Abstract
A detailed phylogeny of bumblebees is urgently needed to understand speciation and biogeographic diversification in the Neotropical region. We sequenced autosomal and mtDNA loci from nine Brazilian bumblebee species and compiled it with the data already available to obtain highly resolved phylogenetic trees with fossil-calibrated dates. The ancestral Bombus lineage was estimated to diversify between 47.08 and 34.27 million years ago (Ma) in the Holarctic region, but largely restricted to the eastern Old World. The Neotropical region was initially colonized in the Late Miocene, where bumblebee diversification was shown to be consistent with geologic and climatic events of the Late Cenozoic. Neotropical bumblebees likely originated from Nearctic lineages, which dispersed towards South America after 29 Ma.
Keywords:
Bombus; historical biogeography; BioGeoBears; LTT; bumblebee biogeography; diversification 1. Introduction
Bees are one of the major pollinating insects that are widely studied in both basic and applied research [1]. However, the evolutionary history of diversification of many subfamilial lineages is still poorly understood, particularly in the Neotropical region [2,3].
Corbiculate bees (Apini sensu Roig-Alsina in [4]) constitute a clade characterized by a spoon-shaped hind tibiae with reduced scopae forming pollen baskets known as corbiculae. This clade is composed of four monophyletic extant groups (subtribes Euglossina, Bombina, Meliponina, Apina sensu Roig-Alsina in [4]) and three extinct groups (subtribes Melikertina, Electrapina, and Electrobombina) [1].
Among the extant corbiculate bees, Bombina, consisting of about 270 living species (http://www.nhm.ac.uk/bombus, accessed on 3 March 2017) of the genus Bombus Latreille, 1802, is the only subtribe with a high diversity of species in temperate and cold environments (120 species in the Palearctic region, 113 species in the Oriental region, 57 species in the Nearctic region, 29 species in the Arctic region, and 23 species in Japan). In the Neotropics, Bombus is represented by 32 species, of which 20 are endemic [5] (updated at http://www.nhm.ac.uk/bombus, accessed on 3 March 2017), living in both forests and open areas under different climates. Among these, eight South American species are the only bumblebees known to inhabit tropical lowlands (below the altitude of 1000 m) in the world [1,6]. Williams [7] divided the Neotropics in four faunal regions on the basis of bumblebee distributions (http://www.nhm.ac.uk/bombus, accessed on 3 March 2017): (1) Northern (Central America, with nine species); (2) Eastern (South America east of the Andes down to northern Argentina, plus northern Chile; 13 species); (3) Southern (Southern Chile and southern Argentina; five species); and (4) Western (Northern South America down, through the western slopes of the Andes, to northern Chile and Argentina; 20 species).
Both morphological and molecular data have been used to infer the relationships among Bombus species [8,9]. According to Williams [8], the Bombus ancestor has a most-likely Asian origin, with some of its earliest descendant species initially dispersing to Europe and North America and, much later, from North to South America. This scenario was later supported by molecular evidence [10,11].
Hines [11] suggested an initial divergence for Bombus species at around 40–25 million years ago (Ma), when central Asian species dispersed, colonizing all the Palearctic region, including Europe. This period comprises a dramatic global cooling at the Eocene–Oligocene boundary, which may have favored bumblebees due to their adaptation to cold climates [8,12]. Hines [11] also suggested a subsequent bumblebee dispersal towards North America (Palearctic to Nearctic) at about 20 Ma and the dispersal of a few taxa in the opposite direction (Nearctic to Palearctic) at about 4 Ma. Bumblebees would have eventually reached South America at about 7.5 Ma, with early regional divergence events taking place between 7.5 and 6 Ma, and most diversification leading to current species occurring in the last 3.5 Ma [11].
Hines [11] presented also some lineage through-time (LTT) plots showing an increased rate of diversification in recent times. However, her results may be biased by the fact that lineages arising in the recent past are more likely to be represented in the phylogeny than lineages arising in the distant past. This is called “pull of the present” and is a property of the birth–death model [13,14,15,16].
Our understanding of the phylogenetic relationships and diversification rates within Bombus can be improved with the consideration of data derived from several fossil species of the genus described after Hine’s [11] paper. This would help to understand the geographic distribution and the origins of the extant species worldwide [17]. Thirteen fossil species of Bombus are known from the Oligocene to the Miocene [18,19]. Among these, B. cerdanyensis Dehon, De Meulemeester and Enge, 2014; B. (Bombus) randeckensis Wappler and Engel, 2012; and B. (Cullumanobombus) trophonius Prokop, Dehon, Michez, and Engel, 2017 are the most thoroughly examined fossil species, with the latter two taxa being the only ones with reliable subgeneric classifications, thus providing accurate calibration points [18,19,20,21].
Here, we used new analytical tools and time calibration methods to improve our knowledge on the biogeographic history and diversification of bumblebees, which led to their current distribution. We employed a comprehensive taxon sampling and additional calibration points on the basis of recently described corbiculate fossil species: Eulaema zigrasi Engel, 2014; Oligobombus cuspidatus Antropov, 2014; B. randeckensis; and B. trophonius. We also included the reinterpretation of the fossil Paleoeuglossa melissiflora Poinar, 1998, which belongs to the Eufriesea group [22]. This study aimed at refining our knowledge on bumblebees’ natural history, answering the following questions: (1) Did lineages in different biogeographic regions evolve under different diversification rates? (2) Which were the main geologic processes underlying bumblebee diversification in the Neotropical region?
2. Materials and Methods
2.1. Taxon and Locus Sampling
We employed sequences available on Genbank and BOLD Systems v3 for 255 bumblebee species, 3 Apina species, 6 Euglossina species, and 6 Meliponina species (Table S1). In addition, DNA samples of Brazilian bumblebee species were extracted from one hind leg (dry pinned specimen) or thoracic musculature (fresh specimen) of each specimen using the phenol-chloroform method (see Table S1 for collection locality and voucher information; [23]). The extracted genomic DNA was resuspended in 50 µL of TE buffer. Three mitochondrial genes—cytochrome oxidase I (COI), cytochrome B (CytB), and 16S—and four nuclear genes—arginine kinase (Argk), elongation factor 1-alpha gene (EF1α F2copy), long-wavelength rhodopsin gene (Opsin), and phosphoenolpyruvate carboxykinase (PEPCK)—were sequenced using the primers listed in Table S2. PCR amplification was performed in a total volume of 20 μL with 0.3 units/tube of Platinum® Taq polymerase (Thermo Fisher Scientific, Waltham, MA, USA), 2 mM MgCl2, 1X Platinum® PCR buffer, 0.5 μM of each primer, 200 μM of dNTPs, and approximately 20 ng of genomic DNA. The thermocycler program consisted of 5 min of denaturation at 94 °C, followed by 37 cycles of 30 s at 94 °C, 40 s at 50–57 °C (Table S3), 90 s at 72 °C, and a final extension for 10 min at 72 °C. PCR products were visualized in a 2% agarose gel. All material produced by PCRs generating a single product were purified using polyethylene glycol 20% (PEG) precipitation [24] and further sequenced in both directions with the same amplification primers in an ABI 3130xl Genetic Analyzer using BigDye Terminator v3.1, following the manufacturer’s recommendations (Thermo Fisher Scientific, Waltham, MA, USA). The consensus sequences were obtained using SeqScape® Software v2.6. Here, 48 new sequences were generated (nine 16S, four CytB, eight Argk, nine EF1α F2copy, nine Opsin, and nine PEPCK sequences) for five species belonging to the subgenus Thoracobombus: Bombus bahiensis (five sequences), Bombus brasiliensis (five sequences), Bombus brevivillus (28 sequences), Bombus morio (five sequences), and Bombus pauloensis (five sequences). All inputs and outputs of the analysis described below, as well as the complete data sets, are available in the Supplementary Material.
