Abstract
The narrow-headed vole complex includes two cryptic species, Lasiopodomys raddei and L. gregalis, and three allopatrically-distributed lineages with obscure taxonomic ranks within the latter. Based on the RNA-seq data of 12 specimens, the current study aims to find the molecular mechanisms of intraspecies differentiation and, in particular, reproductive isolation between analyzed groups. According to the results of the GO-enrichment analysis, about a hundred biological processes associated with genes with contrasting SNPs for L. gregalis and L. raddei were identified. Among them, processes of interspecific interactions, defense responses, responses to external stimuli, and the perception of chemical stimuli and smell were identified, indicating the likely existence of pre-copulatory behavioral and physiological mechanisms that contribute to reproductive isolation between cryptic species. An evaluation of the ratio of non-synonymous substitutions to synonymous ones showed evidence of selection in L. raddei compared to L. gregalis for a large part of the analyzed genes. Among the analyzed genes, genes with both weakening and intensifying selection were found.
1. Introduction
Narrow-headed vole Lasiopodomys (Stenocranius) gregalis Pallas, 1779 is a wide-ranged Palearctic species inhabiting various open landscapes in tundra, steppe, and alpine zones. Many studies considered morphological differentiation among populations of this species [1,2,3], and more than a dozen subspecies have been described [4,5,6], but, before the use of molecular markers, the integrity of the species was not questioned.
Cytochrome b analysis identified four major mitochondrial (mt) lineages of the narrow-headed vole (named as A, B, C, and D) with genetic distances between 6.9–11.4% [7], with lineage D from Southeastern Transbaikalia being the most distant. However, a thorough study of the craniometric characters of the narrow-headed voles from the Transbaikal Region [8] revealed no differences between representatives of lineages D (Southeastern Transbaikalia) and B (neighboring populations of Northwestern Transbaikalia and Eastern Mongolia), but the following comprehensive study based on the multilocus analysis of six nuclear markers, morphological analysis of first lower (m1) and third upper (M3) molar dental traits, and breeding experiments [9] showed that the populations from Southeastern Transbaikalia (lineage D) represent a cryptic species within the narrow-headed vole complex, Lasiopodomys raddei Poljakov, 1881.
Although only a few hybridization experiments have been performed, no offspring were obtained in any variant when crossing L. raddei with representatives of lineage A (the nominative subspecies of L. gregalis) and lineage C. In various reciprocal combinations of three mt lineages (A, B, and C) of L. gregalis sensu stricto and between the lineage B of L. gregalis and L. raddei, offspring were obtained, but in several combinations compared with the control groups, the fertility and survival of the cubs were decreased [9,10].
According to the results of a microsatellite loci analysis, three mt lineages A, B, and C of L. gregalis s. str. were well isolated from each other genetically (gene flow between them was strongly limited and traces of hybridisation were found in only the contact zone of lineages B and C). A species distribution modeling analysis predicted the presence of barriers between the lineages that correspond to landscape heterogeneity and only a small number of potential contact zones [11]. These results suggested that these three genetic lineages of L. gregalis s. str. may be recognised as a separate taxa, probably of the lowest taxonomic rank.
Speciation mainly occurs due to the accumulation of mechanisms of prezygotic and postzygotic isolation between genetically divergent lineages [12]. Distinct populations may be subject to various selective forces, and these differences may contribute to ecological and adaptive divergence and, possibly, further reproductive isolation between populations [13,14]. The genetic mechanisms that underpin the initial stages of species differentiation and trigger reproductive isolation and subsequent speciation remain obscure despite being the focus of many studies [15,16,17,18,19,20]. The development of next-generation sequencing (NGS) tools provides an opportunity to search for traces of these processes at the genomic level. Genome-wide association studies (GWAS) can highlight the mechanisms of interspecific differentiation, showing which biological processes involve multiple genes with contrasting species-specific SNPs [21,22,23].
The application of landscape genomics approaches makes it possible to infer the local adaptation of populations under varying environmental and selective pressures [24,25,26,27,28]. At the molecular level, traces of the selective pressure can be detected by comparing the number of non-synonymous substitutions to synonymous ones [29,30].
In the current study, we analyzed transcriptomic data to seek out signatures of speciation with particular attention to one that may facilitate reproductive isolation among the narrow-headed vole species complex including L. raddei and three lineages of L. gregalis. We searched for species-specific SNPs with special attention for genes that are potentially involved in lineage differentiation. We also looked for genes with traces of selection changes in each of the cryptic species.
2. Materials and Methods
2.1. Sampling
We used 12 individuals for the transcriptome analysis—three specimens of L. raddei and nine of L. gregalis (three per each genetic lineage). The representatives of L. raddei were collected from the laboratory colony maintained at St. Petersburg State University, which were bred from animals caught in two localities, Adon-Chelon and Nizhny Tsasuchey (Borzinsky District, Transbaikal Region, Russia). The representatives from L. gregalis lineage A were taken from the laboratory colony maintained in the Severtsov Institute of Ecology and Evolution RAS, which were bred from animals caught in the vicinity of Novosibirsk (Novosibirsk Region, Russia). Animals from lineages B and C were caught in the wild from two localities, Kuduktug-Khem River and Tapsa River in the Tuva Republic, Russia—see Figure 1 and Table S1 for details. Both males and females were analyzed for each lineage. Tissue mix (muscles, liver, heart, lungs, and testes for males) was fixed in an intactRNA buffer (Evrogen, Russia) to avoid RNA degradation. The belonging of animals to certain lineages was checked using mt cytochrome b genotyping. The study was conducted according to the guidelines of the Declaration of Helsinki and was approved by the Ethics Committee of the Zoological Institute of the Russian Academy of Sciences (permission #1-17, 7 April 2022).
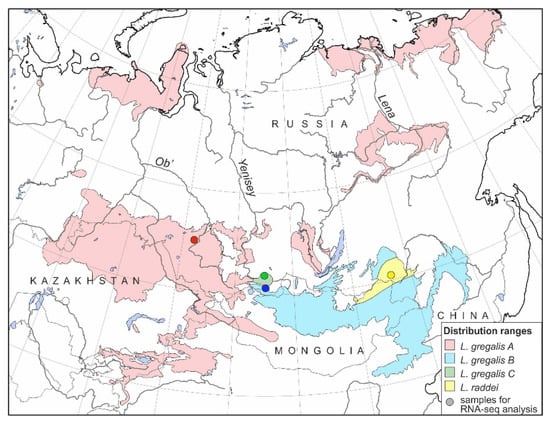
Figure 1.
Distribution ranges of L. raddei and three lineages of L. gregalis (according to Batsaikhan et al. [31] with changes). Localities where animals for RNA-seq were sampled are marked with colored circles.
2.2. RNA Isolation, Library Preparation, and Sequencing
The total RNA was isolated from the tissue mix with an RNeasy mini kit (Qiagen) using the animal cells/spin protocol. RNA quality was quantified using a Bioanalyzer2100 (Agilent Genomics) with a minimum of 7.0 for the RNA Integrity Number score. A combined protocol of the NEBNext Poly(A) mRNA Magnetic Isolation Module and the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina was used to isolate intact poly(A)+ RNA from the total RNA fraction and further prepare the DNA libraries. Sample concentrations were measured using a Qubit fluorometer. Libraries were sequenced to generate paired-end reads with an average length of 75 bp on the Illumina HiSeq 4000 sequencing platform. RNA isolation, library preparation, and sequencing were performed using Skoltech Genomics Core Facility resources (https://www.skoltech.ru/research/en/shared-resources/gcf-2/, accessed on 3 June 2022).
Raw sequence data are available at the NCBI Sequence Read Archive: SRR12765436–SRR12765441, SRR17971092–SRR17971097 (BioProject № PRJNA591473).
2.3. De Novo Transcriptome Assembly
The sequence quality of each RNA-seq sample was assessed by FastQC [32]. Low-quality bases were trimmed in Trimmomatic [33], Illumina adapters were cut with a fastp tool [34], and reads with contamination were filtered with Bowtie 2.3.5.1 [35]. We have assembled the transcriptome de novo in several variants: (1) 12 individual transcriptomes; (2) 4 united transcriptomes per species or lineage (combining reads from 3 specimens) for the dN/dS calculation (see Section 2.6), and (3) a hybrid assembly of the subgenus, Stenocranius, with all 12 libraries from both L. raddei and L. gregalis for subsequent use as a reference for the SNP calling (see Section 2.4). The transcriptomes were assembled using Trinity [36] with default settings.
