Abstract
It is often of interest to biologists to evaluate whether two populations are alike with respect to a similarity index; assessing the numbers of shared species is one way to do this. In this study, we propose two Turing-type estimators for the probability of discovering new shared species and two jackknife-type estimators for the number of shared species in two populations. We use computer simulation and empirical data analysis to evaluate the proposed approach. The jackknife-type estimators provide stable and reliable estimates, for both the probability of discovering new shared species and the number of shared species. We also compare the jackknife-type estimates with that of using sample coverage to estimate the number of shared species. The estimate of using sample coverage has better performance in the case of even populations, while the jackknife-type estimates have smaller bias in the case of unbalanced populations. When combined with a stopping rule based on the probability of observing new shared species, confidence intervals based on the proposed jackknife-type estimators can provide better coverage probability for the true number of shared species. The jackknife-type estimates can provide coverage probability close to 0.95 in all examples.
1. Introduction
Species diversity is a feature often used to compare populations. Among all measures, the number of species is a simple descriptor but its estimation is remarkably challenging. Indeed, there were over 550 papers on the topic as of 1991, as summarized by Bunge and Fitzpatrick [1]. Our primary interest in this paper is to study and evaluate the estimators of the number of shared species in two communities, borrowing ideas from the estimators of number of species in one population.
Good proposed an elegant idea for estimating the probability of discovering new species (Turing’s estimator) [2], using only the information of species observed exactly once in the sample. Following Good’s idea, Burnham and Overton applied a jackknife technique to obtain a nonparametric estimator of the number of species in one population based on the distribution of observed species frequency [3]. Chao and Lee proposed an alternative nonparametric estimator based on the concept of sample coverage [4], and Chao et al. later modified this estimator using the information of species appearing not more than 10 times in the sample [5].
The estimation of the number of shared species in two populations can be generalized from the species richness in one population. Using the information of sample coverage, Chao et al. proposed a nonparametric estimator of the number of shared species [6] and Chuang et al. developed three different types of jackknife estimators [7]. However, neither of these approaches takes advantage of jackknifing the sample and we don’t know if there are enough observations to make the final decision. In a different approach, Yue and Clayton modified Good’s idea and proposed an estimator for the probability of observing new shared species in two populations [8]. They used this probability as an indicator to stop collecting more observations, which can lower overall study cost, in comparing species similarity between two populations. Therefore, in addition to developing two jackknife-type estimators for the number of shared species and comparing them to that by Chao et al. [6], we also evaluate if it is possible to use stopping indicator for estimating the number of shared species.
Note that, in addition to the proposed two jackknife-type estimators of the number of shared species in two populations, we also consider the feasibility of using the probability of observing new shared species as stopping rule. In the next section we briefly review the concept behind jackknife estimators, including Turing-type estimates of the probability of discovering new shared species. We then develop two nonparametric estimators for the number of shared species in two populations and discuss the variances of those estimators. We will use computer simulations and empirical analysis of varies data sets to evaluate the proposed approach.
2. Methodology
Suppose there are two populations and let and denote the species proportions of the two populations, where s is the number of distinct species in the pooled communities. In other words, if we randomly select a single sample, then the probabilities of observing the species i are pi and qi (1 ≤ i ≤ s) in populations 1 and 2, respectively. Let be the number of shared species and, without loss of generality, let the species 1, 2, …, and be the shared species in both populations. Also, let and denote the numbers of times of species i is observed based on n observations from each of populations 1 and 2, respectively, and let denote the number of observed shared species from n (pairs of) observations.
The probability of observing a previously unseen species (which is listed) in a single sample draw from population 1 can be expressed as where is indicator function [9]. The Turing estimate for the probability of discovering new species is based on the number of species appearing exactly once in the sample, i.e., where is the number of singletons [2]. However, Turing’s estimate has a positive bias since is larger than [9].