2.2. Phylogenetic Inference
All sequence alignments for each loci were carried out separately using MAFFT v7.017 [25], implemented in Geneious 8.1.8 [26]. The introns of the nuclear genes Argk, EF1α, Opsin and PEPCK were identified and separated from exons using Mega 7.0.14 [27]. Regions of alignment of genes 12S and 16S, and intronic regions of nuclear genes were removed using gblocks 0.91 b [28,29]. We used the default settings, except for the “allowed gap positions” option, which was set to “with half”. After that, the “12 markers” alignments were concatenated using the software SequenceMatrix v1.8 [30]. We explored the best partitioning schemes and substitution models simultaneously, using PartitionFinder v.2.1.1 [31] under a Bayesian information criterion for the entire matrix.
The aligned full-length concatenated data were employed in the program RaxML version 8 [32] in order to identify rogue taxa, taxa that can change the evolutionary relationships among sets of taxa when included in a phylogeny [33,34]. Bayesian inference (BI) was conducted in MrBayes v.3.2.2 [35] through the Markov chain Monte Carlo (MCMC) method. The BI analyses were made using PartitionFinder results with 300 million generations, 2 runs, 4 chains and with temperature set to 0.05. Convergence of the runs was assessed using the following statistics: standard deviation of split frequencies, potential scale reduction factor (PSRF), and estimated sample size (ESS) for each parameter.
We did not use the results of maximum parsimony (MP) and maximum likelihood (ML) analyses here because in the analysis of large molecular data sets, these methods are outperformed by BI methods [36,37], which accurately reconstruct the position of taxa with up to 95% of missing data, provided that the overall number of loci in the analysis is large [36,38]. Moreover, MP analysis has been shown to present decreased accuracy with large amounts of missing data [39,40]. Despite this, MP and ML analyses were also performed and are described in the Supplementary File S1—methods and results of maximum parsimony (MP) and maximum likelihood (ML) analyses.
2.3. Divergence Time Estimation
Divergence time analyses based on our “aligned full-length concatenated data”, including partitioning scheme and models used in our previous phylogenetic analyses, were performed using BEAST v.2.4.4 [41] implemented on the Sagarana high-performance computer cluster of the bioinformatics facility of the Universidade Federal de Minas Gerais (UFMG). An uncorrelated lognormal relaxed clock model (UCLN) was employed in order to allow for clock rate variation among branches. To apply fossil ages as node calibration points and to optimize the search for optimal ages by starting at a high likelihood in topology space, we constrained the monophyly of Bombus subgenera according to our BI topology (shown in Supplementary File S2—priors for divergence time estimation, and Figures S1 and S2 for BI and BEAST analyses, respectively). Analyses were carried out for 500 million generations with sampling every 1000 generations. Convergence of Markov Chains was assessed in Tracer 1.6 [42] by inspecting the trace and ESS of the parameters. A burn-in of 20% was applied after checking the log-likelihood curves and a maximum clade credibility (MCC) tree with median ages, and their 95% highest posterior densities (HPDs) were subsequently generated using TreeAnnotator 2.4.3 [41].
2.4. Biogeographic Analyses
In order to infer the biogeographic history of bumblebees, we used the R package BioGeoBEARS [43]. The MCC tree obtained from divergence time analyses was used to map the ancestral area probability (Table S4). The R package “ape”—analyses of phylogenetics and evolution [44]—was applied to this tree to remove the outgroup, species synonyms, and species represented by more than one individual. Two sets of distribution areas were employed in the analyses: (1) the biogeographic regions by Williams [7] based on Bombus distribution and available in https://www.nhm.ac.uk/research-curation/research/projects/bombus/regions.html, accessed on 3 March 2017, and (2) an alternative set of distribution areas in which subdivisions of the Nearctic region were ignored and the Neotropics was subdivided according to Morrone’s [45] major geographic regions (see Figure 1), of which, two, “Northwestern South America” and “South American Transition Zone”, were merged. On the basis of the modified MCC tree (with the exclusion of the outgroups, synonyms, and repeated species) and in the shapes of the two distribution areas described above, we carried out unconstrained analyses employing the following models: (1) DEC [46]; (2) DIVA-like [47]; (3) BayArea-like [48]; and a “+j” (a free parameter accounting for the possibility of a founder-event speciation) version of each of them. All six models were compared, and the best fit model was chosen using the corrected Akaike information criterion (AICc) [48,49].

Figure 1.
Map of world bumblebee species by region. Left: biogeographic regions of the world defined for bumblebees modified from Willians [7], and right: subdivisions of the Neotropical region modified from Morrone [45]. A—northern and western regions of the Andes (northwestern South America + South American Transition Zone [45]); B—Amazonian region; C—Chaco region; D—Parana region; E—Andean region; F—Palearctic region; G—Nearctic region; H—Oriental region; I–Sumatra region. The Sumatran region was not represented by any species in the analyses.
3. Results
3.1. Phylogenetic Inference
The full-length concatenated alignment data employed for phylogenetic inference had a total length of 6188 bp (see Table S5 for specimens used and amount of missing data). The best partitioning schemes and substitution models are presented in Table S6. Although two terminals (B. (Psithyrus) coreanus and B. (Thoracobombus) anachoreta) have been shown to behave as rogue taxa, we decided to keep them in our analyses, since their presence did not affect the relationships among the subgenera and other taxa of interest.
All subgenera recognized in Williams’ [50] simplified classification of Bombus were recovered as monophyletic, although some of them were supported by low posterior probabilities (PP). Contrary to previous studies in which Mendacibombus was the sister group to the remaining subgenera [8,9,10], our results indicate Mendacibombus + Bombias to be the sister group of all other Bombus subgenera (Figure S2 and Supplementary File S1), except for BI in MrBayes and MP analyses (Figure S1 and Supplementary File S1).
The relationships among species within some subgenera were not well resolved or were weakly supported (Figure S1). Kallobombus was found to be the sister group of the long-faced (LF) + short-faced (SF) bumblebees. Among the SF the subgenera, Alpinobombus, Bombus s.str., and Pyrobombus formed a monophyletic group (Figures S1 and S7–S10), and the subgenus Melanobombus is sister to a clade including Alpigenobombus, Sibiricobombus, and Cullumanobombus (Figure S1). The relationships among LF bumblebees were the same as those found in previous works [9,10].
Among the Neotropical species, Psithyrus (one species in the northern Neotropical borders), Thoracobombus (15 species in the Neotropics), and Cullumanobombus (12 species) were all well supported subgenera, but the relationships among their species were not fully resolved. Pyrobombus (one species in the Neotropics) was found as a highly supported clade (PP = 1), and the relationships among its species were fully resolved, mostly with high PP values.
Bombus applanatus was recovered as a well-supported lineage within B. brevivillus (high PP), a result also supported by morphologic data. Comparison of the holotype of B. brevivillus with the taxonomic description, two paratypes, and three specimens used for the molecular delimitation of B. applanatus [51] showed the two species to be identical. Thus, B. applanatus is considered here to be a junior synonym of B. brevivillus (Figure S3).