Assembly quality was assessed in BUSCO [37]. ORFs in the assembled transcriptomes were predicted by Transdecoder (https://github.com/TransDecoder/TransDecoder/, accessed on 3 June 2022). The hybrid assembly was aligned against the nr NCBI database (downloaded March 2020) by DIAMOND [38]. The contigs that matched (the only one match with the best score was chosen) to any Mammalian gene with E-value < 10 were set as the reference for the SNP calling. Orthologous genes were detected with Proteinortho [39].
2.4. SNP Calling and Search of Associations
Pair-end trimmed filtered reads from 12 individuals were separately aligned against the reference transcriptome by the bwa-mem algorithm [40] with default options. The resulting bam-files were sorted and filtered in Picard (http://broadinstitute.github.io/picard/, accessed on 3 June 2022). Low-quality SNPs with a read depth below 3 or a genotype quality below 20 were removed from the final vcf-file. Only contrasting SNPs (those where, at a certain position, all specimens from the first group have one nucleotide in homozygous state and all specimens of the second group have an alternative nucleotide in homozygous state) were left. Therefore, the intralineage variation was left behind the scope of this study. The search for associations was performed in the GATK HaplotypeCaller [41]. The SNPs were annotated using eggNOG mapper [42]. GO terms annotation of contigs having associations (p-value < 0.05) was performed using the Gene Ontology Resource in PANTHER software [43]. Correction for multiple testing was carried out by the Bonferroni and FDR methods.
2.5. Phylogenetic Reconstruction
To further assess the selective pressure, we constructed a phylogeny of the narrow-headed vole species complex using single-copy orthologs that were found in 12 individual transcriptomes of L. raddei and L. gregalis. Individual ortholog files were generated from assemblies using a Python script followed by alignment using MAFFT 7.222 [44]. Multiple alignments were edited (gene-like sequences were cropped according to the position of start- and stop-codons) using a Python script (https://github.com/mkviatkovskii/bioutils, accessed on 3 June 2022).
Phylogenetic reconstruction based on the concatenated alignment of 1340 single-copy orthologs with a total length of 1039,800 bp was performed with a Maximum Likelihood (ML) analysis using the IQ-TREE web server [45] with 10,000 ultrafast bootstrap replicates [46].
2.6. Natural Selection Estimation
Since we sought to identify genes that may be involved in species differentiation, we estimated the ratio (⍵) of non-synonymous (dN) and synonymous (dS) substitutions in separate genes. We used both pooled transcriptomes for each lineage, because such assemblies are more complete, and individual transcriptomes, which provide us with more accurate results. Selective pressures were estimated using the classical codeml program [47,48] with the ete-toolkit interface [49]. The branch model assumes significance of the tree (background). The branch model-based approach allows for the estimation of the selection level for separate species compared with phylogenetically close taxa. We marked L. gregalis as the foreground branches and L. raddei as the background branches (Figure 2).
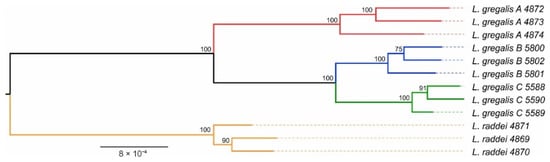
Figure 2.
Phylogenetic reconstruction of the narrow-headed vole species complex. Node labels display ML rapid bootstrap support. The names of the specimens contain tissue numbers, see Table S1 for details.
Also, for each species (and lineages of L. gregalis), an analysis was implemented with a free-branch model, where the foreground and background branches evolve with free rates, and an M0 model, where all the branches evolve at the same rate. A likelihood ratio test was calculated to compare the models; this shows whether the foreground branches differ significantly from the rest of the tree. Values using Bonferroni and Holm methods in R software ver. 3.0.2 (R Core Team, Vienna, Austria) of 999 and 0001 were considered errors. Multiple testing was corrected [50].
RELAX software [51] was employed to test for changes in the selection intensity (relaxation or intensification) in phylogenetic branches. A significant estimate of K > 1 indicates the intensification of selection strength along the test branches, while a significant K < 1 indicates its relaxation. We marked L. gregalis as the foreground branches and L. raddei as the background branches (Figure 2). Multiple testing was corrected using Bonferroni and Holm methods in R software ver. 3.0.2 (R Core Team, Vienna, Austria) [50].
3. Results
3.1. Transcriptome Assembly
Between 34750–165598 contigs per species/genetic lineage were generated, and for individual transcriptomes, the number of contigs ranged from 35,774–98,867 ones (Table S2). The N50 length consisted of 2542–3571 bp for pooled assemblies and 1392–2835 bp for individual ones, whereas the mean contig length was about 1340 and 990 bp, respectively.
The software BUSCO identified between 34.5–69.7% of the complete orthologs out of the 9226 mammalian genes for individual assemblies. For the pooled assemblies, the number of complete orthologs varied from 5271 (57.1%) in L. gregalis lineage A to 7154 (77.6%) in L. gregalis lineage B, and reached 8290 (89.9%) in the hybrid Stenocranius assembly (Table S2).
3.2. Phylogenetic Reconstruction
Phylogeny based on the concatenated alignment of 1340 single-copy orthologs with a total length of 1039800 bp was identical to those constructed on the mitochondrial cytochrome b sequences [7] and on a small set of nuclear genes [9]—lineage A L. gregalis being sister to a pair of B and C lineages (Figure 2).
3.3. Variation Calling
We looked for contrasting SNPs between cryptic species and compared each of the L. gregalis lineages with L. raddei. For combinations of L. raddei and lineages A, B, and C of L. gregalis, 9370, 14069, and 13481 SNPs were detected, respectively. In addition, we compared the combined L. gregalis data with L. raddei, which resulted in 5018 contrasting SNPs.
As variation calling is a more sensitive method than assessing selective pressure, we tried to identify contrasting SNPs within L. gregalis. Comparing three L. gregalis lineages in pairs with each other, we found 5029 SNPs for lineages A and B, 5184 SNPs for lineages A and C, and 3372 SNPs for lineages B and C. According to the results of eggNOG-mapper, 122–159 genes with contrasting SNPs between the three lineages of L. gregalis and L. raddei and 74–101 genes within L. gregalis were revealed (Figure 3).
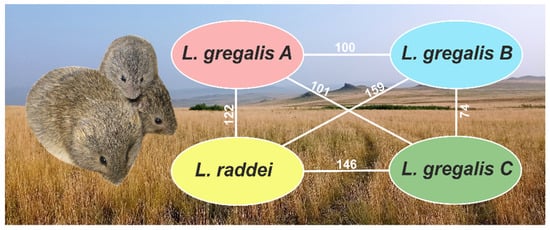
Figure 3.
The number of genes with contrasting SNPs between the compared groups.
Comparing L. raddei with the three lineages of L. gregalis (separately), GO annotation revealed an enrichment in about one hundred terms; including a large number of general regulatory processes, over one-third of the terms were related to various immune system processes (Table S3). Some biological functions potentially underlying species differentiation were also identified, such as the biological processes involved in interspecies interaction between organisms (GO:0044419), defense responses (GO:000695, GO:0098542) and responses to stress (GO:0006950), and responses to different stimuli (GO:0048583, GO:0048584, GO:0050896, GO:0009605, GO:0009607, GO:0043207, GO:0051707), including the sensory perception of chemical stimuli and smell (GO:0007600, GO:0007606, GO:0007608). For genes involved in enriched biological processes listed above, see Table 1. Despite that, we did not observe the enrichment of the categories associated with reproduction, and genes involved in various reproductive processes were found among the genes with SNPs: AATF, CITED1, CNOT6L, EXOC6, FNBP1L, HERC2, KMT2D, NFIX, RPS6KA4, PTK7, STK3, and TAF9B.

Table 1.
The list of genes that could potentially be related to speciation processes found when comparing L. gregalis and L. raddei.