The Turing-type estimator for the probability of discovering new shared species can be derived similarly. First, the probability of discovering new shared species after n observations is
where (p1, p2, …, ps) and (q1, q2, …, qs) are the species proportions of the two populations [8]. We propose two Turing-type estimators, denoted and , based on Equation (1): the first is from [2] and the other is a direct extension from the one-population case. The first estimator is derived from , and is used to replace as in Turing’s estimate. Thus, can be expressed as
Equation (2) is the probability that a shared new species occurs at the nth sample point, given the sample statistics for Since Turing’s estimate has a positive bias, is also biased, as described in the Appendix of [8].
Another Turing-type estimator is to treat the two populations as two independent populations and then the two-population Turing’s estimate is the sum of Turing’s estimates from each population. Specifically, for the new shared species, we only consider the case where they are observed in one population but not yet observed in the other population. The estimator is expressed as
The difference between and is , and thus has the potential to reduce the bias of ; in fact this will be shown to be the case in the next section.
We next develop jackknife-type estimators for the number of shared species similar to those used for the number of species [3]. For a single sample, their (first-order) jackknife estimate of the number of species in a single population is given by: where is the number of observed species and is the number of singletons. A similar idea can be applied to the case of two populations and we can use the number of species appearing once to develop the jackknife type estimate of number of shared species. Let (or ) be the numbers of species appearing exactly once in the first (or second) population, which also appear at least once in the other population. Let be the number of species appearing exactly once in both populations. Then, by analogy of using the singletons and the Equations (2) and (3), the jackknife-type estimators (singleton) for the number of shared species can be expressed as and . The derivation of these two estimators is outlined in Appendix A.
Using techniques similar to those used in the previous study [3], the jackknife-type estimators can also be expressed in the following form,
where
, and is the number of species appearing exactly i times (i ≥ 1) in either population.
One of the advantages of using the jackknife procedure is that the variance of the jackknife-type estimators can be derived easily. The variance of the first estimator is
The second estimator can also be expressed in a form similar to Equation (5):
with variance
where . Since the difference between the two estimators from Equations (2) and (3) for the probability of discovering new shared species is , the difference between the two jackknife-type estimators from Equations (4) and (7) is .
Note that the jackknife-type estimators in Equations (4) and (7) are constructed similar to the form of jackknife estimator for one population, where the estimate of number of species is the sum of the number of observed species with multiplying the number of singletons in the sample. Interestingly, Chao’s estimator for the number of shared species [6] also has the same form as Chao’s estimator for the number of species in one population [4,5]. In particular, using a homogeneous population case as an example, Chao’s estimator for the number of shared species can be expressed as , where is the number of observed rare shared species and is the estimate of sample coverage for the shared species. Using our notation, is the number of observed shared species appearing at most 10 times in both populations (i.e., rarely), and the sample coverage estimate is , with
and
3. Simulation Studies
We first use computer simulation to evaluate the performance of and , especially when used to form stopping rules that lead to estimates of the number of shared species, and compare three nonparametric estimators of the number of shared species in two populations: , , and Chao’s estimate [6]. As pointed out in the previous study [8], the probability of observing new shared species can be used as a stopping indicator for sampling. We shall extend its role to develop the estimate for the number of shared species, and use the probability as a stopping indicator.
Similar to Yue and Clayton [8,10], we use geometric distributions to model the distribution of species within each population. That is, we assume that and likewise for . In addition, we assume that the shared species are dominant in both populations [10,11]. We shall first evaluate the performance of estimators for the probability of discovering new shared species and , using v(n) as a benchmark. Note that the computer simulations conducted in this study are based on an Intel-based PC, using the statistical software R, version 2.12.0. All results are from 1000 simulation replications for each case.
Example 1. Suppose that the species proportions of the two populations follow geometric distributions and with α = 0.9, 0.8, 0.7, and 0.6. Note that a larger α indicates a more even (or balanced) population structure, while a smaller α means that some species are dominant and the population structure is more unbalanced. Let the numbers of species in the two populations be 100, the number of shared species be 20 or 50, and the shared species are the most dominant species in each population. The results are each based on 1000 simulation runs.