3.2. Divergence Time Estimation and Biogeographic History
The best-fitting model for BioGeoBEARS analysis was BAYAREALIKE+J (see Tables S4, S7 and S8), which suggests “dispersal”, “extinction”, and “jump dispersal” as important factors on the biogeographic history of bumblebees. According to this model, the most likely biogeographic distribution of the most recent common ancestor (MRCA) of all bumblebees was in the Eurasian (Palearctic + Oriental) region.
Our results indicate that the origin of the Bombus occurred at 47.08–34.27 Ma (Figure S2) in the Eurasian region. They also suggest 14 dispersal events from the Palearctic to Nearctic at different periods between the Late Paleogene and Late Neogene. Only one dispersal event was found on the opposite direction (Nearctic to Palearctic) between 6.84 and 0.07 Ma (Figure 2, Figure 3, Figures S2 and S4–S6).
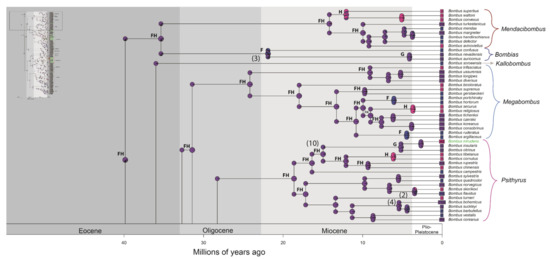
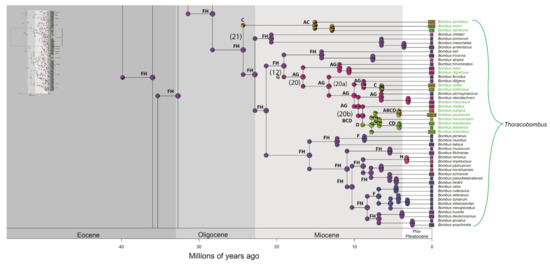
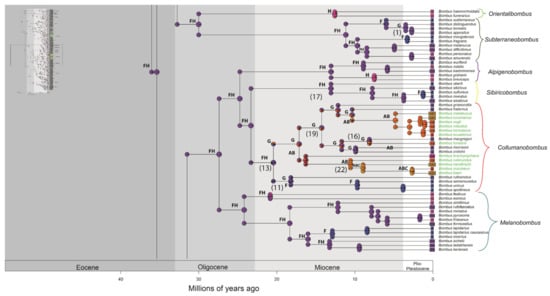
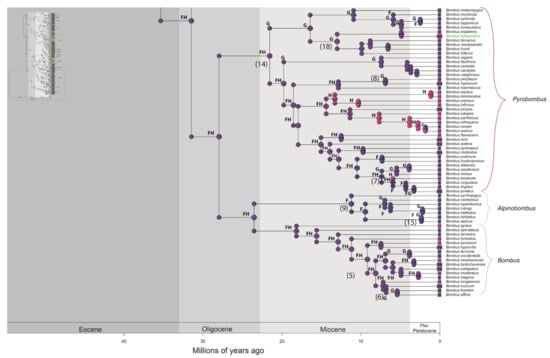
Figure 2.
Distribution of occurrence of data for ancestry inference of bumblebee species. Letters correspond to the areas are shown in Figure 1 (A—northern and western regions of the Andes; B—Amazonian region; C—Chaco region; D—Parana region; E—Andean region; F—Palearctic region; G—Nearctic region; H—Oriental region). Numbers in parentheses correspond to the events shown in Figure 3. Node panels show the probability of each geographic state from BioGeoBEARS analysis. Time tree for bumblebees obtained under an uncorrelated lognormal relaxed clock model calibrated with nine fossils (Supplementary File S2—priors for divergence time estimation, and Figure S2). The names of Neotropical bumblebee species are provided in green.
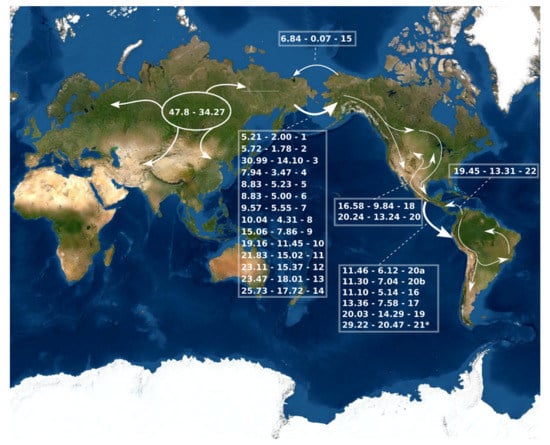
Figure 3.
A simplified summary of the historical dispersal events inferred across Bombus lineages. The timing of these events is presented on HPD 95% (see Figure S2). The continuous arrows refer to the events found in the biogeographic analyses (see Figure 2, Figures S2 and S4–S6). The numbers (1–22) to the right of the date ranges are the events indicated in the phylogenies. * possible artefact of the analyses.
Neotropical Bombus belong to six different lineages, each one representing the invasion event of one ancestor species from the Nearctic region (Figure 2, Figure 3, Figures S2 and S4–S6). These lineages are presented in Table 1 with their estimated dispersal dates. They will be referred to from now on by the names indicated in that table. The oldest of those dispersal events is the one involving the ancestor of the B. morio group, which is suggested by the analyses to have dispersed directly from the Palearctic to the Neotropics. This would have occurred between 29.22 and 20.47 Ma (Figure 2, Figure 3 and Figures S2). One of the species in this lineage, B. morio, subsequently invaded the tropical lowlands of South America (between 17.92 and 8.23 Ma; Figure 2, Figures S2 and S4–S6). The ancestors of the other five lineages of Neotropical bumblebees probably arrived from the Nearctic between 20.24 and 5.14 Ma (Table 1).

Table 1.
Date of invasion, invasion directions, and included extant species of the main Neotropical and South American lineages of Bombus, as recovered in the phylogenetic and biogeographic analyses. Names employed for the lineages are mainly those of https://www.nhm.ac.uk/research-curation/research/projects/bombus/subgenericlist.html, accessed on 3 March 2017. Discrepancies between William’s nomenclature and our lineages are indicated.
Five of the six lineages invading the Neotropical region reached South America between 22.03 and 6.12 Ma. The only exception is B. ephippiatus, which remained in southern North America and northern Central America (Figure 2, Figure 3, Figures S2 and S4–S6). Thoracobombus invaded the Neotropical region twice and reached South America three times. The first invasion event, mentioned above, involved the B. morio group; the second one involved B. opifex + B. bellicosus, between 11.46 and 6.12 Ma; the third one involved the B. pullatus subgroup (Figure 2, Figure 3, Figures S2 and S4–S6). There is also an indication of a reverse dispersal event from South to North America (Table 1).
The ages of the two earliest divergence events in South American groups were estimated between 20.02 and 10.24 Ma (HPD 95%, for the split between B. execellens and the ancestor of B. dahlbomii + B. morio; Table 2) and between 19.45 and 13.31 Ma (HPD 95% for the split between the ancestor of the B. brachycephalus lineage and the ancestor of B. rubicundus + B. baeri + B. coccineus + B. handlirschi lineage). Most of the remaining speciation events appear to have taken place in the last 7 Ma (Table 2).

Table 2.
Dates of diversification for the Neotropical species of Bombus. Tree with the maximum clade credibilities and branch lengths equal to the median ages as calculated from 75,001 post burn-in chronograms (85%). 95% HPD—95% highest posterior density interval. Dates are expressed in million years ago (Ma).