In addition, we compared all three lineages of L. gregalis with each other. In the case of lineages A and B, we did not reveal any enriched GO terms with statistical support, but a search among the GO categories detected terms related to reproductive isolation: male meiosis cytokinesis (SHCBP1L), negative regulation of meiotic nuclear division (RPS6KA2), oocyte growth и oocyte differentiation (KMT2D), regulation of cell differentiation involved in embryonic placenta development (STK3), development of secondary female sexual characteristics (IRF2BPL), and a list of genes associated with cellular responses to chemical stimuli and sensory perception (KMT2D, PDGFC, MYLK, RPS6KA2, SKI, POLR2C, CITED1, PTK7, RPS6KA4, CACNA2D3, MAOA, EHD1, EIF4A3, NRP2, MAP3K1, CES1, MYH7, CD40, TJP2, DAB2IP, POU6F1, and TTC8).
In the case of lineages A and C, there were genes associated with the regulation of the reproductive process (RPS6KA2, FNBP1L, and NFIX), including its positive (FNBP1L) and negative regulation (RPS6KA2); and various developmental processes involved in sexual reproduction (AATF, KMT2D, FNBP1L, CNOT6L, RPS6KA2, EXOC6, CITED1, PTK7, RPS6KA4, TAF9B, and HERC2), including spermatogenesis (HERC2), oocyte development, growth and maturation (KMT2D, RPS6KA2), and the sensory perception of smell (SLC24A3, TTC8), which were identified.
Bonferroni correction revealed only unclassified terms and cellular component biogenesis (GO:0044085) for lineages B and C. However, some genes involved in the reproduction associated processes were identified, such as KMT2D and NUP155, which were involved in oocyte growth and differentiation; PCK1, MAU2, and NLGN4X, which were associated with reproductive behavior; TTC8, which was associated with the sensory perception of smell; as well as a list of others (AATF, FNBP1L, EXOC6, NFIX, SLC24A3, TAF9B, MAOA, NRP2, and POU6F1) that were also associated with reproductive and sensory processes. Of course, it is worth bearing in mind that such results (when special GO terms are not enriched) may be random and result in false positives, especially considering the extremely small amount of material analyzed.
3.4. Signatures of Natural Selection
The complete set of single copy orthologs for the four pooled assemblies was 3786 (and 1340 for the 12 individual assemblies), but the majority of them had omega 0001 and 999 values because of the conservatism of most of the genes analyzed; these genes were removed as erroneous. Thus, 814 genes for pooled assemblies and 234 genes for individual assemblies remained in the two final sets of the genes included in the analysis of the dN/dS ratio estimation. Bonferroni correction for multiple comparisons (taking 814 and 234 genes used as the number of comparisons) showed that all corrected p-values exceeded 0.05.
The difference between M0 and b_free models by LRT (p-value < 0.05) before the multiple testing correction was shown for only 9 genes when using pooled assemblies and for 14 genes when using individual assemblies (Table S4; Figure 4). None of these were corrected for multiple comparisons, thus this result may be a false positive; nevertheless, we considered their functions. In both versions of the analysis, we observed a general trend in the selection level bias towards L. raddei. All the genes were identified, both in the pooled assemblies analysis (ATP2A3, DSG2, EFL1, FBX034, GALNT15, GNL3L, MANBA, MRPS31, and XDH) and in the individual assemblies analysis (AARS, CPT2, DHRS4, FH, GFM1, MTMR6, PITRM1, SAC3D1, TCEA3, TOM1, TRAPPC12, VPS11, VWA8, and ZNF768), which refer to the general regulatory and metabolic processes.
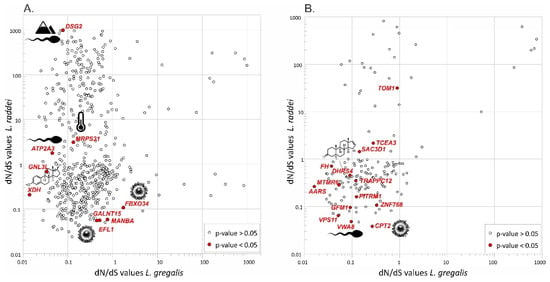
Figure 4.
The dN/dS ratio values for individual genes between L. gregalis and L. raddei. (A) Comparison of four assemblies (3 specimens per lineage). (B) Comparison of 12 individual assemblies. Genes with p-value less than 0.05 (before the Bonferroni correction) are highlighted in red. Genes potentially involved in various reproductive processes and associated with adaptations to environmental conditions are marked with corresponding icons.
A RELAX analysis showed no significant p-values after the Bonferroni and Holm corrections (Table S5) for both variants of assemblies. A p-value < 0.05 before the multiple testing correction was shown for 53 genes (for pooled assemblies) and for 29 genes (for individual assemblies). The number of genes under the intensification of selection (K > 1) tends to be slightly higher than the number of genes under its relaxation (K < 1) (Figure 5). All revealed genes were involved in the general regulatory and metabolic processes.
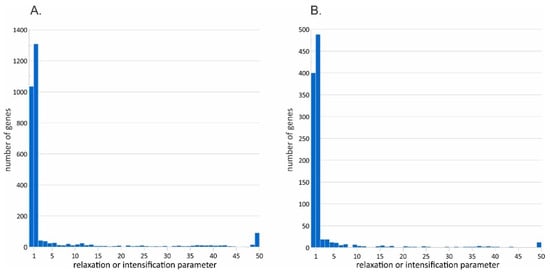
Figure 5.
Relaxation or intensification parameter distribution estimated in the RELAX software. (A) Comparison of four pooled assemblies (3 specimens per lineage). (B) Comparison of 12 individual assemblies.
4. Discussion
4.1. Molecular Mechanisms Underlying Speciation between Cryptic Species L. gregalis and L. raddei
The results of experimental breeding [9,10] showed no posterity when crossing L. raddei and representatives of the A and C L. gregalis lineages. Thus, we aimed to check whether any genes involved in reproductive processes have any signatures of selection. The dN/dS estimation revealed 23 genes with traces of selective pressure changes (Figure 4) for both L. gregalis and L. raddei. However, none of them passed the correction for multiple comparisons. Since the divergence time of L. gregalis and L. raddei is estimated to be approximately 800 thousand years [7], it is possible that there was not enough time for the accumulation of significant selection differences, especially considering the rate of evolution of nuclear genomes [52].
According to the UniProt database, all the genes identified with codeml (which passed the initial test with a p-value < 0.05) are involved in general regulatory and metabolic processes, but seven were also found to be associated with various reproductive processes. According to our data, three genes associated with reproductive processes were under positive selection (with dN/dS > 1): two in L. raddei—DSG2, which is associated with spermatogonial differentiation [53], and ATP2A3, which takes part in the elaborate courtship evolution in Manakin birds, Passeriformes [54] and is also possibly associated with oxidative defense and the semen quality factor in cocks [55]; and one in L. gregalis—FBXO34, which plays a critical role in oocyte meiosis [56]. Two genes under positive selection in L. raddei were associated with different kinds of environmental adaptations—MRPS31, which is probably related to the ability to adapt to temperature gradients [57], and DSG2, which may be helpful for high-altitude adaptation [58] (Figure 4).
Four genes were under weakening of negative selection (dN/dS < 1, but biased between species): two of them in L. raddei—GNL3L, which inhibits estrogen-related receptor activity [59], and DHRS4, which is involved in altering hormone-associated reproductive traits [60,61,62,63,64]; and two in L. gregalis—EFL1, which is associated with oogenesis and early embryogenesis [65], and CPT2, which is involved in oocyte maturation [66,67] and embryo developmental competence [68]. Interestingly, all three genes under selective pressure distinguished for L. gregalis are related to various aspects of oogenesis.
Mostly, the dN/dS values were below 1, indicating the presence of stabilizing selection in both cryptic species (Figure 4). L. raddei is probably characterized by stronger selective pressure than L. gregalis, since the dN/dS values were mostly higher in L. raddei, thereby analyzing both the pooled lineage and individual transcriptome assemblies. We understand the limitations imposed by a small sample size and its geographical representation; however, we hypothesize that this asymmetry may be caused by the fact that L. raddei is the most ancient species within the group and, in addition, is characterized by a very narrow distribution range and extremely low genetic variability [7].
