6 (2.72, 10) long or 17.3–24.3% (21%, 10) of body length; 0.73–1.52 (1.037, 10) width, poorly distinguished from hindbody by width; forebody width to maximum body width ratio: 0.648–0.874 (0.775, 10). Oral sucker round, subventral, 0.73–1.1 (0.9, 10) long, 0.61–1.11 (0.81, 10) wide; larger than ventral sucker. Ventral sucker round, 0.5–0.83 (0.64, 10) long, 0.46–0.83 (0.63, 10) wide. Distance between suckers 0.89–3.5 (1.82, 10). Prepharynx absent, pharynx muscular, moderately thick-walled, with a wide lumen, 0.48–0.78 (0.619, 10) long, 0.3–0.5 (0.41, 10) wide; pharynx width-to-length ratio: 0.545–0.733 (0.637, 10); pharyngeal lumen 0.158–0.228 (0.2, 10) wide, pharyngeal lumen to pharynx width ratio: 0.41–0.6 (0.52, 10). Esophagus very short; intestinal caeca extending dorsal and anteriad to pharynx, arching posterolateral; caeca long, folded along its entire length, nearly reaching to posterior extremity. Reproductive zone (distance from anterior margin of ovary to posterior margin of posterior testis) 1.19–2.52 (1.81, 10) long or 11.8–16.3% (14.2%, 10) of body length. Testes oval or rounded, tandem, anterior testis 0.36–0.69 (0.51, 10) long, 0.53–0.8 (0.68, 10) wide; posterior testis 0.41–0.76 (0.55, 10) long, 0.52–0.89 (0.68, 10) wide; post-testicular region long, 2.49–4.66 (3.54, 10) or 23–35.2% (27.9%, 10) of body length; vasa efferentia not evident. Prostatic sac medial, immediately anterior and dorsal to ventral sucker, 0.246–0.5 (0.35, 8) long, 0.29–0.52 (0.36, 8) wide; seminal vesicle tubular, highly convoluted, thin-walled. Pars prostatica in middle or in anterior part of prostatic sac, 102–144 (114, 8) long, 91–109 (99, 8) wide; metraterm long 153–660 (403, 6), dorsal to ventral sucker. Terminal genitalia of male and female confluent within sinus organ. Genital atrium contains large number of eggs, 0.28–0.88 (0.5, 10) long, 0.39–0.89 (0.55, 10) wide when completely filled with eggs, with wall 13–62 (30, 10) thick; genital pore anterior to ventral sucker, large, 51–103 (73, 5) long, 12–53 (30, 5) wide. Ovary semicircular, irregular oval or slightly triangular in shape, 0.21–0.51 (0.33, 10) long, 0.49–0.7 (0.58, 10) wide. Ovary length to width ratio: 0.42–0.79 (0.58, 10); anterior margin of ovary straight or slightly concave, distance between ovary and anterior testis 0.001–0.301 (0.094, 10). Oviduct emanating dorso-anteriorly from ovary. Vitelline reservoir ventral to ovary, 99–214 (160, 8) long, 46–81 (64, 8) wide; common vitelline duct very short. Oötype lateral, anterior to ovary. Uterine seminal receptacle large, 0.202–0.385 (0.298, 9) long, 0.089–0.0256 (0.165, 9) wide, contiguous with ovary anterior and lateral. Other elements of ovarian complex not visible. Vitellarium follicular, 3.67–8.16 (5.47, 10) long or 38.6–49% (42.5%, 10) of body length, distribute extra-caecal in two bilaterally symmetrical or asymmetrical lateral fields, terminating in post-testicular space, not reaching posterior extremity of body at 1.7–3.87 (2.67, 10). Vitelline follicles oval, subglobular, elongated or wedge-shaped, sometimes slightly lobed, 0.057–0.067 (0.064, 10) long, 0.072–0.127 (0.093, 10) wide. Number of follicles 82–148 (114, 8) in left row, 96–145 (126, 8) in right row. Uterus loops intercaecal, close and transverse. Eggs numerous, 0.041–0.055 (0.047) long, 0.018–0.027 (0.022) wide. Excretory pore terminal. Excretory vesicle tubular, bifurcating at level of posterior testis, its excretory ducts extend anteriorly to level of oral sucker.
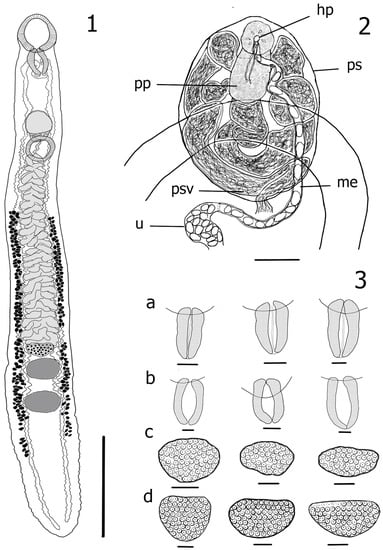
Figure 1.