Three lineages left the Andes towards the lowlands of South America. The first one is B. morio, as presented above. Of the other two lineages, one is composed of the sister species B. opifex and B. bellicosus, which originated in the southern portion of the Chaco region between 9.46 and 3.8 Ma. The last of these lineages to reach the lowlands of South America was the B. pullatus subgroup, which originated in the Parana region, between 11.3 and 7.04 Ma, and is composed of six species (Table 1). Of these, B. pullatus currently inhabits the Andean region, marginally reaching northwestern Amazonia (Figure 2 and Figure S2). The five other species occur along the different lowland ecosystems of South America, including tropical forests, savanna, and the semi-desertic Caatinga. An internal clade of the B. pullatus subgroup include three species closely associated with forest environments: B. transversalis, endemic to the Amazonian Forest, and B. bahiensis, endemic to the coastal evergreen forests of southern Bahia and northern Espírito Santo states in Brazil [6,24]. The third species, B. brasiliensis, is mostly associated with semideciduous, riparian, and evergreen forests in southeastern Brazil and surrounding countries, although it is also able to forage in nearby open areas (J.E.S.J. and F.A.S., personal observations).
The most likely biogeographic distributions estimated for the MRCA of the different clades containing Neotropical bumblebees were (1) the Nearctic Region for the ancestors of B. (Psithyrus) intrudens, B. (Pyrobombus) ephippiatus, and Cullumanobombus clades; (2) the Nearctic region and the northern and western regions of the Andes for the ancestor of the Thoracobombus clades, except the one composed by the species B. morio, B. excellens, and B. dahbomii, see below for more information (Figure 2 and Figures S4–S6, Table S4).
Neotropical species included in primarily Holarctic subgenera, B. (Psithyrus) intrudens and B. (Pyrobombus) ephippiatus, diverged between 4.8 and 0.97 Ma, and 7.69 and 2.63 Ma, respectively (Figure S2). The subgenus Cullumanobombus includes 13 Neotropical species, of which 12 are represented in our data. The B. brachycephalus group forms a weakly supported clade in BI analysis (Figure S1), which diverged between the Oligocene and the Miocene (20.03–14.29 Ma; Figure S2). The clade formed by the B. robustus group was well supported in all phylogenetic analyses, with divergence estimated to have occurred between 6.78 and 3.05 Ma (Figure S2).
The subgenus Thoracobombus has 14 Neotropical species. The B. morio group diverged between 20.02 and 10.24 Ma (Figure S2). The other monophyletic clade in this subgenus, formed by the B. pullatus subgroup + B. medius, diverged between 11.81 and 7.45 Ma. This clade and its relationships with other species are not equally recovered in all analyses (see Figures S1 and S2). Bombus bellicosus and B. opifex (11.46–6.12 Ma) also formed a Neotropical monophyletic subgroup as well as B. digressus and B. weisi (15.79–8.27 Ma). Bombus mexicanus grouped with non-Neotropical species (Figures S1 and S2). The diversification of most extant Neotropical bumblebee species occurred during the Middle Miocene and the Pliocene (Figure S2). The latest pairs of sister species to diverge within the Plio-Pleistocene period are Bombus baeri + B. coccineus (4.76–1.02 Ma), B. ecuadorius + hortulanus (2.31–0.39 Ma), and B. robustus + B. vogti (1.91–0.32 Ma) (Figure S2). Diversification times for the Neotropical species of Bombus are summarized in Table 2.
Our results support the hypothesis that the current restrict range and low diversity of Cullumanobombus and Thoracobombus in the Nearctic region may reflect range contraction during their evolution (Figure 2 and Figures S4–S6), due to the extinction of Nearctic ancestral populations.
4. Discussion
4.1. Phylogenetic Inference
Differences between our phylogenetic results and those of previous studies [8,9,10] involve weakly supported clades. Thus, the relationships between and within them should be taken with caution, until future analyses, including more loci and taxa.
Among the Neotropical and especially South American subgenera, our results also support the general relationships among the clades previously recovered (e.g., [8,9,10]), as well as the monophyly of the subgenera recognized by Williams (2008) in his simplified classification. However, the higher resolution of our tree allowed to infer detailed aspects of the biogeographic and evolutionary history of South American species, especially those dwelling in the tropical lowlands, as discussed bellow
4.2. Divergence Time Estimation and Biogeographic History
The factors indicated by BioGeoBEARS to be important on the biogeographic history of bumblebees can be examined in some cases. Dispersal is required, for instance, to explain the range expansion from Eurasia to North America, and from North America to Central and South Americas. This sequence of long-distance dispersal events was likely favored by climate cooling [54,55,56], and was made possible by the behavior and long-distance flight abilities of queen bumblebees [57,58,59,60]. The occupation of those new geographic areas and the diversification in each of them could, then, be made through “jump dispersal” (founder events—Futuyma and Kirkpatrick [61]). Of course, vicariant events were also important in the diversification of the various bumblebee lineages in each of the new geographic regions they reached through dispersal. Vicariance, probably caused by the elevation of the Andes and marine transgressions in South America [55,56,62], may be responsible, for example, for the split of the common ancestor of B. pullatus and B. pauloensis, and of the common ancestor of B. opifex and B. bellicosus, as well as among the species of the B. morio group (Table 2). Moreover, extinction is the most likely explanation for the absence of certain clades in intermediate portions of their ranges, as, for example, the absence of representatives of the B. morio group in North and Central Americas, given its origin in the Palearctic region and present occurrence restricted to South America.
Contrary to the decline in diversification rates observed here (Supplementary File S3), Hines [11] and Condamine and Hines [63] found a fast lineage divergence between 25 and 10 Ma, followed by an even faster diversification towards the present. Their results, however, were likely biased by what has been named the “pull of the present” [13], and that can be avoided by increasing the sample of “recently” diverging species in the data [14,15,64].
Other two explanations for the results by Hines [11] and Condamine and Hines [63], besides sampling artifact [13], could be (i) the time interval for all diversification to occur being smaller since the ages they estimated for each group are shorter than ours, yielding higher diversification rates [65], and (ii) diversity dependence—species speciation/extinction rates are not constant [66,67]. Nevertheless, we do not exclude the possibility of diversity-dependent diversification [67,68]. Here, we only regarded speciation as a more parsimonious explanation than that proposed by Hines [11] and Condamine and Hines [63].
The use of additional fossil species together with newly developed and more powerful statistical methods to calibrate ages of diversification in our phylogeny resulted in reduction of the confidence intervals of date estimates, as compared to those obtained by Hines [11]. Our datings were generally older and not influenced by the amount of missing data (see Supplementary File S4 and Figure S2 for analyses and results on the influence of missing data in topology and in divergence time estimates). For example, the origin of Bombus was estimated by us to be about 4 million years older than previously estimated (47 to 34 Ma; Figure 3). Despite this, an early diversification of the genus is still explainable by the same general climatic cooling of the planet [69,70,71].
As previously suggested by Williams [8] and Hines [11], our results also indicate that dispersal between the Old and the New Worlds occurred solely through the Bering Strait. Both Hines [11] and Kawakita et al. [10] found an asymmetry in the dispersion between the two continents, as we did, with the vast majority of the events occurring from the Palearctic to the Nearctic. However, our results indicate only 14 dispersal events from the Palearctic to the Nearctic, as opposed to 18 events found by Hines [11], between the Late Paleogene and Late Neogene (Figure 3). Accordingly, while she found three events of inverse dispersal, from the Nearctic to the Palearctic (<4 Ma), only one such inverse dispersal event was found by us (6.84– 0.07 Ma). The two dispersion events from the Nearctic to the Palearctic that are not indicated by our results involve the clade including B. lapponicus, B. sylvicola, B. bimaculatus, and B. monticola in the subgenus Pyrobombus (see Figures S5 and S6). According to Hines [11], those species may represent a hard polytomy involving vicariance speciation processes occurring at approximately 3 Ma. However, our results partially solved this polytomy, indicating that B. lapponicus and B. sylvicola are sister species (PP = 1). The other two species, however, still alternated as the sister to the rest of the clade. According to our results, all speciation events in this clade occurred between 8.5 and 1.2 Ma and the origin of B. lapponicus and B. sylvicola occurred between 4.3 and 1.2 Ma. Thus, the number of invasions of the Palearctic, from the Nearctic, by species in this lineage is unclear, but whatever may have happened, it was probably directed by vicariance events due to land mass separations in the Bering Strait around 3.5 Ma [54].