Table 1 lists the probability and its estimates of discovering new shared species given that n observations are taken from each population and that the species proportions follow the geometric distributions stated above. As expected, the estimate has a larger bias, especially in the cases of smaller sample sizes. On the other hand, the estimate performs better in terms of bias for all cases and it is not influenced by the population structure (i.e., even or unbalanced). It seems that the deduction of from is reasonable since has a positive bias [8], although it looks like could be under-biased from Equation (3). Nonetheless, based on these simulation results, it appears that the estimate is a better estimate for the probability of discovering new shared species.

Table 1.
Probability of Discovering New Shared Species. Numbers of species in two populations are & , the number of shared species is or , and species proportions follow Geom(α).
We shall continue the comparison of estimators for the number of shared species, despite the fact that the estimate is over-biased. Note that both the original and modified versions of Chao’s estimates are considered in this study. However, we will only show the modified Chao’s estimate (denoted as for the rest of this study) since it performs better than the original Chao’s estimate. In the next example, we compare two jackknife-type and Chao’s estimators for the number of shared species in two populations.
Example 2. We now consider the comparison of estimates for the number of shared species using the same settings as in Example 1 and show the averages and variances of estimates from 1000 simulation runs. In addition, we also include the case where the species proportions follow the Zipf’s law, similar to that in [6]. We assume that with δ = 1, 1.5, and 2, and show only the averages of estimates. In general, more observations are required in the case of more unbalanced populations (i.e., smaller α and larger δ). To simplify the discussion, the cases where with α = 0.9 and 0.7 will be used. The details of the simulation results can be found in Appendix B and Appendix C.
We first show the comparison of two jackknife-type and Chao’s estimators for the number of shared species (Figure 1 and Figure 2). In the even population case, Chao’s estimate has the best performance for both = 20 and 50. It converges much faster and does not have larger bias like the jackknife-type estimates. On the other hand, for the unbalanced population cases, the jackknife-type estimators (especially ) have a smaller bias, for both = 20 or 50. But all estimators converge very slowly in the case of larger and unbalanced populations. It seems that, by analogy, the overbiased property of also carries over to the estimation of number of shared species in . In particular, since the behaviors of singletons can be very discrete in the cases of unbalanced populations, it is reasonable to be conservative and choose a slightly overbiased estimator.
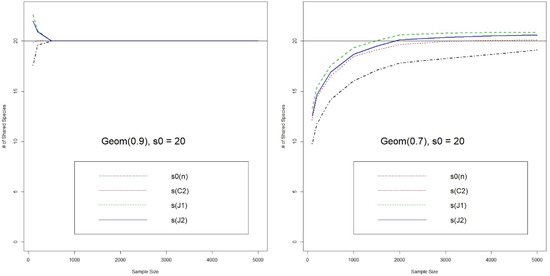
Figure 1.
Estimates for the Number of Shared Species (Numbers of species in two populations are & , and the number of shared species is ; J1 & J2: 1st & 2nd Jackknife estimates, C2: Chao’s estimate, s0(n): number of observed shared species).
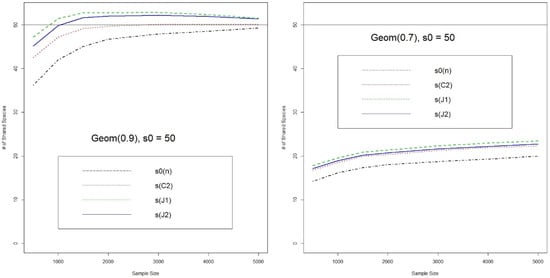
Figure 2.
Estimates for the Number of Shared Species (Numbers of species in two populations are & , and the number of shared species is ; J1 & J2: 1st & 2nd Jackknife estimates, C2: Chao’s estimate, s0(n): number of observed shared species).