1. Azygia lucii from the Volga River, adult worm ventral; 2. Terminal genitalia, ventral view: prostatic sac (ps), proximal region of seminal vesicle (psv), pars prostatica (pp), metraterm (me), hermaphroditic pore (hp), uterus (u); 3. Morphological variations of pharynx (a,b) and ovary (c,d): (a) Azygia sibirica n. sp., (b) A. lucii, (c) Azygia sibirica n. sp., (d) A. lucii. Scale-bars: 1—3 mm, 2—100 μm, 3—200 μm.
3.2. Description of Azygia Sibirica Vainutis, Voronova et Zhokhov, n. sp. (Figure 2)
Taxonomic Summary
Type host: Esox lucius (Linnaeus, 1758).
Type locality: The Ob River, from Nizhnevartovsk to Priobye, Russia (60°93′ N, 76°55′ E–62°54′ N, 65°64′ E).
Localization: Stomach.
Prevalence: 10 of 11 specimens infected.
Intensity range: 3–20 worms.
Type-deposition: The type-specimens are deposited in the Parasite Collection of the I.D. Papanin Institute for Biology of Inland Waters RAS, Russia: holotype No. 3/45 (1), paratype: No. 3/45 (2–7); e-mail: zhokhov@ibiw.ru. Deposited: 10 July 2022.
Etymology: The species is named after Siberia, the region where the worms were collected.
Genetic data: 28S rRNA gene—OP794043–OP794047; cox1 mtDNA gene—OP804429–OP804436.
Morphology. Based on 16 adult specimens. Body long, flattened, evenly expanding behind ventral sucker. Posterior end bluntly rounded, 8.6–14.7 (12.1, 16) long, maximum width 1.1–1.86 (1.56, 16) at level from anterior margin of vitellarium to posterior margin of vitellarium, body width-to-length ratio: 0.11–0.169 (0.13). Forebody short, moderately-well distinguished from hindbody by width, 1.96–4.11 (2.87, 16) long or 20–28.7% (23.8%, 16) of body length, 0.73–1.3 (1.02, 16) wide; forebody width to maximum body width ratio: 0.521–0.883 (0.66, 16). Oral sucker round, subventral, 0.69–1.13 (0.91, 16) long, 0.67–0.95 (0.8, 16) wide, larger than ventral sucker. Prepharynx absent, pharynx thick-walled, muscular, 0.46–0.74 (0.56, 16) long, 0.24–0.39 (0.32, 16) wide, pharynx width to length ratio: 0.497–0.78 (0.58, 16), length of pharynx larger than width, with narrow lumen: 0.04–0.11 (0.082, 16), pharyngeal lumen to pharynx width ratio: 0.077–0.389 (0.178, 16). Esophagus very short; intestinal caeca extending anteriad and dorsally to pharynx, arching posterolateral; caeca wide, long, strongly folded along its entire length, reaching posterior extremity. Ventral sucker rounded, 0.51–0.76 (0.64, 16) long, 0.46–0.8 (0.64, 16) wide, smaller than oral sucker, distance between suckers 1.18–3.11 (1.98, 16). Reproductive zone 1.22–2.56 (1.79, 16) long or 12–18.2% (14.8%, 16) of body length. Testes tandem, transversely oval, anterior testis 0.17–0.62 (0.42, 16) long, 0.21–0.83 (0.63, 16) wide, posterior testes 0.17–0.65 (0.49, 16) long, 0.21–0.92 (0.65, 16) wide; post-testicular space 1.75–3.82 (3.07, 16) or 20.3–34.8% (25.3%, 16) of body length; vasa efferentia not evident. Prostatic sac medial, immediately anterior and dorsal to ventral sucker, 0.19–0.42 (0.29, 13) long, 0.22–0.45 (0.28, 13) wide; seminal vesicle tubular, highly convoluted, thin-walled. Pars prostatica in middle or in anterior part of prostatic sac, dorsally, 76–107 (91, 7) long, 74–124 (98, 7) wide; metraterm not visible. Terminal genitalia of male and female confluent within sinus organ. Genital atrium contains low number of eggs; 0.16–0.495 (0.26, 13) long, 0.193–0.54 (0.298, 13) wide when completely filled with eggs, with wall 12–33 (20, 6) thick; genital pore anterior to ventral sucker, large, 42–86 (60, 7) long, 37–50 (28, 7) wide. Ovary slightly transversely elongate, 0.12–0.42 (0.29, 16) long, 0.2–0.62 (0.47, 16) wide, anterior margin of ovary slightly prominent, ovary length to width ratio: 0.47–0.73 (0.62, 16), distance between ovary and anterior testis: 0.077–0.61 (0.24, 16). Vitelline reservoir ventral to ovary, 0.063–0.29 (0.154, 9) long, 0.029–0.052 (0.04, 9) wide, uterine seminal receptacle present, 0.199–0.25 (0.228, 4) long, 0.9–0.142 (0.109, 4) wide, contiguous with ovary anterior and lateral. Other elements of ovarian complex not visible. Vitellarium follicular, lateral and extra-caecal, distributing in two bilaterally symmetrical or asymmetrical fields, terminating well posterior to posterior testis, not reaching posterior extremity of body at distance 1.44–3.53 (2.31, 16), one vitelline field 3.45–6.56 (5.12, 16) long or 36.4–50.4% (42.3%, 16) of body length. Vitelline follicles oriented transversely, usually elongated-oval, wedge-shaped, seldom irregular in shape, 0.033–0.09 (0.058, 16) long, 0.054–0.152 (0.102, 16) wide. Number of follicles 91–146 (120, 10) in the left row, 93–141 (114, 10) in right row. Uterus intercaecal, forming loops in space between uterine seminal receptacle and sinus organ. Eggs numerous, 0.037–0.053 (0.045) long, 0.016–0.025 (0.02) wide. Excretory pore terminal. Excretory vesicle tubular, bifurcating at level of posterior testis; excretory ducts long, extending to posterior margin of oral sucker.