Generally, the dispersion events toward North America initiated and intensified with the drastic decline in global temperature and the related vegetations substitution at ca. 28 Ma [54]. The trans-Beringean dispersion of bumblebees between 23 and 5 Ma was dominated by boreal species, as in other taxa [54,71]. Some examples of such trans-Beringean dispersion involve the ancestors of cold-adapted sister species, of which one extant taxon is Palearctic and the other Nearctic [10,11], as B. perplexus (Nearctic) and B. hypnorum (Oriental and Palearctic), between 10.4 and 8 Ma (Figure 2, Figure 3, Figures S2 and S4–S6), and between B. affinis + B. franklini (Nearctic) and B. lucorum (Oriental and Palearctic), between 8.8 and 6 Ma (Figure 3). After this period, in Late Pliocene, the dispersion events slowed down (Figure 3).
Our results indicate anachronic dispersals of six different lineages to South America, although with great overlap of timings (Table 1, Figure 3). Most importantly, we observed much older dates for the colonization of South America—while the oldest possible date indicated by Hines [11] is 15–7 Ma, the oldest one indicated by our analysis is 29–20 Ma. However, this oldest event is relative to the B. morio group, which is now extinct in Central and North America. The current presence of a representative of this lineage in the intervening regions might have led to her estimation of a more recently arrival in South America. Despite this, even the second oldest lineage to reach South America according to our data (20–14 Ma, for the B. brachycephalus group) is much older than the oldest arrival suggested by Hines [11]. Another great discrepancy involves the age of the reversal dispersion from South to Central America, which is estimated to have occurred less than 1 Ma by Hines [11] and between 19 and 13 Ma by our results.
Our results suggest the colonization of South America by bumblebees to have been initiated before the Panama Isthmus was even only partially uplifted (between 15 and 13 Ma [72]). However, in this period, the sea level was lower and continental masses were closer, possibly allowing for faunistic dispersal [73,74]. Bumblebees are known to be capable of dispersing long distances through continuous corridors of suitable habitat, but they appear to be poor travelers at long distances across wide barriers of severely unsuitable habitats to establish populations in previously unoccupied areas [75]. Notwithstanding, some existing data indicate that B. morio individuals are able to cross relatively wide water barriers, between islands and continents [76]. Thus, a small set of islands present before the complete uplifting of the isthmus may have been sufficient as stepping-stones for the crossing of bumblebees from Central to South America. Coincidently or not, B. morio belongs to the first bumblebee lineage to enter South America.
Our dating analysis also indicated an early diversification of South American lineages to have started between 20.02 and 10.24 Ma (Table 2), coinciding with the uplift of the Andes (mid Miocene), which may have created orographic and/or climatic barriers, the latter due to changes in the geographic configuration of South America [62,77]. Most of the remaining speciation events appear to have taken place in the last 7 Ma (Table 2).
Two different lineages colonized the tropical lowlands of South America. This was accomplished in two different moments by the B. morio and B. pullatus groups, both belonging in Thoracobombus (Table 1). The B. morio group has its probable ancestral area in the Andes (Figures S5 and S6), between 29 and 20 Ma, a period marked by the beginning of the modifications in the Andean arch (ca. 25 Ma), and by a glaciation event [71] and lowering of sea levels [78]. Once it colonized the Chaco region (Figure 1, Figure 2 and Figure S4), the ancestor population of the B. morio group may have been isolated by one or more of the different transgressions of the Atlantic Ocean (three of them occurred between ca. 17 and 15 Ma), which isolated small islands and three large land masses: eastern Amazonia, the Andean region, and the Parana region plus part of the Chaco (Figure 6 of [78]). The isolation of such regions may have caused, in different periods, both the divergence of B. excellens from the common ancestor of B. morio + B. dahlbomii, and the divergence between B. morio and B. dahlbomii.
The origin of the South American tropical lowland bumblebees in the B. pullatus subgroup is best explained by a dispersion event from the Andes to the Parana Region during the Miocene (ca. 18 to 7 Ma), which was followed by range expansion to the Chaco and Amazonian regions still during the Miocene (Figure 1, Figure 2 and Figure 3).
According to our results, the ancestral range of the B. pullatus subgroup is the Parana Region (11.3–7.04 Ma; Figure 1, Figure 2 and Figure S4), from which different species would have invaded the Amazonian Forest and the Chaco region. This would imply an improbable direct jump between the Andean region and the eastern forests of South America (accordingly, this ancestral range was recovered with low probability). It seems reasonable to expect that the ancestor of the B. pullatus subgroup has left the Andes, reaching the Atlantic Forest after occupying the Chaco region. The populations of this subgroup originally occupying the Chaco region may have been extinct due to the Atlantic Ocean transgressions, which flooded most of this region (Figure 6 of [78]). The biogeographic history of the B. pullatus subgroup recovered in our results began after the Atlantic Ocean transgressions, when the previously submerged areas were exposed, forming large plains (ca. 11–3 Ma). The origin and diversification of the B. pullatus subgroup occurred during a period of intense diastrophism (late Miocene–Pliocene), marked by a general climatic cooling, interrupted at times by warmer periods [71,78]. Considering the ecological similarities of the species and the biogeographic analyses, we infer that the MRCA of the B. pullatus sub-group occupied the open areas in a large region, including Amazonia, Chaco, and Parana regions, between 9 and 5 Ma, when submerged areas were being replaced by plains [62,78].
Bombus brevivillus, the first species to diverge in the B. pullatus subgroup, is currently distributed in open areas of the Chaco, Amazonia, and Parana regions (Figure 1 and Figure 2) [79,80]. The ancestor of the remaining species posteriorly split in two lineages, one composed of species also related to open areas (B. pullatus, B. pauloensis) [79,80] and the other by species related to tropical forests (B. transversalis, B. bahiensis, and B. brasiliensis) [6,24,81]. The climatic oscillations and the expansion and retraction dynamics of open areas and forests [78] are possible causes of the speciation events in the group. Santos Júnior et al. [6] were able to show, for instance, that the divergence between B. brasiliensis and B. bahiensis was associated to forest retraction in the Plio-Pleistocene boundary. An exception would be the cladogenetic event producing the species B. pullatus and B. pauloensis, which is inferred (Table 2) to be contemporary to and maybe consequence of the final uplift of the Andes, ca. 5 Ma [62]. Currently, B. pullatus is restricted to southern Central America and northwestern South America, while B. pauloensis is a widely distributed species in South America [79,80]. It is relevant that several taxonomists suspect that the populations of B. pauloensis inhabiting across the Andes in northern Colombia do not belong in the same species as those in the tropical lowlands of Brazil and surrounding countries. This issue is probably associated to misidentification or mislabeled specimens (see Moure and Sakagami [52] and Milliron [53]).