Note that, although we found that Chao’s estimate performs well in the even population case, it can still produce undesirable results. For example, assume that the species proportions satisfy and , and that the number of shared species is 80. Under this setting, there will be no observed rare shared species once the sample size is big enough. As shown in Table 2, we cannot compute Chao’s estimate since all observed shared species appear more than 10 times. On the other hand, the jackknife-type estimators converge to the true number of shared species as the sample size increases.

Table 2.
Estimates for the Number of Shared Species (Numbers of species in two populations are & , the number of shared species is , and species proportions are from Geom(0.99) and Geom(0.9); J1 & J2: 1st & 2nd Jackknife estimates, C2: Chao’s estimate, s0(n): number of observed shared species).
Next we compute the Monte Carlo variance of the two jackknife-type and Chao’s estimators, and also the variance of jackknife-type estimators from Equations (6) and (8). Since all estimators converge to the true value fairly fast in the even population case (α = 0.8 & 0.9), we will focus on the case of α = 0.7. (Appendix B shows the details of simulation results for all cases with α = 0.9, 0.8, 0.7, and 0.6). Figure 3 shows the sample variances of two jackknife-type and Chao’s estimators from 1000 runs. On average, the jackknife-type estimators have smaller and smoother variances ( the smallest). The variance of Chao’s estimate jumps up and down even when there are 2000 or more observations, which might indicate that Chao’s estimate can still be unstable even when there are a lot of observations.
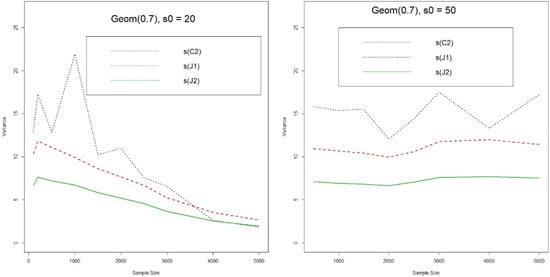
Figure 3.
Variance of different Estimates for the Number of Shared Species (J1 & J2: 1st & 2nd Jackknife estimates, C2: Chao’s estimate, s0(n): number of observed shared species).
We shall also check whether Equations (6) and (8) can provide reliable approximation to the variance of jackknife-type estimators, by using the sample variance from Monte Carlo simulation as the baseline. Figure 4 shows the variances from Equations (6) and (8) and those from Monte Carlo simulations which are marked with “Monte Carlo”. Similar to the overbias in estimating the number of shared species, the variance of from Equation (6) is always larger than that from Monte Carlo simulation. In contrast, the variance Equation (8) for is a good approximation to that of Monte Carlo simulation. In any case, the variance formulae for the jackknife-type estimators provide fairly reliable approximations.
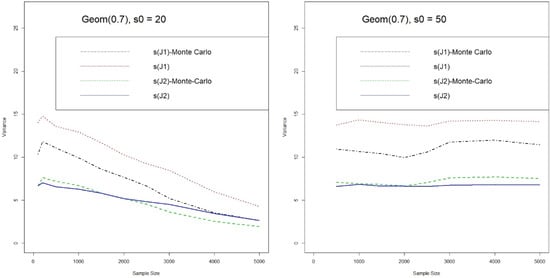
Figure 4.
Variance Comparison (Sample vs. Monte Carlo) of Jackknife-type Estimates for the Number of Shared Species (J1 & J2: 1st & 2nd Jackknife estimates).
4. Empirical Studies
In addition to the simulations of the previous section, we also use empirical data to evaluate the three estimates of shared species. Four data sets are considered in this study: the first two are data on wild birds and on crabs [8], the third one is based on forest data, and the last one comes from Chinese literature. Also, we consider the case of sampling with replacement since there are finitely many observations in all data sets. In other words, we are using these data sets as representing the true populations, and our sampling emulates sampling from these populations.