Remarks
The morphologically new species is very similar to Azygia lucii, but it can be distinguished by the following combination of features: (1) body form (in A. lucii the body is narrower); (2) in A. sibirica Vainutis, Voronova et Zhokhov, n. sp. The maximum body width is at the level of the vitellarium, whereas in A. lucii, the maximum body width is at the level of the ovary or anterior testis; (3) pharyngeal lumen in A. sibirica n. sp. is very narrow (0.082 vs. 0.2 at A. lucii); (4) anterior margin of ovary is slightly prominent, whereas in A. lucii anterior margin straight or slightly concave; (5) ovary in A. sibirica n. sp. is not very close to the anterior testis (0.24 vs. 0.094 at A. lucii); (6) and ovary, testes and prostatic sac size distinctly smaller than in A. lucii.
Azygia sibirica n. sp. differs from A. mirabilis by an oral sucker larger than the ventral sucker (in A. mirabilis the suckers equal in size), its pharynx width-to-length ratio: (0.58 vs. 1.08 at A. mirabilis) [34], its tandem testes (in A. mirabilis anterior testes is displaced relative to the median), the smaller size of its vitelline follicles (in A. mirabilis vitelline follicles 0.076–0.109 long, 0.164–0.185 wide) [34] and its smaller size of eggs.
Azygia sibirica n. sp. differs from A. robusta by smaller body and organs sizes (8.6–14.7 vs. 40–80 long at A. robusta), oval pharynx (in A. robusta the pharynx is round).
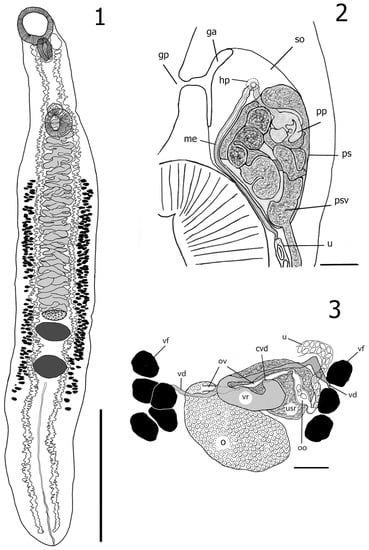
Figure 2.
1. Azygia sibirica Vainutis, Voronova et Zhokhov, n. sp. from the Ob River, adult worm ventral, holotype; 2. Terminal male genitalia, lateral view: prostatic sac (ps), genital atrium not filled with eggs (ga), genital pore (gp), proximal region of seminal vesicle (psv), pars prostatica (pp), metraterm (me), hermaphroditic pore (hp), sinus organ (so); 3. female reproductive system: ovary (o); ootype (oo); oviduct (ov); vitelline reservoir (vr); uterine seminal receptacle (usr); vitelline duct (vd); vitelline follicles (vf); uterus (u); common vitelline duct (cvd). Scale-bars: 1—3 mm, 2, 3—100 μm.
3.3. Multivariate Analysis
The results of the multidimensional analysis of morphological data are shown in Figure 3. Azygia lucii and A. sibirica Vainutis, Voronova et Zhokhov, n. sp. did not intersect with each other in the first three principal component spaces (Figure 3a). The first three principal components of the 23 conceded characters explained 64.35% of the total variance (Table 2). This allowed us to consider the distribution in Figure 3a as representative, reflecting the distribution of individuals in the 23-dimensional space of the initial morphological parameters with sufficient completeness and accuracy. The main difference with a noticeable hiatus occurred along PC3. The three characters with the highest vector loadings in PC3 were: pharyngeal lumen/pharynx width, pharyngeal lumen/BL and forebody width/maximum body width (Table 2). The PCA of these three characters are shown in Figure 3b, where the species did not intersect along PC1. The vector loadings were as follows: pharyngeal lumen/pharynx width = 0.629, pharyngeal lumen/BL = 0.641, and forebody width/maximum body width = 0.440. There was also no intersection of species in Figure 3c, where the PCA was made according to the two characters with the largest vector loadings in PC1. The analysis showed that two characters (pharyngeal lumen/BL and pharyngeal lumen/pharynx width) were sufficient for the preliminary separation of individuals of the two studied species, and the use of the third one (forebody width/maximum body width) allowed us to do this with sufficient confidence. In the Figure 3d, individuals of A. lucii and A. sibirica n. sp. were separated into two different clusters on the dendrogram, corresponding to their species status.