The increased number of calibration points, the use of powerful analytical tools, and the addition of many Neotropical taxa indicated an older origin for Bombus as well as for the main dispersion and diversification events in this taxon. Our results confirm that invasion of the New World by bumblebees occurred exclusively through the Bering Strait, which was accomplished by the ancestors of six lineages in the subgenera Cullumanobombus, Psithyrus, Pyrobombus, and Thoracobombus. Six lineages in two subgenera, Thoracobombus and Cullumanobombus, colonized South America, and two Thoracobombus lineages were able to invade the tropical lowlands in this continent. We suggest that transgression of the Atlantic Ocean in these lowlands are the main responsible for the diversification in the B. pullatus and B. morio subgroups of species, while the final uplift of the Andes may be responsible for the vicariance event, leading to the origin of B. pullatus and B. pauloensis.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/d14040238/s1, Figure S1: Phylogenetic relationships among bumblebees using Bayesian inference–BI and all species available for each subgenus; Figure S2: Estimated divergence times using Unncorrelated Lognormal Relaxed Clock model; Figure S3: Head in profile (a and b), dorsal view of the mesoscutum (c), frontal view of the head (d), hind tibia (e) and labels of the Holotype of Bombus brevivillus Franklin, 1913; Figure S4: Global ancestral state estimates under the unconstrained BAYAREALIKE+J; Figure S5: Distribution of occurrence of data for ancestry inference of bumblebee species; Figure S6: Global ancestral state estimates under the unconstrained BAYAREALIKE+J; Figure S7: Strict consensus tree of Maximum Parsimony–MP analysis of bumblebees’ species; Figure S8. Strict consensus tree of Maximum Parsimony–MP analysis of bumblebees’ species; Figure S9. Maximum Likelihood tree–ML search obtained with RAxML; Figure S10. Maximum Likelihood tree–ML search obtained with IQ-TREE; Figure S11: Bumblebee species chronogram with shading of branches reflective of estimated diversification rates (see scale at right) estimated in BAMM; Figure S12: Macroevolutionary cohort matrix for diversification in bumblebees; Figure S13: Estimates of speciation rate variability across the bumblebees (red line), with average global climate overlain; Figure S14: Phylogenetic relationships among bumblebees using Bayesian inference–BI and all species available for each subgenus; Figure S15: Estimated divergence times using Uncorrelated Lognormal Relaxed Clock model (BEAST v2.4.4); Table S1: Specimens sequenced for the genetic analyses with their geographic origins and specimens belonging to GenBank and BOLD Systems; Table S2: List of primers used to amplify mitochondrial and nuclear genes; Table S3: PCR primer pairs and annealing temperatures used for analysis (See Supplementary Table S2 for primer sequences used); Table S4: Table with distribution for each bumblebee species; Table S5: Data of full-length concatenated alignment data used on phylogenetic analysis; Table S6: Best partitioning schemes and substitution models; Table S7: BioGeoBEARS results for each model implemented in the analysis (The information of distribution areas used in the analyses followed the available in http://www.nhm.ac.uk/research-curation/research/projects/bombus, accessed on 1 January 2022), Table S8: BioGeoBEARS results for each model implemented in the analysis (The information of distribution areas used in the analyses followed the available in http://www.nhm.ac.uk/research-curation/research/projects/bombus with few modified for the Nearctic, Neotropical and Palaearctic species. See the Figure 1 to the modifications); Table S9: Basic information on the two data sets used in analyses about the impacts of missing data; Table S10: Data of full-length concatenated alignment data used on phylogenetic analysis; Table S11: Best partitioning schemes and substitution models; Supplementary File S1: Methods and results of maximum parsimony (MP) and maximum likelihood (ML) analyses; Supplementary File S2: Priors for divergence time estimation; Supplementary File S3: Methods and results of diversification patterns; Supplementary File S4: Methods and results of missing data analyses.
Author Contributions
J.E.S.J., F.A.S. and F.R.S. designed the study. J.E.S.J. performed the laboratory work. J.E.S.J., C.A.R.D. and P.F. analyzed the data primarily, with contributions from P.H.W., R.T.F.C. and D.P.C. J.E.S.J. and F.A.S. drafted the manuscript, and all authors provided subsequent input and contributions. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by Brazilian grants from the Fundação de Amparo à Pesquisa de Minas Gerais (FAPEMIG–APQ-03625-17 and APQ-05368-18), the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq–421303/2017-4), Coordenação de Aperfeiçoamento do Ensino Superior (CAPES), and ICMBio to F.R.S. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. J.E.S.J. also thank FAPEMIG for conceding the scholarship, and CAPES and the Programa de Pós-Graduação em Genética of UFMG for a post-doctorate fellowship.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The new sequences obtained during this research were deposited in GenBank under the accession codes OM649762-OM649809.
Acknowledgments
We thank Fernanda Fontes Trancoso (UFMG) for editing some figures. We are also grateful to Betina Blochtein, Carmen Lucia Yurrita Obiols, David Ward Roubik, Eduardo Almeida, Gabriel Augusto Rodrigues Melo, Lucio Antônio de Oliveira Campos, Laurence Parker, Elaine Françoso, Maria Cristina Arias, Maria Cristina Gaglianone, Marcio Luiz de Oliveira, Cristiane Krug, João Steiner, and Yasmine Antonini for the loan of specimens, and to Laurence Packer for providing DNA sequences of some Bombus specimens. We acknowledge the Instituto Chico Mendes de Biodiversidade (ICMBio) for the collecting permit granted to J.E.S.J.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Michener, C.D. The Bees of the World, 2nd ed.; Johns Hopkins University Press: Baltimore, MD, USA, 2007; ISBN 978-0-8018-8573-0. [Google Scholar]
- Danforth, B.N.; Cardinal, S.; Praz, C.; Almeida, E.A.B.; Michez, D. The Impact of Molecular Data on Our Understanding of Bee Phylogeny and Evolution. Annu. Rev. Entomol. 2013, 58, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Peters, R.S.; Krogmann, L.; Mayer, C.; Donath, A.; Gunkel, S.; Meusemann, K.; Kozlov, A.; Podsiadlowski, L.; Petersen, M.; Lanfear, R.; et al. Evolutionary History of the Hymenoptera. Curr. Biol. 2017, 27, 1013–1018. [Google Scholar] [CrossRef] [PubMed]
