Abstract
We report for the first time the occurrence of at least two species of the phylum Gnathostomulida in the Southern Ocean, along the shores of the Ross Sea in Antarctica. At least one species for each of the orders of the phylum (Filospermoidea and Bursovaginoidea) was found using both morphological inspection and DNA metabarcoding of the shallow marine sediments collected with a Van Veen grab or by scuba diving in the area facing the Italian research station “Mario Zucchelli”.
1. Introduction
Gnathostomulida Ax, 1956, microscopic worms unique for their monociliated epithelium, were described from detritus-rich, shallow marine sand and are among the most recently discovered animal phyla [1,2,3]. More than 100 species in two orders (Filospermoidea and Bursovaginoidea) are known to date, mostly from the tropics, but many with a global distribution, from as far north as Disko Island, Greenland and Murmansk, Russia (about 69° N) to Dunedin, New Zealand (about 46° S). Knowledge of this phylum is very limited, and their occurrence is often overlooked in studies of marine microscopic animals [4].
The aim of the current note is to provide unambiguous evidence of the previously unnoticed occurrence of Gnathostomulida as far south as the shores of Antarctica in the Southern Ocean. This record represents the highest absolute latitude reached by the phylum, which is now reported down to 74° S.
2. Materials and Methods
Sampling activities were carried out during the Austral summer 2018–2019 in the framework of the Italian National Antarctic Program (PNRA) project “TNB-CODE” (Terra Nova Bay barCODing and mEtabarcoding of Antarctic organisms from marine and limno-terrestrial environments). Sediment samples were collected either with a van Veen grab deployed from a dinghy or by SCUBA divers in the area nearby the Italian Research Station “Mario Zucchelli” along the Terra Nova Bay shores (Ross Sea) (Figure 1, Table 1). Different types of substrates were targeted for sampling (e.g., gravel, sand, silt, spicule mat, etc.), based on available sedimentological maps [5] and previous knowledge, at depths ranging from the intertidal down to 70 m. Sediment samples were not meant to provide a quantitative estimate of abundances, but only to provide a preliminary survey of marine meiofauna in the area. Samples were temporarily preserved in plastic buckets or jars and immediately brought back to the laboratory in the “Mario Zucchelli” research station. Part of the sample was inspected visually under a stereomicroscope to sort meiofauna, take photos of the living organisms, and store them in ethanol as vouchers. Another part of the sample was passed through sieves of mesh size from 500 to 20 μm, with the organisms passing the filtering at 500 µm but not the 20 µm stored in ethanol at −20 °C for DNA metabarcoding. Mesh sizes were selected to collect all organisms belonging to the ecological guild of microscopic animals of the meiofauna [6]. All samples, both for morphology and for DNA metabarcoding, were processed with a preliminary extraction using magnesium chloride, as it is common for soft-bodied meiofauna [7].
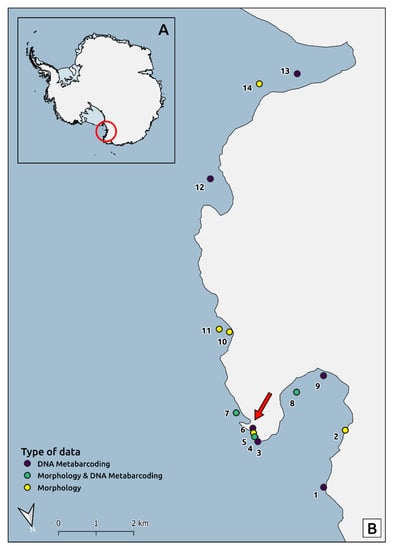
Figure 1.
Location of Terra Nova Bay (Ross Sea) in Antarctica (A) and study area with sampled stations (B). The red arrow indicates the position of Mario Zucchelli research station. Sampling sites that appear to lie on the coastline represent very shallow, near shore sites. Numbers refer to sampling stations listed in Table 1 (field “STATION-ID”). This map was produced using the collection of datasets “Quantarctica” [8] and QGIS [9].

Table 1.
Number of Gnathostomulida specimens found from morphological inspection and number of reads from DNA metabarcoding (“MORPHOLOGY” and “DNA METABARCODING” fields, respectively) of marine sediment samples collected around the Italian Research Station “Mario Zucchelli” in the Ross Sea in Antarctica. The table includes fields referring to the museum voucher codes of the specimens inspected by morphological examination and preserved at the Italian National Antarctic Museum (“MNA#”), the sampling stations’ labels as reported in Figure 1 (“STATION-ID”), the name of the sampling station (“EVENT-ID”), type of habitat (“HABITAT”), sampling gear or methodology adopted (“GEAR”), depth (m), sampling date and geographic coordinates in WGS84.