The RELAX analysis revealed a large number of genes with a p-value < 0.05 before the multiple testing correction (Table S5). It is noteworthy that among these genes, there were DSG2 and VWA8 genes that passed the first confidence threshold during the codeml analysis, and EXOC6 was revealed as a result of the SNP calling. MRPS9 was revealed in both variants of the RELAX analysis.
In both variants of the assemblies, a large number of genes associated with reproductive processes were identified, demonstrating the intensification of selection. ABCD3, EHBP1L1, IDE, IP6K3, NSUN2, and MSH2 were potentially involved in spermatogenesis [69,70,71,72,73,74,75]; ASMT was involved in oocyte maturation [76,77]; and GGH was associated with the biosynthesis of folic acid regulation [78].
The relaxation of selection was demonstrated with CST3, DNAJB4, DSG2, MRTO4, MFSD14A, NOSTRIN, PSPC1, and VWA8, which were potentially involved in various aspects of spermatogenesis [53,79,80,81,82,83,84,85]; ITGB5, which played a role in male fertility [86,87,88]; IPO13, which was involved in meiosis of germ cells [89]; and GTPBP1, MRPS5, and PSPC1, which were involved in oogenesis [90,91,92]. CST3 regulated proteolysis in the male reproductive system [93], NOSTRIN was involved in placenta development [94], CA4 and ITM2B were differentially expressed in female and male reproductive tissues [95,96,97,98].
The list of genes with contrasting SNPs associated with different reproductive processes included CITED1 (participates in the development of the placenta, as well as in the growth and survival of embryos [99]), CNOT6L (involved in the meiotic cell cycle in mouse oocytes [100]), FNBP1L (associated with male fertility [101]), HERC2 (mutations in this gene lead to sperm acrosome defects, other developmental abnormalities, and juvenile lethality of mice [102]), KMT2D (expressing in male germ cells and regulating spermatogonial differentiation [103]), NFIX (involved in the formation of the synaptonemal complex during male meiosis [104]), PTK7 (involved in the sexually dimorphic development of the genital tract [105]), STK3 (involved in the regulation of dynamic uterine epithelium during the estrous cycle [106]), and TAF9B (involved in mice embryonic germ cell development [107]).
According to the results of the GO-enrichment analysis of genes with contrasting SNPs, a huge number of various immune system processes were revealed that could be caused by the fact that immune genes are characterized by a higher evolutionary rate in various animal groups [108,109,110]. Albeit no biological processes associated with reproduction were detected with GO-enrichment analysis, the list of enriched biological processes included such categories as the sensory perception of chemical stimuli, perception of smell, interspecies interaction between organisms, defense responses, and responses to stress (Table S3). Thus, we may suggest that reproductive isolation between cryptic species L. gregalis and L. raddei is formed not only by the occurrence of any violations of the reproductive processes itself, but also at the behavioral level. Our results are consistent with the Species Recognition Concept [111], focusing on a specific mate recognition system (SMRS), which is responsible for finding a suitable mating partner using prezygotic mechanisms such as gamete design features, co-adapted signals and receivers of mating partners (in particular, chemical communication), and their co-adapted gamete delivery and reception organs.
Interestingly, the maximum number of genes with SNPs (Figure 3), which can be considered a simplified measure of the genetic distance between groups, was found for a pair of parapatric L. gregalis lineage B and L. raddei (Figure 1). Although the samples we analyzed were not directly from the zone of parapatry, a similar pattern was observed when analyzing CYTB differentiation using a wide geographical sample [7], with the maximum p-distances between L. raddei and adjacent populations of L. gregalis lineage B from Northwestern Transbaikalia and Eastern Mongolia compared with more distant subclades of L. gregalis [112], which was consistent with the theory of Reinforcement (the so-called Wallace effect), thereby implying the emerging selection against hybrids in groups formed in allopatry during the further formation of the secondary contact zone [113]. A subsequent study of the structural changes in the identified genes is necessary to determine the specific mechanisms of isolation.
4.2. Traces of Molecular Differentiation at the Early Stages of Speciation within L. gregalis
The results of experimental breeding showed the presence of posterity in all combinations of A, B, and C L. gregalis lineages; however, it was shown that not every reciprocal combination had posterity that was numerous and viable [9,10]. Using a microsatellite loci analysis [11] we found traces of hybridisation in the two most phylogenetically close lineages, B and C, which, according to our estimates [7], separated about 250 thousand years ago. Despite the absence of continuous geographical barriers in nature between the lineage distribution ranges, we did not find any traces of hybridization from lineage A with lineages B and C, separation from which probably occurred about 400 thousand years ago. The number of detected SNPs, as expected, correlates with the age of separation of the lineages—so for comparisons of lineages A and B and A and C, it is about 5000, and for sister lineages B and C, it is a half as less.
Within L. gregalis, EggNOG-mapper revealed the list of genes involved in processes of reproduction (including male meiosis cytokinesis, spermatogenesis, negative regulation of meiotic nuclear division, oocyte development, growth and maturation, regulation of cell differentiation involved in embryonic placenta development, development of secondary female sexual characteristics, and male courtship behavior) and those associated with responses to chemical stimuli and the sensory perception of smell. No enriched GO categories were found for the reproduction processes, which can be explained by the low number of genes with contrasting SNPs. The genes that were revealed were mostly the same as those found when comparing L. gregalis and L. raddei.
It is assumed that the degree of development of reproductive isolation mechanisms is related to the time of taxa divergence. A study of speciation on a phylogenetically close group—hamsters of the subfamily Cricetinae [114]—showed that the sterility of F1 males and conspecific preferences were formed in Phodopus species (separated about 0.8–1.0 mya), wherein postzygotic isolation mechanisms and specific preferences have been formed partially, although fertile F1 and F2 hybrids were obtained in the laboratory in Allocricetulus species (diverged 0.3–0.4 mya), while the reproductive barriers were less pronounced between groups of the superspecies Cricetullus barabensis sensu lato (which diverged only 160–200 kya). This relationship of the degree of reproductive isolation on the age of the group is very similar to the pattern that we observe in the subgenus Stenocranius. In the previous experiments, the narrow-headed voles mated, but they were not given the choice of a conspecific partner. Further work on experimental hybridization is needed by taking into account the possible existence of prezygotic isolation mechanisms.
5. Conclusions
Based on the RNA-seq data of the narrow-headed vole species complex, we revealed the list of biological processes associated with various pre-copulatory behavioral and physiological mechanisms that may contribute to the emergence of reproductive isolation between cryptic species. The analysis of the selective pressure at individual orthologs has also detected the list of genes that may be potentially involved in reproductive processes and ecological adaptations. Although at this stage it is premature to draw conclusions on the molecular mechanisms of reproductive isolation between L. raddei and L. gregalis (and especially between lineages with obscure taxonomic rank within the latter), based on transcriptomic data, we can outline a direction for further molecular and physiological research.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d14070512/s1, Table S1: Material used in the study, Table S2: Quality of de-novo assembled transcriptomes, Table S3: Genes potentially related to speciation processes detected by comparison L. gregalis and L. raddei, Table S4: Estimation of omega values in ete-toolkit using a branch model, Table S5: Estimation of natural selection intensity using the RELAX approach.
Author Contributions
Conceptualization, T.P., O.B. and N.A.; Methodology, O.B. and M.S.; Formal Analysis, T.P. and M.S.; Investigation, T.P., M.S. and A.K.; Data Curation, T.P. and N.A.; Writing—Original Draft Preparation, T.P., M.S., A.K., O.B. and N.A.; Writing—Review & Editing, T.P., O.B. and N.A.; Visualization, T.P. and O.B.; Supervision, N.A.; Project Administration, N.A.; Funding Acquisition, N.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Science Foundation (RSF) N 19-74-20110.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of Zoological Institute Russian Academy of Sciences (permission # 1-17, 7 April 2022).
Data Availability Statement
Raw sequence data are available at the NCBI SRA: SRR12765436–SRR12765441, SRR17971092–SRR17971097 (BioProject № PRJNA591473), see Table S1 for details.