- Roig-Alsina, A.; Michener, C.D. Studies of the Phylogeny and Classification of Long-Tongued Bees. Univ. Kans. Sci. Bull. 1993, 55, 123–173. [Google Scholar]
- Williams, P. An Annotated Checklist of Bumble Bees with an Analysis of Patterns of Description (Hymenoptera: Apidae, Bombini). Bull. Nat. Hist. Mus. 1998, 67, 79–152. [Google Scholar]
- Santos Júnior, J.E.; Silveira, F.A.; Oliveira, U.; Dias, C.A.R.; Santos, F.R. Conservation and Historical Distribution of Two Bumblebee Species from the Atlantic Forest. Syst. Biodivers. 2019, 17, 22–38. [Google Scholar] [CrossRef]
- Williams, P.H. Mapping Variations in the Strength and Breadth of Biogeographic Transition Zones Using Species Turnover. Proc. R. Soc. Lond. B Biol. Sci. 1996, 263, 579–588. [Google Scholar]
- Williams, P.H. A Preliminary Cladistic Investigation of Relationships among the Bumble Bees (Hymenoptera, Apidae). Syst. Entomol. 1985, 10, 239–255. [Google Scholar] [CrossRef]
- Cameron, S.A.; Hines, H.M.; Williams, P.H. A Comprehensive Phylogeny of the Bumble Bees (Bombus). Biol. J. Linn. Soc. 2007, 91, 161–188. [Google Scholar] [CrossRef]
- Kawakita, A.; Sota, T.; Ito, M.; Ascher, J.S.; Tanaka, H.; Kato, M.; Roubik, D.W. Phylogeny, Historical Biogeography, and Character Evolution in Bumble Bees (Bombus: Apidae) Based on Simultaneous Analysis of Three Nuclear Gene Sequences. Mol. Phylogenetics Evol. 2004, 31, 799–804. [Google Scholar] [CrossRef]
- Hines, H.M. Historical Biogeography, Divergence Times, and Diversification Patterns of Bumble Bees (Hymenoptera: Apidae: Bombus). Syst. Biol. 2008, 57, 58–75. [Google Scholar] [CrossRef]
- Williams, P.H.; Lobo, J.M.; Meseguer, A.S. Bumblebees Take the High Road: Climatically Integrative Biogeography Shows That Escape from Tibet, Not Tibetan Uplift, Is Associated with Divergences of Present-Day Mendacibombus. Ecography 2018, 41, 461–477. [Google Scholar] [CrossRef]
- Nee, S.; May, M.R.; Harvey, P.H. The Reconstructed Evolutionaty Process. Philos. Trans. R. Soc. B Biol. Sci. 1994, 344, 305–311. [Google Scholar]
- Jablonski, D.; Roy, K.; Valentine, J.W.; Price, R.M.; Anderson, P.S. The Impact of the Pull of the Recent on the History of Marine Diversity. Science 2003, 300, 1133–1135. [Google Scholar] [CrossRef]
- Nee, S. Birth-Death Models in Macroevolution. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 1–17. [Google Scholar] [CrossRef]
- Etienne, R.S.; Rosindell, J. Prolonging the Past Counteracts the Pull of the Present: Protracted Speciation Can Explain Observed Slowdowns in Diversification. Syst. Biol. 2012, 61, 204–213. [Google Scholar] [CrossRef]
- Engel, M.S. A Monograph of the Baltic Amber Bees and Evolution of the Apoidea (Hymenoptera). Bull. Am. Mus. Nat. Hist. 2001, 259, 1–192. [Google Scholar] [CrossRef]
- Wappler, T.; De Meulemeester, T.; Murat Aytekin, A.; Michez, D.; Engel, M.S. Geometric Morphometric Analysis of a New Miocene Bumble Bee from the Randeck Maar of Southwestern Germany (Hymenoptera: Apidae). Syst. Entomol. 2012, 37, 784–792. [Google Scholar] [CrossRef]
- Prokop, J.; Dehon, M.; Michez, D.; Engel, M.S. An Early Miocene Bumble Bee from Northern Bohemia (Hymenoptera, Apidae). ZooKeys 2017, 710, 43–63. [Google Scholar] [CrossRef]
- Dehon, M.; Michez, D.; Nel, A.; Engel, M.S.; De Meulemeester, T. Wing Shape of Four New Bee Fossils (Hymenoptera: Anthophila) Provides Insights to Bee Evolution. PLoS ONE 2014, 9, e108865. [Google Scholar] [CrossRef][Green Version]
- Dehon, M.; Engel, M.S.; Gérard, M.; Aytekin, A.M.; Ghisbain, G.; Williams, P.H.; Rasmont, P.; Michez, D. Morphometric Analysis of Fossil Bumble Bees (Hymenoptera, Apidae, Bombini) Reveals Their Taxonomic Affinities. Zookeys 2019, 891, 71–118. [Google Scholar] [CrossRef]
- Engel, M.S.; Grimaldi, D.A.; Gonzalez, V.H.; Hinojosa-Díaz, I.A.; Michener, C.D. An Exomalopsine Bee in Early Miocene Amber from the Dominican Republic (Hymenoptera: Apidae). Am. Mus. Novit. 2012, 3758, 1–16. [Google Scholar] [CrossRef][Green Version]
- Sambrook, J.; Russel, D.W. Molecular Cloning. A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Santos Júnior, J.E.; Santos, F.R.; Silveira, F.A. Hitting an Unintended Target: Phylogeography of Bombus Brasiliensis Lepeletier, 1836 and the First New Brazilian Bumblebee Species in a Century (Hymenoptera: Apidae). PLoS ONE 2015, 10, e0125847. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of Phylogenies after Removing Divergent and Ambiguously Aligned Blocks from Protein Sequence Alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation Software for the Fast Assembly of Multi-Gene Datasets with Character Set and Codon Information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Pattengale, N.; Aberer, A.; Swenson, K.; Stamatakis, A.; Moret, B. Uncovering Hidden Phylogenetic Consensus in Large Data Sets. IEEE/ACM Trans. Comput. Biol. Bioinform. 2011, 8, 902–911. [Google Scholar] [CrossRef] [PubMed]
- Westover, K.M.; Rusinko, J.P.; Hoin, J.; Neal, M. Rogue Taxa Phenomenon: A Biological Companion to Simulation Analysis. Mol. Phylogenetics Evol. 2013, 69, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian Inference of Phylogenetic Trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Wiens, J.J.; Morrill, M.C. Missing Data in Phylogenetic Analysis: Reconciling Results from Simulations and Empirical Data. Syst. Biol. 2011, 60, 719–731. [Google Scholar] [CrossRef]
- Guillerme, T.; Cooper, N. Effects of Missing Data on Topological Inference Using a Total Evidence Approach. Mol. Phylogenetics Evol. 2016, 94, 146–158. [Google Scholar] [CrossRef]
- Wiens, J.J.; Moen, D.S. Missing Data and the Accuracy of Bayesian Phylogenetics. J. Syst. Evol. 2008, 46, 307–314. [Google Scholar]
- Dunn, K.A.; McEachran, J.D.; Honeycutt, R.L. Molecular Phylogenetics of Myliobatiform Fishes (Chondrichthyes: Myliobatiformes), with Comments on the Effects of Missing Data on Parsimony and Likelihood. Mol. Phylogenetics Evol. 2003, 27, 259–270. [Google Scholar] [CrossRef]
- Simmons, M.P. Misleading Results of Likelihood-Based Phylogenetic Analyses in the Presence of Missing Data. Cladistics 2012, 28, 208–222. [Google Scholar] [CrossRef]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.-H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef]
- Rambaut, A.; Suchard, M.A.; Drummond, A.J. Tracer v1.6: Molecular Evolution, Phylogenetics and Epidemiology. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 1 January 2022).