DNA was extracted from ethanol-stored samples using the commercial PowerSoil extraction kit (Qiagen). The primers used for Illumina sequencing targeted a DNA fragment of approximately 450 base pairs corresponding to the V1–V2 regions of the nuclear small subunit rRNA gene (18S rDNA). They were based on the commonly used 18SF04 (5′-GCTTGTCTCAAAGATTAAGCC-3′) for V1-V2 region of 18S [10,11,12] and a modified version of 18SR22, called 18SR22 (5′-GCCTGCTGCCTTCCTTRGA-3′) as in Martínez [13]. Sequencing was performed on the NovaSeq Illumina platform using the 2 × 250 paired-end (PE) approach. The pipeline to obtain unique sequences called amplicon sequence variants (ASV) was that of Martínez [13], including a maximum number of nucleotides allowed to differ in the merging operation set to 10 and a minimum length of merged sequences corresponding to 250 bp following the UPARSE pipeline [14]. Adapters and primer removal was previously performed using cutadapt [15], whereas the clustering procedure was conducted using the UNOISE algorithm implemented in USEARCH [16]. The sequences that passed the bioinformatics quality filtering steps were blasted in GenBank for an initial identification to the phylum level, and all sequences that had a potential hit to the phylum Gnathostomulida were retained. The retained sequences (Supplementary File S1) were then aligned with a reference set of clean gnathostomulid 18S sequences downloaded from GenBank. The aligned dataset, trimmed to the length of the DNA sequences from metabarcoding, was used to obtain a visual representation of the phylogenetic relationships through a neighbor-joining tree, from the R v4.1.3 [17] package ape v5.6.2 [18].
All gnathostomulid samples are part of the collections of the Italian National Antarctic Museum (MNA, Section of Genoa, voucher codes MNA-14061 to MNA-14085 and MNA-14743 to MNA-14747).
3. Results
The two analyses conducted in parallel, i.e., morphological inspection and DNA metabarcoding, both supported the presence of Gnathostomulida in the analyzed samples. The morphological inspection allowed isolating a total of 47 individual organisms belonging to the phylum Gnathostomulida obtained from 10 sampling stations (Table 1) by visually sorting the collected sediment. Organisms of both orders, Filospermoidea and Bursovaginoidea, were found (Figure 2). Similarly, the DNA metabarcoding analyses supported the occurrence of organisms belonging to two separate clades, one in each order of Gnathostomulida (Figure 3), by producing 10,717 sequences, which clustered in 10 unique sequences (ASV) from 18 sampling events (Table 1). Interestingly, in only three sampling sites, the occurrence of the phylum was supported both from morphological inspection and DNA metabarcoding (Table 1).
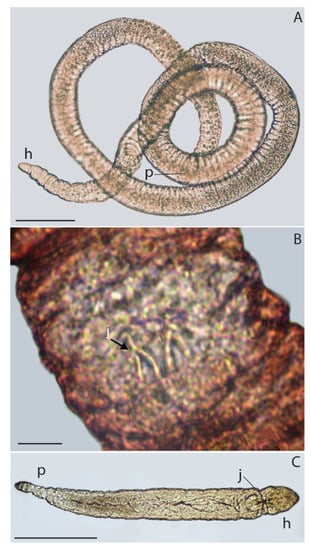
Figure 2.
Light micrographs with live specimens of Gnathostomulida from Antarctica. (A) A filospermoid, with a pointed anterior and a rounded posterior. (B) Jaws of a red filospermoid, Haplognathia cf. ruberrima (Sterrer, 1966). (C) A juvenile bursovaginoid (Austrognathiidae), with rounded anterior and tailed posterior. Abbreviations: head (h), jaws (j), posterior end (p). Scale bars: (A) = 100 µm, (B) = 10 µm, (C) = 200 µm.