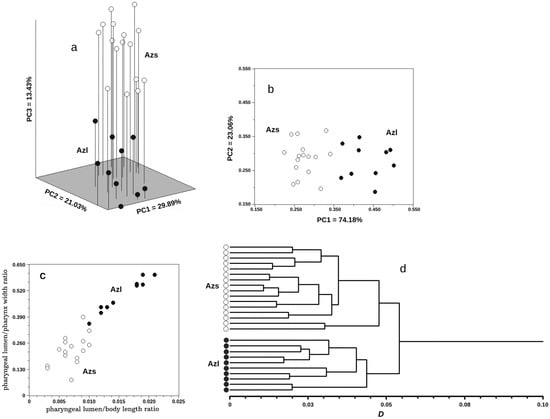
Figure 3.
Results of the multidimensional analysis. The distribution of the individuals in the first three principal components space according to: of studied characters (a) three principal components space of 23 characters; (b) two principal components space of 3 characters; (c) space of 2 characters; (d) Dendrogram of cluster similarity. Abbreviations: D—Manhattan distance; Azl—Azygia lucii; Azs—Azygia sibirica n. sp.

Table 2.
Morphological characters, eigenvectors, and eigenvalues used for performing the multivariate analysis, including species Azygia lucii and A. sibirica n. sp.
3.4. Molecular Genetic Analyses
3.4.1. 28S rRNA Gene
In total, 21 partial sequences of 28S rDNA of three Azygia species had a length of 1708 bp, including the complete 5′-end of the gene. The intraspecific variability of A. lucii from the Volga and Ob Rivers, A. hwangtsiyui and A. sibirica Vainutis, Voronova et Zhokhov, n. sp. was 0% (Table 3). The genetic distances varied from minimum values of 0.091 ± 0.001 (between A. hwangtsiyui and A. sinipercae) and 0.156 ± 0.001% (between A. lucii from the Volga and Ob Rivers), to a maximum interspecific of 4.853 ± 0.006% (between A. longa and A. hwangtsiyui). Generally, intergeneric distances within the family Azygiidae varied from the minimum of 4.84–6.88%, when comparing Azygia spp. And Proterometra spp., to the maximum of 12.99–13.37%, between Proterometra and Otodistomum (Table 3).

Table 3.
Genetic p-distances estimated for eight species of the family Azygiidae based on the 28S gene sequences. Lower diagonal—genetic distances, in %; upper diagonal—standard error.
Both phylogenetic trees based on the ML and BI methods showed identical clusterization. The resulting tree was subdivided into two major clades, A (outgroup) and B (Azygiidae) (Figure 4). Clade A consisted of the families Gonocercidae (four species of Gonocerca and Hemipera manteri) and Derogenidae (Thometrema lotzi). The subdivision of clade B was well statistically supported. This clade was formed of three monophyletic subclades: I—Otodistomum; II—Proterometra; III—Azygia (terminal). Subclade III was successively subdivided into four separated groups. Group 1 comprised only Azygia longa. Group 2 included A. sinipercae from China and A. hwangtsiyui from the Primorsky region. Azygia sibirica n. sp. from the Ob River nested in Group 3. Group 4 was sister to Group 3 and included A. lucii from the Ob River (near Surgut, Priobye, and Khanty-Mansiysk) and from the Volga River.
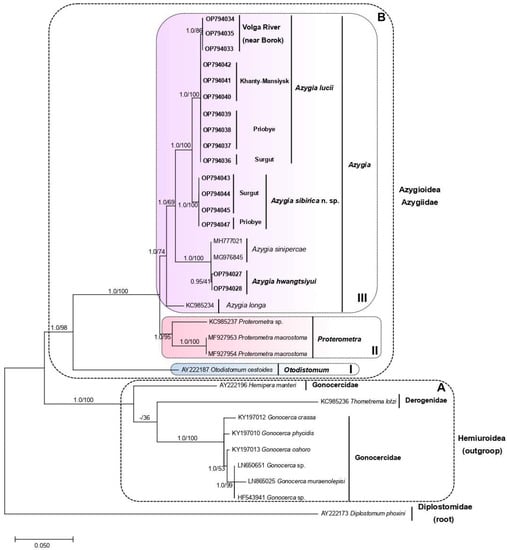
Figure 4.
The consensus phylogenetic tree (BI/ML), reconstructed for eight azygiid species (analyzed group) and six hemiuroid species (outgroup), based on the 28S rRNA gene fragment. The values of the bootstrap support are indicated on the branches. The tree was rooted with Diplostomum phoxini. Symbols on the figure: A, B—clades; I, II, III—subclades.
3.4.2. cox1 mtDNA Gene
In total, 60 partial sequences of the cox1 gene of three Azygia species with different lengths, depending on species (828 bp—Azygia sibirica Vainutis, Voronova et Zhokhov, n. sp.; 828 bp—A. lucii (the Ob River); 852 bp—A. lucii (the Volga River); 858 bp—A. hwangtsiyui), were obtained. The values of the intraspecific variability were observed for A. hwangtsiyui: 0% within the individuals from the Bolotnaya River and Dongting Lake (China) and 0–0.12% within the worms from the Ivnyanka River. The genetic distances between individuals of A. hwangtsiyui were as follows: 0.12–0.24% when comparing “Bolotnaya” and “Ivnyanka”; 1.32% when comparing “Bolotnaya” and “Dongting”; and 1.44–1.56% when comparing “Ivnyanka” and “Dongting”. The intraspecific variability within A. sibirica n. sp. was in the range of 0–0.73%; within A. lucii from the Volga River, it was 0–0.85%; within A. lucii from the Ob River, it was 0–0.48%. The divergence between individuals of A. lucii from the Ob and Volga Rivers overlapped with intraspecific values for both, with a range of 0.12–0.85%.