Acknowledgments
We are grateful to Kowalskaya Yu.M. (Severtsov Institute of Ecology and Evolution RAS, Moscow) and Smorkacheva A.V. (St. Petersburg State University, Saint-Petersburg) for providing animals for the analysis, to Chash U-M.G. (State Nature Reserve “Ubsunurskaya Kotlovina”, Kyzyl), Bodrov S.Yu. and Tursunova L.S. (ZIN RAS, Saint-Petersburg) for help during the field work. We thank Ezhova M.A. and Logacheva M.D. (Genomics Core Facility of Skolkovo Institute of Science and Technology, Moscow) for NGS library preparation and sequencing, Rasskazov D.A. (ZIN RAS, Saint-Petersburg) for technical support with calculations, Barbitoff Y.A. (Bioinformatics Institute, Saint-Petersburg) for valuable comments and advice and Kviatkovskii M.O. (EPAM Systems, Life Sciences Department) for help with scripts for the analysis, Genelt-Yanovskiy E.A. (ZIN RAS, Saint-Petersburg) for valuable discussion and four anonymous reviewers for their helpful and critical comments.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dupal, T.A. Geographical Variation and Subspecies Systematics of Narrow-Headed Vole Microtus (Stenocranius) Gregalis (Rodentia, Cricetidae). Zool. Zhurnal 2000, 79, 851–858. [Google Scholar]
- Golenishchev, F.N.; Petrovskaya, N.A. Geographic Variation of Narrow-Headed Vole Microtus (Stenocranius) Gregalis Pall., 1779. Theriol. Issled. 2002, 1, 17–34. [Google Scholar]
- Dupal, T.A.; Abramov, S.A. Intrapopulation Morphological Variability of Narrow-Headed Vole (Microtus Gregalis, Rodentia, Arvicolidae). Zool. Zhurnal 2010, 89, 850–861. [Google Scholar]
- Gromov, I.M.; Polyakov, I.Y. Voles (Microtinae); Brill Academic Publishers: Boston, MA, USA, 1992; Volume 3. [Google Scholar]
- Ognev, S.I. Zveri SSSR I Prilezhashhih Stran (The Mammals of the USSR and Adjacent Countries). Vol VII. Gryzuny (Rodentia); Publ. Acad. Nauk USSR: Leningrad, Russia, 1950. [Google Scholar]
- Gromov, I.M.; Erbaeva, M.A. Mammals of the Fauna of Russia and Adjacent Regions. Lagomorphs and Rodents; Zoological Institute RAS: Saint Petersburg, Russia, 1995. [Google Scholar]
- Petrova, T.V.; Zakharov, E.S.; Samiya, R.; Abramson, N.I. Phylogeography of the Narrow-Headed Vole Lasiopodomys (Stenocranius) Gregalis (Cricetidae, Rodentia) Inferred from Mitochondrial Cytochrome b Sequences: An Echo of Pleistocene Prosperity. J. Zool. Syst. Evol. Res. 2015, 53, 97–108. [Google Scholar] [CrossRef]
- Lissovsky, A.A.; Obolenskaya, E.V.; Petrova, T.V. Morphological and Genetic Variation of Narrow-Headed Voles Lasiopo-Domys Gregalis from South-East Transbaikalia. Russ. J. Theriol. 2013, 12, 83–90. [Google Scholar] [CrossRef]
- Petrova, T.V.; Tesakov, A.S.; Kowalskaya, Y.M.; Abramson, N.I. Cryptic Speciation in the Narrow-Headed Vole Lasiopodomys (Stenocranius) Gregalis (Rodentia: Cricetidae). Zool. Scr. 2016, 45, 618–629. [Google Scholar] [CrossRef]
- Kowalskaya, Y.M.; Petrova, T.V.; Abramson, N.I. Preliminary Results of Experimental Hybridization of Mitochondrial Forms of the Narrow-Headed Vole Lasiopodomys (Stenocranius) Gregalis (Rodentia, Arvicolinae); KMK Scientific Press: Moscow, Russia, 2015; p. 92. [Google Scholar]
- Petrova, T.V.; Genelt-Yanovskiy, E.A.; Lissovsky, A.A.; Chash, U.-M.G.; Masharsky, A.E.; Abramson, N.I. Signatures of Genetic Isolation of the Three Lineages of the Narrow-Headed Vole Lasiopodomys Gregalis (Cricetidae, Rodentia) in a Mosaic Steppe Landscape of South Siberia. Mamm. Biol. 2021, 101, 275–285. [Google Scholar] [CrossRef]
- Coyne, J.A.; Orr, H.A. Speciation; Vancouver: Sunderland, MA, USA, 2004. [Google Scholar]
- McKinnon, J.S.; Rundle, H.D. Speciation in Nature: The Threespine Stickleback Model Systems. Trends Ecol. Evol. 2002, 17, 480–488. [Google Scholar] [CrossRef]
- Räsänen, K.; Hendry, A.P. Disentangling Interactions between Adaptive Divergence and Gene Flow When Ecology Drives Diversification: Adaptive Divergence and Gene Flow. Ecol. Lett. 2008, 11, 624–636. [Google Scholar] [CrossRef]
- Orr, H.A. The Genetic Basis of Reproductive Isolation: Insights from Drosophila. Proc. Natl. Acad. Sci. USA 2005, 102, 6522–6526. [Google Scholar] [CrossRef]
- Moehring, A.J.; Llopart, A.; Elwyn, S.; Coyne, J.A.; Mackay, T.F.C. The Genetic Basis of Prezygotic Reproductive Isolation Between Drosophila Santomea and D. Yakuba Due to Mating Preference. Genetics 2006, 173, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Rogers, S.M.; Bernatchez, L. The Genetic Basis of Intrinsic and Extrinsic Post-Zygotic Reproductive Isolation Jointly Promoting Speciation in the Lake Whitefish Species Complex (Coregonus Clupeaformis). J. Evol. Biol. 2006, 19, 1979–1994. [Google Scholar] [CrossRef] [PubMed]
- Howard, D.J.; Marshall, J.L.; Hampton, D.D.; Britch, S.C.; Draney, M.L.; Chu, J.; Cantrell, R.G. The Genetics of Reproductive Isolation: A Retrospective and Prospective Look with Comments on Ground Crickets. Am. Nat. 2002, 159, S8–S21. [Google Scholar] [CrossRef] [PubMed]
- Pogson, G.H. Studying the Genetic Basis of Speciation in High Gene Flow Marine Invertebrates. Curr. Zool. 2016, 62, 643–653. [Google Scholar] [CrossRef] [PubMed]
- Poelstra, J.W.; Ellegren, H.; Wolf, J.B.W. An Extensive Candidate Gene Approach to Speciation: Diversity, Divergence and Linkage Disequilibrium in Candidate Pigmentation Genes across the European Crow Hybrid Zone. Heredity 2013, 111, 467–473. [Google Scholar] [CrossRef]
- Janoušek, V.; Wang, L.; Luzynski, K.; Dufková, P.; Vyskočilová, M.M.; Nachman, M.W.; Munclinger, P.; Macholán, M.; Piálek, J.; Tucker, P.K. Genome-Wide Architecture of Reproductive Isolation in a Naturally Occurring Hybrid Zone between Mus Musculus Musculus and M. m. Domesticus: Reproductive Isolation in House Mouse. Mol. Ecol. 2012, 21, 3032–3047. [Google Scholar] [CrossRef]
- Turner, L.M.; Harr, B. Genome-Wide Mapping in a House Mouse Hybrid Zone Reveals Hybrid Sterility Loci and Dobzhansky-Muller Interactions. eLife 2014, 3, e02504. [Google Scholar] [CrossRef]
- Malinsky, M.; Challis, R.J.; Tyers, A.M.; Schiffels, S.; Terai, Y.; Ngatunga, B.P.; Miska, E.A.; Durbin, R.; Genner, M.J.; Turner, G.F. Genomic Islands of Speciation Separate Cichlid Ecomorphs in an East African Crater Lake. Science 2015, 350, 1493–1498. [Google Scholar] [CrossRef]