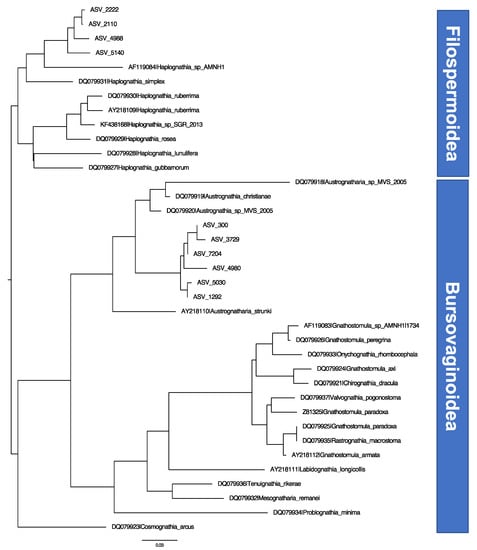
Figure 3.
Neighbor-joining tree of the 10 unique sequences found from DNA metabarcoding of the analyzed samples (named as ASV followed by an identification number) and the reference library of 18S sequences from each available species, obtained from GenBank (named as accession number followed by species name).
4. Discussion
This is the first record of the phylum from the Southern Ocean along the shores of Antarctica, and it represents the most extreme latitudinal record for both hemispheres by reaching 74° S. Although the collected material has yet to be analyzed in detail, morphology and DNA metabarcoding concur in supporting that the samples from Antarctica contain at least one species of each order, expanding the geographic coverage of the phylum to also include the only continent where Gnathostomulida were not known to occur.
The type of substrate where gnathostomulids were found ranges from silt to sand and gravel but also includes spicule mats and epilithic biofilms with encrusting bryozoans (Table 1). Such habitats represent the usual ones where organisms of the phylum are known to live (i.e., sand with detritus), but finding them in spicule mats has not been reported previously [3,4].
We have to acknowledge that some of the samples where only one or few sequences were found from DNA metabarcoding could represent instances of contamination [19]. However, samples wherein hundreds of sequences were found should be considered reliable.
The discrepancy between the detection of gnathostomulids based on visual inspection and based on DNA metabarcoding underlines the difficulties of comparing results from different approaches. On the one hand, it is surely difficult to retrieve live specimens of meiofauna from samples in Antarctica: it is highly likely that for some samples, the visual inspection was not carefully performed, making it possible that animals were not seen, even if present. On the other hand, DNA metabarcoding is known to produce false positives, especially in the samples with very few reads: in our datasets, the sites where only one or two reads were found may represent false positives (Table 1). Sites such as TETH 6, with thousands of reads, may be considered reliable.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d14050382/s1, Supplementary File S1: Alignment of the sequences used to obtain the tree in Figure 3, in fasta format.
Author Contributions
Conceptualization, D.F. and S.S.; methodology, W.S., M.V.S., M.C., A.M., R.S., E.M.E., D.F. and S.S.; formal analysis, W.S., M.V.S., M.C., A.M., R.S., E.M.E., D.F. and S.S.; data acquisition M.C., A.M., R.S. and E.M.E.; data curation, M.C., A.M., R.S. and E.M.E.; writing—original draft preparation, W.S., M.V.S., D.F. and S.S.; writing—review and editing, M.C., A.M., R.S. and E.M.E.; funding acquisition, S.S. All authors have read and agreed to the published version of the manuscript.
Funding
Sampling activities were performed within the PNRA project “TNB-CODE” (Terra Nova Bay barCODing and mEtabarcoding of Antarctic organisms from marine and limno-terrestrial environments) (PNRA 16_00120, PI: Stefano Schiaparelli). Authors are grateful to the Italian National Antarctic Scientific Commission (CSNA) and the Italian National Antarctic program (PNRA) for the endorsement of the Special Issue initiative and to the Italian National Antarctic Museum (MNA) for the financial support.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
In accordance with FAIR principles, the DNA sequence dataset (Supplementary File S1) is available in the Supplemental Materials.
Acknowledgments
We are indebted with Ellenico Francesco and Gainnoni Paolo (Gruppo Operativo Subacquei COMSUBIN, Marina Militare Italiana) and Corti Ramon (Gruppo Operativo Incursori, COMSUBIN, Marina Militare Italiana) for logistic support during diving operations and sampling at sea. This paper is an Italian contribution to the Commission for the Conservation of Antarctic Marine Living Resources CONSERVATION MEASURE 91-05 (2016) for the Ross Sea Region Marine Protected Area, specifically, addressing the priorities of Annex 91-05/C.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; and in the decision to publish the results.