Interspecific distances between A. sibirica n. sp. and A. lucii were 13.41–14.25%; the interspecific values between A. hwangtsiyui and A. lucii were 20.29–20.89%; the highest interspecific variability was observed between A. hwangtsiyui and A. sibirica n. sp., with a range of 22.34–22.71%.
Auxiliary techniques (ABGD, GMYC, bPTP) for defining interspecific boundaries revealed a different number of groups that could potentially be assigned a species rank (Supplementary Material Figure S1). In ABGD analysis, four groups were found that corresponded to the following clusters: (1) A. lucii, which included worms from the Volga and Ob Rivers; (2) A. sibirica n. sp.; and (3) two groups within A. hwangtsiyui, which included worms from Dongting Lake (China) and the Bolotnaya and Ivnyanka Rivers. GMYC and bPTP analyses identified three potential species: A. lucii, A. sibirica n. sp., and A. hwangtsiyui.
4. Discussion
Azygiids are a poorly studied group of trematodes in Russia, and little known about their molecular epidemiology, population ecology and phylogeny. Based on the integration of morphological and molecular genetic data, we firstly explored the species diversity of Azygia spp. from three Russian rivers; these were related to different geographical regions of the country.
The morphology and morphometry of the studied worms from the Volga and Ob Rivers completely corresponded to those of Azygia lucii, described by different, mostly European, authors [17,35,36,37,38]. Genetic data also confirmed the conspecificity of these worms, which were characterized with low divergence based on the 28S (0.156%) and cox1 (0.12–0.85%) genes. In one study, Pavlov [17] described the subspecies A. lucii johanseni from Rutilus rutilus lacustris in the Ob River, but Skrjabin and Guschanskaja [2] did not accept the validity of this subspecies.
Based on the 28S rRNA gene distances, the differentiation between Azygia sinipercae and A. hwangtsiyui was at the supported intraspecific level of 0.091%. Given this, we suggest synonymizing these species, and according to the principle of priority [39], A. sinipercae should be considered as a junior synonym of a valid A. hwangtsiyui. Chen [40] described A. hwangtsiyui from Channa argus and A. sinipercae from Siniperca chuatsi; both species differed in their size of body (7.96–14.6 mm vs. 5.4–6.1 mm), diameters of oral and ventral suckers (0.63–1.06 mm and 0.58–0.82 mm vs. 0.59–0.63 mm and 0.4–0.58 mm), and the testes of A. hwangtsiyui were larger; however, the size of the pharynx and ovary overlapped in both species. The general topology of the internal organs, and the extension of the uterus and vitellarium, were the same in both species [40]. In addition, according to the original description of Zmejev [10] and the study of Chen [40], Azygia amuriensis from the Russian Far East is synonymous with A. hwangtsiyui. Both species have similar morphological features, described as follows: an elongated body, the same extension of the vitellarium, the posterior border reaching the posterior end of the body and the anterior border being posterior to the ventral sucker. The morphometric values of A. amuriensis overlap with the minimum values of A. hwangtsiyui, or the values are larger than the minimum. Such features reveal the intraspecific variability of A. hwangtsiyui, presumably depending on the definitive host. Therefore, A. hwangtsiyui has two junior synonyms: A. amuriensis and A. sinipercae. The species A. sinipercae and A. amuriensis have morphological correspondence, in terms of their organ shape and topology, with A. hwangtsiyui; however, the latter usually has larger morphometric values. Thus, in addition to distribution in China, A. hwangtsiyui inhabits the rivers of the South of the Russian Far East: the Amur River (Jewish Autonomous Oblast, Khabarovsk region [10], the Arsenyevka [14], the Bolotnaya [15], and the Ivnyanka Rivers (original genetic data) (Primorsky Krai). It is probable that the whole of the Primorsky and Khabarovsk regions are widely inhabited with A. hwangtsiyui, infecting Perccottus glenii, Tilapia zillii, Channa argus, Siniperca chuatsi, and S. scherzeri.
The genetic differentiation of A. sibirica Vainutis, Voronova et Zhokhov, n. sp., in relation to A. lucii, A. longa, and A. hwangtsiyui, corresponds to interspecific features within the genus Azygia (1.25–4.46%) and a hit in the range, according to 28S, known for the most species of trematodes [41,42]. A phylogenetic tree, based on the 28S gene, revealed the sister relationships of A. sibirica n. sp. and A. lucii, supported by genetic distances estimated for the 28S (1.25–1.41%) and cox1 (13.41–14.25%) genes. Finally, according to the results of the multivariate analysis (Figure 3) and three methods of species unit delimitation (Figure S1), both A. lucii and A. sibirica n. sp. were divided into groups, supported well statistically.