- Nosil, P.; Egan, S.P.; Funk, D.J. Heterogeneous Genomic Differentiation between Walking-Stick Ecotypes: “Isolation by Adaptation” and Multiple Roles for Divergent Selection. Evolution 2008, 62, 316–336. [Google Scholar] [CrossRef]
- Schmidt, P.S.; Serrão, E.A.; Pearson, G.A.; Riginos, C.; Rawson, P.D.; Hilbish, T.J.; Brawley, S.H.; Trussell, G.C.; Carrington, E.; Wethey, D.S.; et al. Ecological Genetics in the North Atlantic: Environmental Gradients and Adaptation at Specific Loci. Ecology 2008, 89, S91–S107. [Google Scholar] [CrossRef]
- Stinchcombe, J.R.; Hoekstra, H.E. Combining Population Genomics and Quantitative Genetics: Finding the Genes Underlying Ecologically Important Traits. Heredity 2008, 100, 158–170. [Google Scholar] [CrossRef]
- Vincent, B.; Dionne, M.; Kent, M.P.; Lien, S.; Bernatchez, L. Landscape Genomics in Atlantic Salmon (Salmo Salar): Searching for Gene-Environment Interactions Driving Local Adaptation. Evolution 2013, 67, 3469–3487. [Google Scholar] [CrossRef] [PubMed]
- Manthey, J.D.; Moyle, R.G. Isolation by Environment in White-Breasted Nuthatches (Sitta Carolinensis) of the Madrean Archipelago Sky Islands: A Landscape Genomics Approach. Mol. Ecol. 2015, 24, 3628–3638. [Google Scholar] [CrossRef] [PubMed]
- Delport, W.; Scheffler, K.; Seoighe, C. Models of Coding Sequence Evolution. Brief. Bioinform. 2008, 10, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, M.; Kosiol, C. Investigating Protein-Coding Sequence Evolution with Probabilistic Codon Substitution Models. Mol. Biol. Evol. 2009, 26, 255–271. [Google Scholar] [CrossRef]
- Batsaikhan, N.; Tsytsulina, K.; Formozov, N.; Sheftel, B. Microtus Gregalis. In The IUCN Red List of Threatened Species 2016; The International Union for Conservation of Nature (IUCN): Cambridge, UK, 2016. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 April 2020).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De Novo Transcript Sequence Reconstruction from RNA-Seq Using the Trinity Platform for Reference Generation and Analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Lechner, M.; Findeiß, S.; Steiner, L.; Marz, M.; Stadler, P.F.; Prohaska, S.J. Proteinortho: Detection of (Co-)Orthologs in Large-Scale Analysis. BMC Bioinform. 2011, 12, 124. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.A.; O’Connor, B.D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra; O’Reilly Media: Sebastopol, CA, USA, 2020. [Google Scholar]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. EggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.-P.; Mushayamaha, T.; Thomas, P.D. PANTHER Version 16: A Revised Family Classification, Tree-Based Classification Tool, Enhancer Regions and Extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Yang, Z.; Nielsen, R.; Goldman, N.; Pedersen, A.-M.K. Codon-Substitution Models for Heterogeneous Selection Pressure at Amino Acid Sites. Genetics 2000, 155, 431–449. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Wertheim, J.O.; Murrell, B.; Smith, M.D.; Kosakovsky Pond, S.L.; Scheffler, K. RELAX: Detecting Relaxed Selection in a Phylogenetic Framework. Mol. Biol. Evol. 2015, 32, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.V.; Culver, M.; Stephens, J.C.; Johnson, W.E.; O’Brien, S.J. Rates of Nuclear and Cytoplasmic Mitochondrial DNA Sequence Divergence in Mammals. Mol. Biol. Evol. 1997, 14, 277–286. [Google Scholar] [CrossRef]
- Von Kopylow, K.; Kirchhoff, C.; Jezek, D.; Schulze, W.; Feig, C.; Primig, M.; Steinkraus, V.; Spiess, A.-N. Screening for Biomarkers of Spermatogonia within the Human Testis: A Whole Genome Approach. Hum. Reprod. 2010, 25, 1104–1112. [Google Scholar] [CrossRef]
- Pease, J.B.; Driver, R.J.; de la Cerda, D.A.; Day, L.B.; Lindsay, W.R.; Schlinger, B.A.; Schuppe, E.R.; Balakrishnan, C.N.; Fuxjager, M.J. Layered Evolution of Gene Expression in “Superfast” Muscles for Courtship. Proc. Natl. Acad. Sci. USA 2022, 119, e2119671119. [Google Scholar] [CrossRef]
- Elokil, A.A.; Abouzaid, M.; Magdy, M.; Xiao, T.; Liu, H.; Xu, R.; Li, S. Testicular Transcriptome Analysis under the Dietary Inclusion of L-Carnitine Reveals Potential Key Genes Associated with Oxidative Defense and the Semen Quality Factor in Aging Roosters. Domest. Anim. Endocrinol. 2021, 74, 106573. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.-W.; Sun, S.-M.; Xu, K.; Li, Y.-Y.; Lei, W.-L.; Li, L.; Liu, S.-L.; Ouyang, Y.-C.; Sun, Q.-Y.; Wang, Z.-B. FBXO34 Regulates the G2/M Transition and Anaphase Entry in Meiotic Oocytes. Front. Cell Dev. Biol. 2021, 9, 647103. [Google Scholar] [CrossRef] [PubMed]
- Wollenberg Valero, K.C.; Garcia-Porta, J.; Irisarri, I.; Feugere, L.; Bates, A.; Kirchhof, S.; Jovanović Glavaš, O.; Pafilis, P.; Samuel, S.F.; Müller, J.; et al. Functional Genomics of Abiotic Environmental Adaptation in Lacertid Lizards and Other Vertebrates. J. Anim. Ecol. 2021, 91, 1163–1179. [Google Scholar] [CrossRef]
- Wu, D.-D.; Yang, C.-P.; Wang, M.-S.; Dong, K.-Z.; Yan, D.-W.; Hao, Z.-Q.; Fan, S.-Q.; Chu, S.-Z.; Shen, Q.-S.; Jiang, L.-P.; et al. Convergent Genomic Signatures of High-Altitude Adaptation among Domestic Mammals. Natl. Sci. Rev. 2020, 7, 952–963. [Google Scholar] [CrossRef]
- Yasumoto, H.; Meng, L.; Lin, T.; Zhu, Q.; Tsai, R.Y.L. GNL3L Inhibits Activity of Estrogen-Related Receptor γ by Competing for Coactivator Binding. J. Cell Sci. 2007, 120, 2532–2543. [Google Scholar] [CrossRef]
- Gewiss, R.; Topping, T.; Griswold, M.D. Cycles, Waves, and Pulses: Retinoic Acid and the Organization of Spermatogenesis. Andrology 2020, 8, 892–897. [Google Scholar] [CrossRef] [PubMed]
- Grindflek, E.; Berget, I.; Moe, M.; Oeth, P.; Lien, S. Transcript Profiling of Candidate Genes in Testis of Pigs Exhibiting Large Differences in Androstenone Levels. BMC Genet. 2010, 11, 4. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, Y.; Li, W.; Zhang, Y.; Yang, Y.; Li, H.; Geng, Z.; Ao, H.; Zhou, R.; Li, K. NRDR Inhibits Estradiol Synthesis and Is Associated with Changes in Reproductive Traits in Pigs: LIU et al. Mol. Reprod. Dev. 2019, 86, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, T.; Endo, S.; Maeda, S.; Ishikura, S.; Tajima, K.; Tanaka, N.; Nakamura, K.T.; Imamura, Y.; Hara, A. Characterization of Human DHRS4: An Inducible Short-Chain Dehydrogenase/Reductase Enzyme with 3β-Hydroxysteroid Dehydrogenase Activity. Arch. Biochem. Biophys. 2008, 477, 339–347. [Google Scholar] [CrossRef]