Example 3. The Taiwan Bird data [11] contain two communities of wild birds consisting of 184 different species and 144,963 observations. There are 155 and 149 species in population 1 and 2, respectively, and 111 shared species (more than half are shared species). The shared species are dominant in each population, similar to the setting in the previous section. We therefore expect that the results of the jackknife-type estimates to be similar to those in the previous section.
Table 3 shows the estimates of the probability of discovering new shared species and the estimates of the number of shared species as a function of sample size. Moreover, we also calculate the coverage probability for the number of shared species; that is, the probability that the confidence interval covers the true number of shared species. We expect this interval to behave approximately like a 95% confidence interval and so this coverage probability is intended to verify whether the estimate can be used in building confidence intervals. Note that is the estimate for the number of shared species, and its variance is calculated via 1000 simulation runs. Note that we can also use the variances via Equations (6) and (8) to compute the coverage for jackknife-type estimators (and the results of coverage probability are fairly close). However, the variance of Chao’s estimator can only be computed via Monte Carlo simulation, and we shall compute the variances all based on simulation.

Table 3.
Taiwan’s Bird Data (Numbers of species in two populations are & and the number of shared species is ; J1 & J2: 1st & 2nd Jackknife estimates, C2: Chao’s estimate, s0(n): number of observed shared species).
From the table we can see that, for the probability of discovering new shared species, again is a better estimate for small and large samples, and is always over-biased. The first jackknife-type estimate of the number of shared species again is the largest among the three estimates, but, unlike the over-biasedness of , it is still smaller than the true when the sample drawn is large. Its variance decreases gradually as the sample size increases and becomes stable when the sample size is around 50,000, where the coverage probability is about 95%. The second jackknife-type estimate has a similar behavior but it requires a larger sample to become stable.
Chao’s estimate , on the other hand, does not reach the true number of shared species when the sample size is 51,000, and it might need considerably more samples to reach the true number. It seems that is more conservative in estimating the number of shared species, and its coverage probability is too small even when there are 51,000 observations from each population (about 70% of the original sample size 144,963).
Example 4. The Panama Crab data [12] were collected in two coral communities at two locations in Panama. There are 55 and 50 species in populations 1 and 2, respectively, and 31 shared species, accounting for 74 different species and 5831 observations. Unlike the Taiwan Bird data, the shared species in the crab data are not so dominant and the number of shared species is less than half of the total species.
Among all the examples in these empirical analyses, the crab data have the smallest numbers of shared species and total observations. Because the smaller population in the crab data has about 1100 observations in total, we start with 110 observations from each population and consider only the case where the sample size is a multiple of 110 for computational simplicity (Table 4). Once again, is shown to be better than for estimating the probability of discovering new shared species, no matter what the sample size is. For the number of shared species, has the largest averages and is the smallest. Also, Chao’s estimate performs the best in coverage probability.

Table 4.
Panama’s Crab Data (Numbers of species in two populations are & and the number of shared species is ; J1 & J2: 1st & 2nd Jackknife estimates, C2: Chao’s estimate, s0(n): number of observed shared species).
The jackknife-type estimates never reached 90% of the coverage probability, although their estimates increase gradually and their variances are more stable. The reason why the jackknife-type estimates have smaller coverage probability is the variance, since the averages of are smaller than those of and (and smaller than ). This matches the result that has the smallest variance and smallest coverage probability. However, since has a larger estimate of variance via Equation (6), would have a better coverage probability if its variance were computed from Equation (6).
Example 5. Barro Colorado Island’s Forest Data (We would like express our appreciation to Professor T.J. Shen, Department of Applied Mathematics, National Chung Hsing University, Taiwan, for providing this data set) are collected around the Gatun Lake area in Panama. The forest is separated into 4 regions (or populations): A, AB, D, and P. We choose regions A and AB in this study, containing 308 and 207 species, respectively. The reason for choosing this combination is that there are 207 shared species, i.e., AB can be treated as a sub-population of A, and the number of shared species in the two populations is equivalent to the number of species in AB. Also, the number of observations in region A is 242,083, much larger than that in region AB (5883).