It is remarkable that A. sibirica n. sp. and A. lucii, occupying the same niche in a habitat and infecting the same fish hosts (Esox lucius), were able to diverge and maintain species individuality through time; this is notwithstanding the opportunity to migrate between the basins of the Volga and Ob Rivers through an invasion corridor that was created at the beginning of the 20th century [43]. Given the terminal position on the phylogenetic tree (Figure 4), it is highly likely that the divergence path of A. lucii and A. sibirica n. sp. was relatively short compared to other species of Azygia. Moreover, the divergence of these trematodes was not associated with the sub-divergence of Esox lucius. Two independent studies [44,45] have estimated that the differentiation of the northern pike into the three different lineages across the Holarctic took place in the middle Pleistocene, 300–200 Mya. At the same time, pike hosts, from which the samples studied in this work were collected, represented a single panmictic population of Northern pike with a circumpolar lineage; this included fish from the rivers Volga, Ural, Kura, Danube, Anadyr and Huron [44,45].
5. Conclusions
Based on the analysis of the 28S rRNA and cox1 mtDNA gene variability, we established that the genus Azygia included four valid species: A. lucii, A. longa, A. hwangtsiyui, and A. sibirica Vainutis, Voronova et Zhokhov, n. sp. The integrative approach, taking into account morphology, genetic divergence, and host specificity data, helped us to verify the presence of A. lucii in the largest Russian rivers: the Volga and the Ob. The newly described species A. sibirica n. sp. from the Ob River is well morphologically and genetically distinguished from three valid species: A. lucii, A. longa, and A. hwangtsiyui. To conclude, A. lucii infects Esox lucius in the western and northern parts of Russia, A. sibirica n. sp. also infects Esox lucius in the North, while A. hwangtsiyui infects Perccottus glenii in the South of East Asia. The provided reconstructions will simplify the phylogenetic analyses in further studies with the inclusion of more azygiid representatives, especially when evaluating genetic relationships between populations.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d15010119/s1, Figure S1. Model to infer putative species (A. lucii, A. sibirica Vainutis, Voronova et Zhokhov, n. sp., A. hwangtsiyui) boundaries on a given bPTP phylogenetic input tree based on cox1 gene sequences. ABGD/GMYC/bPTP results are shown in color blocks.
Author Contributions
Conceptualization—K.S.V. and A.E.Z.; Methodology—K.S.V., A.N.V., A.N.M., O.N.Z. and A.E.Z.; Formal Analysis—K.S.V., A.N.V. and A.E.Z.; Investigation—K.S.V., A.N.M., O.N.Z. and A.E.Z.; Visualization—K.S.V., A.N.V., A.N.M. and A.E.Z.; Software—A.N.V. and A.N.M.; Resources—O.N.Z. and A.E.Z.; Validation—A.N.V. and A.E.Z.; Writing—Original Draft Preparation—K.S.V. and A.E.Z.; Writing—Review and Editing—A.N.V.; Supervision—A.E.Z.; Project Administration—A.E.Z. All authors have read and agreed to the published version of the manuscript.
Funding
The study was carried out within State Assignments for the Institute of Ecology and Evolution, Russian Academy of Sciences (no. 0109-2018-0076 AAAA-A18-118042490059-5), the Institute for Biology of Inland Waters, Russian Academy of Sciences (no. 121051100100-8), and Federal Scientific Center of the East Asia Terrestrial Biodiversity, Far Eastern Branch of the Russian Academy of Sciences (no. 121031000154-4). This research was funded by the Tyumen Oblast Government, as part of the West-Siberian Interregional Science and Education Center’s project No. 89-DON (2).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Data is contained within this article and its Supplementary Material.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Jadhav, S.; Humbe, A.; Padwal, N. Histopathological changes of the intestine of the freshwater fish, Channa gachua (Hamilton, 1822) due to infection of metacercaria of trematode parasites. Int. J. Fish Aquat. Stud. 2019, 7, 279–281. [Google Scholar]
- Skrjabin, K.I.; Guschanskaja, L.K. Family Azygiidae Odhner, 1911. In Trematodes of Animals and Man. Principles of Trematodology; Skrjabin, K.I., Ed.; Akademiya Nauk Press: Moscow, Russia, 1958; Volume 14, pp. 671–788. (In Russian) [Google Scholar]