- Ran, X.; Hu, F.; Mao, N.; Ruan, Y.; Yi, F.; Niu, X.; Huang, S.; Li, S.; You, L.; Zhang, F.; et al. Differences in Gene Expression and Variable Splicing Events of Ovaries between Large and Small Litter Size in Chinese Xiang Pigs. Porc. Health Manag. 2021, 7, 52. [Google Scholar] [CrossRef]
- Chi, W.; Reinke, V. Promotion of Oogenesis and Embryogenesis in the C. Elegans Gonad by EFL-1/DPL-1 (E2F) Does Not Require LIN-35 (PRB). Development 2006, 133, 3147–3157. [Google Scholar] [CrossRef]
- Paczkowski, M.; Schoolcraft, W.B.; Krisher, R.L. Fatty Acid Metabolism during Maturation Affects Glucose Uptake and Is Essential to Oocyte Competence. Reproduction 2014, 148, 429–439. [Google Scholar] [CrossRef]
- Sharma, M. Mitochondrial Fatty Acid Transport System and Its Relevance to Ovarian Function. J. Infertil. Reprod. Biol. 2018, 6, 1–3. [Google Scholar] [CrossRef]
- Gentile, L.; Monti, M.; Sebastiano, V.; Merico, V.; Nicolai, R.; Calvani, M.; Garagna, S.; Redi, C.A.; Zuccotti, M. Single-Cell Quantitative RT-PCR Analysis of Cpt1b and Cpt2 Gene Expression in Mouse Antral Oocytes and in Preimplantation Embryos. Cytogenet. Genome Res. 2004, 105, 215–221. [Google Scholar] [CrossRef]
- Nenicu, A.; Lüers, G.H.; Kovacs, W.; Bergmann, M.; Baumgart-Vogt, E. Peroxisomes in Human and Mouse Testis: Differential Expression of Peroxisomal Proteins in Germ Cells and Distinct Somatic Cell Types of the Testis. Biol. Reprod. 2007, 77, 1060–1072. [Google Scholar] [CrossRef]
- Hussain, S.; Tuorto, F.; Menon, S.; Blanco, S.; Cox, C.; Flores, J.V.; Watt, S.; Kudo, N.R.; Lyko, F.; Frye, M. The Mouse Cytosine-5 RNA Methyltransferase NSun2 Is a Component of the Chromatoid Body and Required for Testis Differentiation. Mol. Cell. Biol. 2013, 33, 1561–1570. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Rojas, T.; Chin, A.C.; Cheng, W.; Bernstein, I.A.; Albacarys, L.K.; Wright, W.W.; Snyder, S.H. Multiple Aspects of Male Germ Cell Development and Interactions with Sertoli Cells Require Inositol Hexakisphosphate Kinase-1. Sci. Rep. 2018, 8, 7039. [Google Scholar] [CrossRef] [PubMed]
- Meneses, M.J.; Borges, D.O.; Dias, T.R.; Martins, F.O.; Oliveira, P.F.; Macedo, M.P.; Alves, M.G. Knockout of Insulin-Degrading Enzyme Leads to Mice Testicular Morphological Changes and Impaired Sperm Quality. Mol. Cell. Endocrinol. 2019, 486, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Hekim, N.; Gunes, S.; Asci, R.; Henkel, R.; Abur, U. Semiquantitative Promoter Methylation of MLH1 and MSH2 Genes and Their Impact on Sperm DNA Fragmentation and Chromatin Condensation in Infertile Men. Andrologia 2021, 53, e13827. [Google Scholar] [CrossRef]
- Venditti, M.; Donizetti, A.; Aniello, F.; Minucci, S. EH Domain Binding Protein 1-like 1 (EHBP1L1), a Protein with Calponin Homology Domain, Is Expressed in the Rat Testis. Zygote 2020, 28, 441–446. [Google Scholar] [CrossRef]
- Venditti, M.; Minucci, S. Differential Expression and Localization of EHBP1L1 during the First Wave of Rat Spermatogenesis Suggest Its Involvement in Acrosome Biogenesis. Biomedicines 2022, 10, 181. [Google Scholar] [CrossRef]
- El-Raey, M.; Geshi, M.; Somfai, T.; Kaneda, M.; Hirako, M.; Abdel-Ghaffar, A.E.; Sosa, G.A.; El-Roos, M.E.A.A.; Nagai, T. Evidence of Melatonin Synthesis in the Cumulus Oocyte Complexes and Its Role in Enhancing Oocyte Maturation In Vitro in Cattle. Mol. Reprod. Dev. 2011, 78, 250–262. [Google Scholar] [CrossRef]
- Sakaguchi, K.; Itoh, M.T.; Takahashi, N.; Tarumi, W.; Ishizuka, B. The Rat Oocyte Synthesises Melatonin. Reprod. Fertil. Dev. 2013, 25, 674. [Google Scholar] [CrossRef]
- Xue, Q.; Li, G.; Cao, Y.; Yin, J.; Zhu, Y.; Zhang, H.; Zhou, C.; Shen, H.; Dou, X.; Su, Y.; et al. Identification of Genes Involved in Inbreeding Depression of Reproduction in Langshan Chickens. Anim. Biosci. 2021, 34, 975–984. [Google Scholar] [CrossRef]
- Huang, D.; Li, J.; He, L.-Q. Influence of Tripterygium Wilfordii on the Expression of Spermiogenesis Related Genes Herc4, Ipo11 and Mrto4 in Mice. Hered. Beijing 2009, 31, 941–946. [Google Scholar] [CrossRef]
- Xiang, W.; Wen, Z.; Hu, L.; Li, H.; Xiong, C. Expression of NOSTRIN in the testis tissue of azoospermia patients. Zhonghua Nan Ke Xue Natl. J. Androl. 2011, 17, 38–42. [Google Scholar]
- Bansal, S.K.; Gupta, N.; Sankhwar, S.N.; Rajender, S. Differential Genes Expression between Fertile and Infertile Spermatozoa Revealed by Transcriptome Analysis. PLoS ONE 2015, 10, e0127007. [Google Scholar] [CrossRef] [PubMed]
- Doran, J.; Walters, C.; Kyle, V.; Wooding, P.; Hammett-Burke, R.; Colledge, W.H. Mfsd14a (Hiat1) Gene Disruption Causes Globozoospermia and Infertility in Male Mice. Reproduction 2016, 152, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.K.-K.; Tseng, H.-C.; Hwu, Y.-M.; Fan, C.-C.; Lin, M.-H.; Yu, J.-J.; Yeh, L.-Y.; Li, S.-H. Expression of Cystatin C in the Female Reproductive Tract and Its Effect on Human Sperm Capacitation. Reprod. Biol. Endocrinol. 2018, 16, 8. [Google Scholar] [CrossRef] [PubMed]
- Major, A.T.; Hogarth, C.A.; Young, J.C.; Kurihara, Y.; Jans, D.A.; Loveland, K.L. Dynamic Paraspeckle Component Localisation during Spermatogenesis. Reproduction 2019, 158, 267–280. [Google Scholar] [CrossRef]
- Gispert, S.; Key, J.; Kohli, A.; Torres-Odio, S.; Koepf, G.; Amr, S.; Reichlmeir, M.; Harter, P.N.; West, A.P.; Münch, C.; et al. ClpP-Deletion Causes Azoospermia, with Meiosis-I Delay and Insufficient Biosynthesis of Spermatid Factors, Due to Mitochondrial Dysfunction with Accumulation of Perrault Proteins ERAL1, PEO1, and HARS2 2022. Preprints 2022, 2022040245. [Google Scholar] [CrossRef]
- Grant, K.E. Molecular and Cellular Correlates of Sperm Viability Associated with Male Fertility. Theses Diss 2013, 3178. Available online: https://scholarsjunction.msstate.edu/td/3178 (accessed on 12 May 2022).