Corresponding to region AB, the largest sample size considered is about two times its number of observations (12,000). As expected, is a good estimate of the probability for discovering new shared species and is always biased (Table 5). The jackknife-type estimates are fairly accurate estimates for the number of shared species, and they also have good coverage probabilities. Their variances decrease smoothly as the sample size increases. On the other hand, Chao’s estimate grows slower, compared to of the jackknife-type estimates. Chao’s estimate does not have a good coverage probability and it is likely that more observations are required.

Table 5.
Barro Colorado Island’s Forest Data (Numbers of species in two populations are & and the number of shared species is ; J1 & J2: 1st & 2nd Jackknife estimates, C2: Chao’s estimate, s0(n): number of observed shared species).
Example 6. The Chinese Novel Data contain two novels from Louis Cha Leung Yung, a famous Chinese writer. He has 10 famous historical novels, written between 1955 and 1972. The two novels chosen are “Fox of Snowy Mountain” (A) and “The Legendary Swordsman Enjoy Itinerant Life” (B) written in 1959 and 1967, respectively. We will treat different Chinese characters as different species. Then, there are 2591 and 3690 species in A and B, and 2457 shared species.
Novels A and B have about 110,000 and 420,000 characters (or observations). Thus, for computational efficiency, the sample size starts at 21,200 observations, about 20% of the observations in Novel A. We found that is a reliable estimate for the probability of discovering new shared species (Table 6). On the other hand, although is slightly over-biased, it is still a good estimate and is about 10% to 20% over-biased.

Table 6.
Chinese Novel Data (Numbers of species in two populations are & and the number of shared species is ; J1 & J2: 1st & 2nd Jackknife estimates, C2: Chao’s estimate, s0(n): number of observed shared species).
Neither Chao’s estimate nor the jackknife-type estimates have desirable results in coverage probability. Unlike the previous three examples, the coverage probability does not stabilize as the sample size increases. The coverage probability of Chao’s estimate is always 0, and those of the jackknife-type estimates decrease to 0 after reaching the maximum. It seems that the jackknife-type estimates can still provide useful information about the number of shared species, but the sample size is a very important factor. This result is similar to the optimal stopping for estimating the similarity index between two populations in Yue and Clayton [8]. Since it is not possible to sample all the individuals in the populations, knowing the appropriate time to stop sampling would be more feasible and cost efficient. Together with the probability of discovering new shared species and , the jackknife-type estimators provide fairly accurate estimates to the number of shared species. For example, it seems that or is a possible candidate for stopping, where the coverage probability of jackknife-type estimators is around 0.95.
5. Conclusions
The rare species are often more important than dominant species in the estimation of the probability of discovering new species and the number of species in a population [13,14,15]. For example, two popular methods, Turing’s and Chao’s estimates, use the information on rare species for estimation of new species. The estimation of shared species in two populations can be directly extended from the methods used in one population. In this study, we establish jackknife-type estimates of shared species and compare it with that developed by Chao et al. [6].
First, we proposed a modified estimate for the probability for discovering new shared species in two populations, in order to reduce the bias of the estimate suggested by Yue and Clayton [8]. Then, based on these two estimates for discovering new shared species, we extended the jackknife-type estimate of Burnham and Overton [3] to obtain two estimates for the number of shared species in two populations. We compare these two jackknife-type estimates with that of Chao et al. [6]. Simulation studies and real examples confirm that the modified estimate has a smaller bias in estimating the probability of discovering new shared species, no matter what the sample size is.
For the number of shared species, the performance of estimates is influenced by the population structure and the sample size. In general, Chao’s estimate has a smaller bias and converges to the true value much faster in the case of more even populations, and the jackknife estimates are better in the case of unbalanced populations (i.e., smaller α and larger δ). In the case of more even populations, all estimates are accurate even when there are not many observations. On the other hand, in the case of unbalanced populations, more observations are required and the jackknife-type estimates have a smaller bias. In addition, the variance of jackknife-type estimates can be approximated by the derived equations, which can be convenient in empirical analyses.