References
- Ax, P. Die Gnathostomulida, Eine rätselhafte Wurmgruppe aus dem Meeressand. Abh. Akad. Wiss. Lit. Mainz. Math. Nat. Klasse 1956, 8, 1–32. [Google Scholar]
- Riedl, R.J. Gnathostomulida from America: This Is the First Record of the New Phylum from North America. Science 1969, 163, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, M.V.; Sterrer, W. Gnathostomulida. In Guide to the Identification of Marine Meiofauna; Schmidt-Rhaesa, A., Ed.; Dr. Friedrich Pfeil Publishing: Munich, Germany, 2020; pp. 199–226. [Google Scholar]
- Sterrer, W.; Sørensen, M.V. Phylum Gnathostomulida. In Handbook of Zoology, Volume 3(2) Gastrotricha and Gnathifera; Schmidt-Rhaesa, A., Ed.; De Gruyter: Berlin, Germany, 2014; pp. 135–196. ISBN 3110274272. [Google Scholar]
- Brambati, A.; Fanzutti, G.P.; Finocchiaro, F.; Simeoni, U. Sediments and Sedimentological Processes in the Ross Sea Continental Shelf (Antarctica): Results and Preliminary Conclusions. Boll Ocean. Teor. Appl. 1989, 7, 159–188. [Google Scholar]
- Giere, O. (Ed.) Sampling and Processing Meiofauna BT—Meiobenthology: The Microscopic Motile Fauna of Aquatic Sediments; Springer: Berlin/Heidelberg, Germany, 2009; pp. 63–86. ISBN 978-3-540-68661-3. [Google Scholar]
- Curini-Galletti, M.; Artois, T.; Delogu, V.; De Smet, W.H.; Fontaneto, D.; Jondelius, U.; Leasi, F.; Martinez, A.; Meyer-Wachsmuth, I.; Nilsson, K.S. Patterns of Diversity in Soft-Bodied Meiofauna: Dispersal Ability and Body Size Matter. PLoS ONE 2012, 7, e33801. [Google Scholar] [CrossRef]
- Matsuoka, K.; Skoglund, A.; Roth, G.; de Pomereu, J.; Griffiths, H.; Headland, R.; Herried, B.; Katsumata, K.; Le Brocq, A.; Licht, K. Quantarctica, an Integrated Mapping Environment for Antarctica, the Southern Ocean, and Sub-Antarctic Islands. Environ. Model. Softw. 2021, 140, 105015. [Google Scholar] [CrossRef]
- QGIS Development Team. QGIS Geographic Information System. QGIS Association: 2022. Available online: https://www.qgis.org (accessed on 21 April 2022).
- Bik, H.M.; Halanych, K.M.; Sharma, J.; Thomas, W.K. Dramatic Shifts in Benthic Microbial Eukaryote Communities Following the Deepwater Horizon Oil Spill. PLoS ONE 2012, 7, e38550. [Google Scholar] [CrossRef]
- Blaxter, M.L.; De Ley, P.; Garey, J.R.; Liu, L.X.; Scheldeman, P.; Vierstraete, A.; Vanfleteren, J.R.; Mackey, L.Y.; Dorris, M.; Frisse, L.M. A Molecular Evolutionary Framework for the Phylum Nematoda. Nature 1998, 392, 71–75. [Google Scholar] [CrossRef]
- Fonseca, V.G.; Carvalho, G.R.; Sung, W.; Johnson, H.F.; Power, D.M.; Neill, S.P.; Packer, M.; Blaxter, M.L.; Lambshead, P.J.D.; Thomas, W.K. Second-Generation Environmental Sequencing Unmasks Marine Metazoan Biodiversity. Nat. Commun. 2010, 1, 98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez, A.; Eckert, E.M.; Artois, T.; Careddu, G.; Casu, M.; Curini-Galletti, M.; Gazale, V.; Gobert, S.; Ivanenko, V.N.; Jondelius, U. Human Access Impacts Biodiversity of Microscopic Animals in Sandy Beaches. Commun. Biol. 2020, 3, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. UPARSE: Highly Accurate OTU Sequences from Microbial Amplicon Reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Edgar, R.C. UNOISE2: Improved Error-Correction for Illumina 16S and ITS Amplicon Sequencing. BioRxiv 2016, 81257. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing 2021; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Paradis, E.; Schliep, K. Ape 5.0: An Environment for Modern Phylogenetics and Evolutionary Analyses in {R}. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Ficetola, G.F.; Taberlet, P.; Coissac, E. How to Limit False Positives in Environmental DNA and Metabarcoding? Mol. Ecol. Resour. 2016, 16, 604–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).