- Gibson, D.I. Superfamily Azygioidea Lühe, 1909. In Keys to the Trematoda; Gibson, D.I., Jones, A., Bray, R.A., Eds.; CAB International: Wallingford, England, 2002; Volume 1, pp. 19–24. [Google Scholar] [CrossRef]
- Zhang, S.; Peng, J.; Chen, J.; Xu, X.; Zhi, T.; Brown, C.L.; Yang, T. Two Native Trematodes, Azygia sinipercae and Pseudophyllodistomum anguilae, Colonized in Invasive Redbelly Tilapia Tilapia zillii (Cichlidae) in South China and Their Ecology; Life Science Academy, Sun Yat-Sen University: Guangdong, China, 2018; Unpublished. [Google Scholar]
- Calhoun, D.M.; Curran, S.S.; Pulis, E.E.; Provaznik, J.M.; Franks, J.S. Hirudinella ventricosa (Pallas, 1774) Baird, 1853 represents a species complex based on ribosomal DNA. Syst. Parasitol. 2013, 86, 197–208. [Google Scholar] [CrossRef]
- Van Steenkiste, N.; Locke, S.A.; Castelin, M.; Marcogliese, D.J.; Abbott, C.L. New primers for DNA barcoding of digeneans and cestodes (Platyhelminthes). Mol. Ecol. Resour. 2015, 15, 945–952. [Google Scholar] [CrossRef]
- Moszczynska, A.; Locke, S.A.; McLaughlin, J.D.; Marcogliese, D.J.; Crease, T.J. Development of primers for the mitochondrial cytochrome c oxidase I gene in digenetic trematodes (Platyhelminthes) illustrates the challenge of barcoding parasitic helminths. Mol. Ecol. Resour. 2009, 9 (Suppl. S1), 75–82. [Google Scholar] [CrossRef]
- Wu, Y.A.; Gao, J.W.; Cheng, X.F.; Xie, M.; Yuan, X.P.; Liu, D.; Song, R. Characterization and comparative analysis of the complete mitochondrial genome of Azygia hwangtsiyui Tsin, 1933 (Digenea), the first for a member of the family Azygiidae. ZooKeys 2020, 945, 1–16. [Google Scholar] [CrossRef]
- Linstow, O.F.B. Zwei neue Distomum aus Lucioperca sandra der Wolga. Ezegodnik zool. Mus. imp. Akad. Nauk. 1907, 12, 201–202. (In German) [Google Scholar]
- Zmejev, G.J. Flukes and tapeworms of fish of the Amur River. Parazitol. Sb. Zool. Inst. Akad. Nauk SSSR 1936, 6, 408–435. (In Russian) [Google Scholar]
- Shcherbina, T.V.; Frolova, E.N. Distribution of Azygia mirabilis (Trematoda, Azygiidae). Parazitologiia 1980, 14, 112–117. (In Russian) [Google Scholar]
- Zhokhov, A.E.; Tyutin, A.V. Parasitic fauna of fishes in the conditions of acidification of lakes. Structure and functioning of acid lake ecosystems. Tr. Inst. Biol. Vnutr. Vod Ross. Akad. Nauk. 1994, 70, 186–201. (In Russian) [Google Scholar]
- Ermolenko, A.V.; Besprozvannykh, V.V.; Shedko, S.V. The Fauna of Parasites of Salmonids (Salmonidae, Salmoniformes) of Primorsky Krai; Dalnauka: Vladivostok, Russia, 1998; p. 89. [Google Scholar]
- Besprozvannykh, V.V. Life cycles of the trematode species Azygia hwangtsiytii and A. robusta (Azygiidae) in Primorsky Territory. Parazitologiia 2005, 39, 278–284. (In Russian) [Google Scholar]
- Vainutis, K.S.; Voronova, A.N. Study of helminth fauna in the Bolotnaya River and floodplain lake near the Solovey-Klyuch village (Nadezhdinsky district, Primorsky region). Vestn. Far East Branch Russ. Acad. Sci. 2021, 1, 94–101. (In Russian) [Google Scholar] [CrossRef]
- Müller, O.F. Zoologiae Danicae Prodromus: Seu Animalium Daniae et Norvegiae Indigenarum; Characteres, Nomina, et Synonyma Imprimis Popularium; Typis Hallageriis: Copenhagen, Denmark, 1776; p. 322, (In Latin). [Google Scholar] [CrossRef]
- Pavlov, N.N. The problem of parasitology in fish in the River Ob. Trudy Daltnevostoch. Pod. In-ta 1931, 5, 35–46. (In Russian) [Google Scholar]
- Dorovskikh, G.N. Results of the study of fishes’ parasites in river basins of the north-east of the European part of Russia. Trematodes (Trematoda). Parazitologiya 1997, 31, 551–563. (In Russian) [Google Scholar]
- Molodozhnikova, N.M.; Zhokhov, A.E. The taxonomic diversity of the parasites of agnathans and fishes in the Volga basin. III. Aspidogastrea Trematoda. Parazitol. 2007, 41, 28–54. (In Russian) [Google Scholar]
- Romanova, N.N.; Golovina, N.A.; Vishtorskaya, N.A.; Golovin, P.P. Fauna of parasite of cyprinids and percoids in the reservoirs of the Moscow Canal. Russ. J. Parasitol. 2021, 15, 32–47. [Google Scholar] [CrossRef]
- Strelkov, Y.A. Trematodes of fish from Amur River basin. Parasitol. Sb. ZIN AN SSSR 1971, 25, 41–76. (In Russian) [Google Scholar]
- Truett, G.E. Preparation of genomic DNA from animal tissues. In The DNA book: Protocols and Procedures for the Modern Molecular Biology Laboratory; Kieleczawa, J., Ed.; Jones and Bartlett Publisher: Sudbury, ON, Canada, 2006; pp. 33–46. [Google Scholar]