The coverage probability calculated in the real examples shows another difference between the jackknife and Chao’s estimates. Applying a normal approximation for a 95% confidence interval, we evaluated the probability of covering the true number of shared species. Except for the Panama Crab data, Chao’s estimate does not have coverage probability near 0.95. In contrast, both jackknife-type estimates can provide coverage probability close to 0.95 in all examples, provided that there are enough observations. Based on our experience, it seems that (or ) is a possible useful indicator for stopping sampling. When the sampling cost the jackknife-type estimate derived from in Yue and Clayton [8] has coverage probability close to 0.95 (except for the Panama Crab data). A similar result holds for another jackknife-type estimator . This is similar to the results in Yue and Clayton [8], although their interest is in the similarity index.
Note that we also conducted supplementary simulations to explore group sampling, group sampling with variable (i.e., random) numbers of observations, and sampling with one group observed sequentially and one group observed through a fixed sample. By and large the conclusions remain the same. It seems that the paired sampling represents the slowest incremental rate of accruing information and provides a useful baseline for examining the estimators.
As an alternative to our approach, using the sample coverage is another feasible approach for estimating species numbers, and there has been considerable success in using that for single populations. Among others, Chao and her colleagues have made important contributions to that topic [4,6]. However, addressing the sample coverage for estimating shared species requires a study separate from the work presented here.
Author Contributions
J.C.Y. proposed the general methodology, designed the simulation study and empirical analysis, and drafted the manuscript. M.K.C. designed the simulation study, interpreted the results, and helped to draft the manuscript. C.-R.H. conducted simulation studies and summarized the research results. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported in part by a grant from the National Science Counsel in Taiwan, NSC 96-2118-M-004-003.
Institutional Review Board Statement
This study does not involve human participants, human tissue, or animal samples. The data used are open accessed and this study does not need ethical approval.
Informed Consent Statement
Not applicable.
Data Availability Statement
There are four data sets used in this study, all are open and accessible to the public.
Acknowledgments
The authors are grateful for the funding from the National Science Counsel in Taiwan, NSC 96-2118-M-004-003.
Conflicts of Interest
The authors declare that they have no competing interest.
Appendix A. Derivation of Jackknife-Type Estimators
The first estimator can be treated as a combination of jackknife-type and moment-type approaches. The jackknife-type estimate of the number of species for a population is the number of observed species plus , where is the probability of observing new species. Then, plugging into the Turing-type estimator for the probability of discovering new shared species , we obtain the jackknife-type estimator of the number of shared species in two populations.
The second estimator is based on jackknife technique noting that there are quite a few approaches to obtain jackknife estimators. Considering all possible combinations (i.e., permutations) is a natural choice, similar to Burnham and Overton [3]. There are two ways for counting possible combinations: one is pair-wise and the other is completely random. For the pairwise case, the observations are drawn in pairs, so (X(i), Y(i)) are chosen together, where X(i) and Y(i) are the ith sample in the first and second populations, i.e., there are n possible jackknife subsamples if one pair of observations are omitted each time. For the completely random case, the observations are drawn randomly, so (X(i), Y(j)) are chosen and it is possible i ≠ j, which means that there are possible jackknife subsamples if one observation is omitted from each population. Since the derivation of jackknife-type estimators are obtained via listing all possible combinations, we will only show the final results.
In the pair-wise case, depending on whether X(i) and Y(i) are both shared species and singletons, the jackknife-type estimator lies between two values
where is the number of species appearing exactly once in first population and at least once in the second population. The definition of is similar. The upper bound in the pair-wise case can be treated as a direct extension of the jackknife estimator of Burnham and Overton in the one population case, and thus we define the upper bound as the second jackknife-type estimator . The derivation of jackknife-type estimators in the completely random case is similar and the jackknife-type estimator equals . Asymptotically, the jackknife-type estimator in the completely random case is very similar to those in the pair-wise case (closer to the upper bound).