- Feugang, J.M.; Kaya, A.; Page, G.P.; Chen, L.; Mehta, T.; Hirani, K.; Nazareth, L.; Topper, E.; Gibbs, R.; Memili, E. Two-Stage Genome-Wide Association Study Identifies Integrin Beta 5 as Having Potential Role in Bull Fertility. BMC Genom. 2009, 10, 176. [Google Scholar] [CrossRef]
- Velho, A.; Wang, H.; Koenig, L.; Grant, K.E.; Menezes, E.S.; Kaya, A.; Moura, A.; Memili, E. Expression Dynamics of Integrin Subunit Beta 5 in Bovine Gametes and Embryos Imply Functions in Male Fertility and Early Embryonic Development. Andrologia 2019, 51, e13305. [Google Scholar] [CrossRef]
- Yamaguchi, Y.L.; Tanaka, S.S.; Yasuda, K.; Matsui, Y.; Tam, P.P.L. Stage-Specific Importin13 Activity Influences Meiosis of Germ Cells in the Mouse. Dev. Biol. 2006, 297, 350–360. [Google Scholar] [CrossRef][Green Version]
- Arraztoa, J.A.; Zhou, J.; Marcu, D.; Cheng, C.; Bonner, R.; Chen, M.; Xiang, C.; Brownstein, M.; Maisey, K.; Imarai, M.; et al. Identification of Genes Expressed in Primate Primordial Oocytes. Hum. Reprod. 2005, 20, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cui, P.; Sun, Q.; Du, Z.; Chen, Z.; Li, Z.; Liu, C.; Cao, Y.; Yang, Z.; Liu, R.; et al. PSPC1 Regulates CHK1 Phosphorylation through Phase Separation and Participates in Mouse Oocyte Maturation. Acta Biochim. Biophys. Sin. 2021, 53, 1527–1537. [Google Scholar] [CrossRef]
- Moura, L.B.S.; Magalhães-Padilha, D.M.; Morais, A.N.P.; Aguiar, F.L.N.; Geisler–Lee, J.; Wischral, A.; Gastal, M.O.; Fonseca, G.R.; Geisler, M.; Figueiredo, J.R. Folliculogenesis-Related Genes Are Differently Expressed in Secondary and Tertiary Ovarian Follicles. Zygote 2021, 29, 503–506. [Google Scholar] [CrossRef]
- Jiborn, T.; Abrahamson, M.; Wallin, H.; Malm, J.; Lundwall, Å.; Gadaleanu, V.; Abrahamsson, P.-A.; Bjartell, A. Cystatin C Is Highly Expressed in the Human Male Reproductive System. J. Androl. 2013, 25, 564–572. [Google Scholar] [CrossRef]
- Chakraborty, S.; Ain, R. NOSTRIN: A Novel Modulator of Trophoblast Giant Cell Differentiation. Stem Cell Res. 2018, 31, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Parkkila, S.; Parkkila, A.K.; Kaunisto, K.; Waheed, A.; Sly, W.S.; Rajaniemi, H. Location of a Membrane-Bound Carbonic Anhydrase Isoenzyme (CA IV) in the Human Male Reproductive Tract. J. Histochem. Cytochem. 1993, 41, 751–757. [Google Scholar] [CrossRef]
- Hynninen, P.; Hämäläinen, J.M.; Pastorekova, S.; Pastorek, J.; Waheed, A.; Sly, W.S.; Tomas, E.; Kirkinen, P.; Parkkila, S. Transmembrane Carbonic Anhydrase Isozymes IX and XII in the Female Mouse Reproductive Organs. Reprod. Biol. Endocrinol. 2004, 2, 73. [Google Scholar] [CrossRef][Green Version]
- Rengaraj, D.; Gao, F.; Liang, X.-H.; Yang, Z.-M. Expression and Regulation of Type II Integral Membrane Protein Family Members in Mouse Male Reproductive Tissues. Endocrine 2007, 31, 193–201. [Google Scholar] [CrossRef]
- Rengaraj, D.; Liang, X.-H.; Gao, F.; Deng, W.-B.; Mills, N.; Yang, Z.-M. Differential Expression and Regulation of Integral Membrane Protein 2b in Rat Male Reproductive Tissues. Asian J. Androl. 2008, 10, 503–511. [Google Scholar] [CrossRef]
- Rodriguez, T.A.; Sparrow, D.B.; Scott, A.N.; Withington, S.L.; Preis, J.I.; Michalicek, J.; Clements, M.; Tsang, T.E.; Shioda, T.; Beddington, R.S.P.; et al. Cited1 Is Required in Trophoblasts for Placental Development and for Embryo Growth and Survival. Mol. Cell. Biol. 2004, 24, 228–244. [Google Scholar] [CrossRef]
- Sha, Q.; Yu, J.; Guo, J.; Dai, X.; Jiang, J.; Zhang, Y.; Yu, C.; Ji, S.; Jiang, Y.; Zhang, S.; et al. CNOT 6L Couples the Selective Degradation of Maternal Transcripts to Meiotic Cell Cycle Progression in Mouse Oocyte. EMBO J. 2018, 37, e99333. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Silva, G.; Caballero-Campo, P.; Chirinos, M. Sperm MRNAs as Potential Markers of Male Fertility. Reprod. Biol. 2022, 22, 100636. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y. The Ancestral Gene for Transcribed, Low-Copy Repeats in the Prader-Willi/Angelman Region Encodes a Large Protein Implicated in Protein Trafficking, Which Is Deficient in Mice with Neuromuscular and Spermiogenic Abnormalities. Hum. Mol. Genet. 1999, 8, 533–542. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dumasia, K.; Kumar, A.; Deshpande, S.; Balasinor, N.H. Estrogen, through Estrogen Receptor 1, Regulates Histone Modifications and Chromatin Remodeling during Spermatogenesis in Adult Rats. Epigenetics 2017, 12, 953–963. [Google Scholar] [CrossRef] [PubMed]
- Davila, R.A.; Spiller, C.; Harkins, D.; Harvey, T.; Jordan, P.W.; Gronostajski, R.M.; Piper, M.; Bowles, J. Deletion of NFIX Results in Defective Progression through Meiosis within the Mouse Testis. Biol. Reprod. 2022, 106, ioac049. [Google Scholar] [CrossRef]
- Linnemannstöns, K.; Ripp, C.; Honemann-Capito, M.; Brechtel-Curth, K.; Hedderich, M.; Wodarz, A. The PTK7-Related Transmembrane Proteins Off-Track and Off-Track 2 Are Co-Receptors for Drosophila Wnt2 Required for Male Fertility. PLoS Genet. 2014, 10, e1004443. [Google Scholar] [CrossRef]
- Moon, S.; Lee, O.-H.; Lee, S.; Lee, J.; Park, H.; Park, M.; Chang, E.M.; Park, K.-H.; Choi, Y. STK3/4 Expression Is Regulated in Uterine Endometrial Cells during the Estrous Cycle. Cells 2019, 8, 1643. [Google Scholar] [CrossRef]
- Gura, M.A.; Mikedis, M.M.; Seymour, K.A.; de Rooij, D.G.; Page, D.C.; Freiman, R.N. Dynamic and Regulated TAF Gene Expression during Mouse Embryonic Germ Cell Development. PLoS Genet. 2020, 16, e1008515. [Google Scholar] [CrossRef]
- Obbard, D.J.; Welch, J.J.; Kim, K.-W.; Jiggins, F.M. Quantifying Adaptive Evolution in the Drosophila Immune System. PLoS Genet. 2009, 5, e1000698. [Google Scholar] [CrossRef]
- McTaggart, S.J.; Obbard, D.J.; Conlon, C.; Little, T.J. Immune Genes Undergo More Adaptive Evolution than Non-Immune System Genes in Daphnia Pulex. BMC Evol. Biol. 2012, 12, 63. [Google Scholar] [CrossRef]
- Shultz, A.J.; Sackton, T.B. Immune Genes Are Hotspots of Shared Positive Selection across Birds and Mammals. eLife 2019, 8, e41815. [Google Scholar] [CrossRef] [PubMed]
- Paterson, H.E.H. The Recognition Concept of Species. In Species and Speciation; Vrba, E., Ed.; Transvaal Museum: Pretoria, South Africa, 1985; pp. 21–29. [Google Scholar]
- Petrova, T.V. Narrow-Headed Vole Lasiopodomys (Stenocranius) Gregalis (Pallas, 1779): Taxonomic Structure, Phylogenetic Position and Evolution; Zoological Institute RAS: Saint Petersburg, Russia, 2017. [Google Scholar]
- Wallace, A.R. Darwinism. An Exposition of the Theory of Natural Selection with Some of Its Applications; Macmillan & Co.: London, UK; New York, NY, USA, 1889. [Google Scholar]
- Feoktistova, N.Y.; Kropotkina, M.V.; Potashnikova, E.V.; Gureeva, A.V.; Kuznetsova, E.V.; Surov, A.V. Speciation in Allopatric Species of the Hamster Subfamily Cricetinae (Rodentia, Cricetidae). Biol. Bull. Rev. 2019, 9, 230–242. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).