- Lockyer, A.E.; Olson, P.D.; Littlewood, D.T.J. Utility of complete large and small subunit rRNA genes in resolving the phylogeny of the Neodermata (Platyhelminthes): Implications and a review of the cercomer theory. Biol. J. Linn. Soc. 2003, 78, 155–171. [Google Scholar] [CrossRef]
- Bowles, J.; Blair, D.; McManus, D.P. A molecular phylogeny of the human schistosomes. Mol. Phylog. Evol. 1995, 4, 103–109. [Google Scholar] [CrossRef]
- Miura, O.; Kuris, A.M.; Torchin, M.E.; Hechinger, R.F.; Dunham, E.J.; Chiba, S. Molecular-genetic analyses reveal cryptic species of trematodes in the intertidal gastropod, Batillaria cumingi (Crosse). Int. J. Parasitol. 2005, 35, 793–801. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F.; Nielsen, R.; Bollback, J.P. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 2001, 294, 2310–2314. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef]
- Fujisawa, T.; Barraclough, T.G. Delimiting species using single-locus data and the generalized mixed yule coalescent approach: A revised method and evaluation on simulated data sets. Syst. Biol. 2013, 62, 707–724. [Google Scholar] [CrossRef]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A General Species Delimitation Method with Applications to Phylogenetic Placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Frolova, E.N.; Shcherbina, T.V. New species in genus Azygia Looss, 1899 (Trematoda, Azygiidae). Parazitologiia 1975, 9, 489–493. (In Russian) [Google Scholar] [PubMed]
- Looss, A. Die Distomen unserer Fische und Frösche. Neue Untersuchungen über Bau und Entwickelung des Distomenkörpers. Bibl. Zool. Stuttg. 1894, 16, 296. (In German) [Google Scholar]
- Markevich, A.P. Parasite Fauna of Freshwater Fishes of the Ukrainian SSR; Publishing House of the Academy of Sciences of the Ukrainian SSR: Kyiv, Ukraine, 1951; p. 375. (In Russian) [Google Scholar]
- Stunkard, H.W. The Morphology and Life-History of the Digenetic Trematode, Azygia sebago Ward, 1910. Biol. Bull. 1956, 111, 248–268. [Google Scholar] [CrossRef]
- Odening, K. Cercaria splendens Szidat, the larva of the trematode Azygia lucii Looss. Parazitologiia 1975, 9, 232–236. (In Russian) [Google Scholar]
- International Commission on Zoological Nomenclature. Article 23. Principle of Priority. In International Code of Zoological Nomenclature., 4th ed.; Adopted by the International Union of Biological Sciences; Publishing House KMK: Moscow, Russia, 2004; p. 64. (In Russian) [Google Scholar]
- Chen, C.L. (Ed.) An Illustrated Guide to the Fish Diseases and Causative Pathogenic Fauna and Flora in the Hubei Province; Science Press: Beijing, China, 1973. (In Chinese) [Google Scholar]
- Vainutis, K.S.; Voronova, A.N.; Urabe, M. Systematics of Crepidostomum species from the Russian Far East and northern Japan, with description of a new species and validation of the genus Stephanophiala. Parasitol. Int. 2021, 84, 102412. [Google Scholar] [CrossRef]
- Vainutis, K.S.; Voronova, A.N.; Duscher, G.G.; Shchelkanov, E.M.; Shchelkanov, M.Y. Origins, phylogenetic relationships and host-parasite interactions of Troglotrematoidea since the cretaceous. Infect. Genet. Evol. 2022, 101, 105274. [Google Scholar] [CrossRef] [PubMed]
- Korlyakov, K.A.; Nohrin, D.Y. Tendencies of occurrence invasion of the corridor Volga-Ob. Vestn. SMUS Chelyabinskoy Obl. 2014, 2, 19–38. (In Russian) [Google Scholar]
- Skog, A.; Vøllestad, L.A.; Stenseth, N.C.; Kasumyan, A.; Jakobsen, K.S. Circumpolar phylogeography of the northern pike (Esox lucius) and its relationship to the Amur pike (E. reichertii). Front. Zool. 2014, 11, 67. [Google Scholar] [CrossRef]
- Takács, P.; Bánó, B.; Czeglédi, I.; Erős, T.; Ferincz, Á.; Gál, B.; Bánó-Kern, B.; Kovács, B.; Nagy, A.A.; Nyeste, K.; et al. The mixed phylogenetic origin of northern pike (Esox lucius Linnaeus 1758) populations in the Middle Danubian drainage. BMC Zool. 2022, 7, 28. [Google Scholar] [CrossRef]
- Blair, P.; Franklin, H.; Kelner, N.; Ke-Lind, P.; Paulmier, M.; Shrestha, M. Targeted rDNA sequence determination from geographically isolated populations of Proterometra macrostoma (Trematoda: Azygiidae). Proc. Indian Acad. Sci. 2018, 127, 82–88. [Google Scholar]
- Olson, P.D.; Cribb, T.H.; Tkach, V.V.; Bray, R.A.; Littlewood, D.T.J. Phylogeny and classification of the Digenea (Platyhelminthes: Trematoda). Int. J. Parasitol. 2003, 33, 733–755. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).