In addition to the previous two jackknife-type estimators, it is also possible to derive other types of two-sample jackknife estimators. For example, Chuang et al. [7] used the jackknifing technique by Schechtman and Wang [16] and proposed a jackknife estimator .
We can see that these jackknife-type estimators have similar form, and only differ in how we weight the singletons. The differences would be more obvious in the case of small samples and are small if there are many observations. Still, there is another reason for choosing and . The proposed estimators are based on the probability of discovering new shared species and , and these probabilities can be used as stopping indicators. A detailed discussion of this can be seen in our empirical study (Section 4).
References
- Bunge, J.; Fitzpatrick, M. Estimating the Number of Species: A Review. J. Am. Stat. Assoc. 1993, 88, 364–373. [Google Scholar]
- Good, I.J. The Population Frequencies of Species and the Estimation of Population Parameters. Biometrika 1953, 40, 237–264. [Google Scholar] [CrossRef]
- Burnham, P.K.; Overton, S.W. Estimation of the Size of a Closed Population When Capture Probabilities Vary among Animals. Biometrika 1978, 65, 625–633. [Google Scholar] [CrossRef]
- Chao, A.; Lee, S.-M. Estimating the Number of Classes via Sample Coverage. J. Am. Stat. Assoc. 1992, 87, 210–217. [Google Scholar] [CrossRef]
- Chao, A.; Ma, M.; Yang, M.C.K. Stopping Rule and Estimation for Recapture Debugging with Unequal Detection Rates. Biometrika 1993, 80, 193–201. [Google Scholar] [CrossRef]
- Chao, A.; Hwang, W.; Chen, Y.; Kuo, C. Estimating the Number of Shared Species in Two Communities. Stat. Sin. 2000, 10, 227–246. [Google Scholar]
- Chuang, C.; Shen, T.; Hwang, W. Estimating the Number of Shared Species by a Jackknife Procedure. Environ. Ecol. Stat. 2015, 22, 759–778. [Google Scholar] [CrossRef]
- Yue, J.C.; Clayton, M.K. Sequential Sampling in the Search for New Shared Species. J. Stat. Plan. Inference 2012, 142, 1031–1039. [Google Scholar] [CrossRef]
- Rasmussen, S.L.; Starr, N. Optimal and Adaptive Stopping in the Search for New Species. J. Am. Stat. Assoc. 1979, 74, 661–667. [Google Scholar] [CrossRef]
- Yue, J.C.; Clayton, M.K. An Overlap Measure based on Species Proportions. Commun. Stat.-Theory Methods 2005, 34, 2123–2131. [Google Scholar] [CrossRef] [Green Version]
- Yue, J.C.; Clayton, M.K.; Lin, F. A Nonparametric Estimator of Species Overlap. Biometrics 2001, 57, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Smith, W.; Solow, A.R.; Preston, P.E. An Estimator of Species Overlap Using a Modified Beta-binomial Model. Biometrics 1996, 52, 1472–1477. [Google Scholar] [CrossRef]
- Gaston, K. The Importance of Being Rare. Nature 2012, 487, 46–47. [Google Scholar] [CrossRef] [PubMed]
- Mi, X.; Swenson, N.G.; Valencia, R.; Kress, W.J.; Erickson, D.L.; Pérez, A.J.; Ren, H.; Su, S.; Gunatilleke, N.; Gunatilleke, S.; et al. The Contribution of Rare Species to Community Phylogenetic Diversity across a Global Network of Forest Plots. Am. Natuarlist 2012, 180, 17–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schechtman, E.; Wang, S. Jackknifing Two-sample Statistics. J. Stat. Plan. Inference 2004, 119, 329–340. [Google Scholar] [CrossRef]
- Shen, T.-J.; Chen, Y. Predicting the Number of Newly Discovered Rare Species: A Bayesian Weight Approach. Conserv. Biol. 2019, 33, 444–455. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).