- Matzke, N.J. Probabilistic Historical Biogeography: New Models for Founder-Event Speciation, Imperfect Detection, and Fossils Allow Improved Accuracy and Model-Testing. Front. Biogeogr. 2013, 5, 242–248. [Google Scholar] [CrossRef]
- Paradis, E.; Schliep, K. Ape 5.0: An Environment for Modern Phylogenetics and Evolutionary Analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Morrone, J.J. Cladistic Biogeography of the Neotropical Region: Identifying the Main Events in the Diversification of the Terrestrial Biota. Cladistics 2013, 30, 1–13. [Google Scholar] [CrossRef]
- Ree, R.H.; Smith, S.A. Maximum Likelihood Inference of Geographic Range Evolution by Dispersal, Local Extinction, and Cladogenesis. Syst. Biol. 2008, 57, 4–14. [Google Scholar] [CrossRef]
- Ronquist, F. Dispersal-Vicariance Analysis: A New Approach to the Quantification of Historical Biogeography. Syst. Biol. 1997, 46, 195–203. [Google Scholar] [CrossRef]
- Landis, M.J.; Matzke, N.J.; Moore, B.R.; Huelsenbeck, J.P. Bayesian Analysis of Biogeography When the Number of Areas Is Large. Syst. Biol. 2013, 62, 789–804. [Google Scholar] [CrossRef]
- Matzke, N.J. Model Selection in Historical Biogeography Reveals That Founder-Event Speciation Is a Crucial Process in Island Clades. Syst. Biol. 2014, 63, 951–970. [Google Scholar] [CrossRef]
- Williams, P.H.; Cameron, S.A.; Hines, H.M.; Cederberg, B.; Rasmont, P. A Simplified Subgeneric Classification of the Bumblebees (Genus Bombus). Apidologie 2008, 39, 46–74. [Google Scholar] [CrossRef]
- Françoso, E.; de Oliveira, F.F.; Arias, M.C. An Integrative Approach Identifies a New Species of Bumblebee (Hymenoptera: Apidae: Bombini) from Northeastern Brazil. Apidologie 2016, 47, 171–185. [Google Scholar] [CrossRef]
- Moure, J.S.; Sakagami, S. As Mamamgabas Sociais Do Brasil (Bombus Latreille) (Hymenoptera, Apoidea). Stud. Entomol. 1962, 51, 65–194. [Google Scholar]
- Milliron, H.E. A Monograph of the Western Hemisphere Bumblebees (Hymenoptera: Apidae; Bombinae. II. The Genus Megabombus Subgenus Megabombus. Mem. Entomol. Soc. Can. 1973, 89, 81–237. [Google Scholar] [CrossRef]
- Sanmartín, I.; Enghoff, H.; Ronquist, F. Patterns of Animal Dispersal, Vicariance and Diversification in the Holarctic. Biol. J. Linn. Soc. 2001, 73, 345–390. [Google Scholar] [CrossRef]
- Uba, C.E.; Strecker, M.R.; Schmitt, A.K. Increased Sediment Accumulation Rates and Climatic Forcing in the Central Andes during the Late Miocene. Geology 2007, 35, 979. [Google Scholar] [CrossRef]
- Retallack, G.J.; Kirby, M.X. Middle Miocene Global Change and Paleogeography of Panama. PALAIOS 2007, 22, 667–679. [Google Scholar] [CrossRef]
- Goulson, D. Bumblebees: Behaviour, Ecology, and Conservation, 2nd ed.; Oxford Biology; Oxford University Press: Oxford, UK; New York, NY, USA, 2010; ISBN 978-0-19-955306-8. [Google Scholar]
- Lepais, O.; Darvill, B.; O’Connor, S.; Osborne, J.L.; Sanderson, R.A.; Cussans, J.; Goffe, L.; Goulson, D. Estimation of Bumblebee Queen Dispersal Distances Using Sibship Reconstruction Method. Mol. Ecol. 2010, 19, 819–831. [Google Scholar] [CrossRef]
- Owen, R.E.; Otterstatter, M.C.; Cartar, R.V.; Farmer, A.; Colla, S.R.; O’Toole, N. Significant Expansion of the Distribution of the Bumble Bee Bombus Moderatus (Hymenoptera: Apidae) in Alberta over 20 Years 1 This Paper Is Dedicated to the Memory of Adolf Scholl. Can. J. Zool. 2012, 90, 133–138. [Google Scholar] [CrossRef]
- Makinson, J.C.; Woodgate, J.L.; Reynolds, A.; Capaldi, E.A.; Perry, C.J.; Chittka, L. Harmonic Radar Tracking Reveals Random Dispersal Pattern of Bumblebee (Bombus Terrestris) Queens after Hibernation. Sci. Rep. 2019, 9, 4651. [Google Scholar] [CrossRef]
- Futuyma, D.J.; Kirkpatrick, M. Evolution, 4th ed.; Sinauer Associates, Inc.: Massachusetts, UK, 2017; ISBN 978-1-60535-696-9. [Google Scholar]
- Hoorn, C.; Wesselingh, F.P.; ter Steege, H.; Bermudez, M.A.; Mora, A.; Sevink, J.; Sanmartin, I.; Sanchez-Meseguer, A.; Anderson, C.L.; Figueiredo, J.P.; et al. Amazonia Through Time: Andean Uplift, Climate Change, Landscape Evolution, and Biodiversity. Science 2010, 330, 927–931. [Google Scholar] [CrossRef]
- Condamine, F.L.; Hines, H.M. Historical Species Losses in Bumblebee Evolution. Biol. Lett. 2015, 11, 20141049. [Google Scholar] [CrossRef] [PubMed]
- Raup, D.M.; Gould, S.J.; Schopf, T.J.; Simberloff, D.S. Stochastic Models of Phylogeny and the Evolution of Diversity. J. Geol. 1973, 81, 525–542. [Google Scholar] [CrossRef]
- Purvis, A.; Orme, C.D.L.; Toomey, N.H.; Pearson, P.N. Temporal Patterns in Diversification Rates. In Speciation and Patterns of Diversity; Butlin, R., Schluter, D., Bridle, J., Eds.; Cambridge University Press: Cambridge, UK, 2009; Chapter 15. [Google Scholar]
- Phillimore, A.B.; Price, T.D. Density-Dependent Cladogenesis in Birds. PLoS Biol. 2008, 6, e71. [Google Scholar] [CrossRef]
- Ricklefs, R.E. Evolutionary Diversification, Coevolution between Populations and Their Antagonists, and the Filling of Niche Space. Proc. Natl. Acad. Sci. USA 2010, 107, 1265–1272. [Google Scholar] [CrossRef] [PubMed]
- Rosindell, J.; Harmon, L.J.; Etienne, R.S. Unifying Ecology and Macroevolution with Individual-Based Theory. Ecol. Lett. 2015, 18, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Hartenberger, J.-L. An Asian Grande Coupure. Nature 1998, 394, 321. [Google Scholar] [CrossRef]
- Meng, J.; McKenna, M.C. Faunal Turnovers of Palaeogene Mammals from the Mongolian Plateau. Nature 1998, 394, 364–367. [Google Scholar] [CrossRef]
- Zachos, J.; Pagani, M.; Sloan, L.; Thomas, E.; Billups, K. Trends, Rhythms, and Aberrations in Global Climate 65 Ma to Present. Science 2001, 292, 686–693. [Google Scholar] [CrossRef]
- Montes, C.; Cardona, A.; Jaramillo, C.; Pardo, A.; Silva, J.C.; Valencia, V.; Ayala, C.; Pérez-Angel, L.C.; Rodriguez-Parra, L.A.; Ramirez, V.; et al. Middle Miocene Closure of the Central American Seaway. Science 2015, 348, 226–229. [Google Scholar] [CrossRef]
- Cozzuol, M.A. The Acre Vertebrate Fauna: Age, Diversity, and Geography. J. South Am. Earth Sci. 2006, 21, 185–203. [Google Scholar] [CrossRef]
- O’Dea, A.; Lessios, H.A.; Coates, A.G.; Eytan, R.I.; Restrepo-Moreno, S.A.; Cione, A.L.; Collins, L.S.; de Queiroz, A.; Farris, D.W.; Norris, R.D.; et al. Formation of the Isthmus of Panama. Sci. Adv. 2016, 2, e1600883. [Google Scholar] [CrossRef]
- Williams, P.; Colla, S.; Xie, Z. Bumblebee Vulnerability: Common Correlates of Winners and Losers across Three Continents. Conserv. Biol. 2009, 23, 931–940. [Google Scholar] [CrossRef]
- Francisco, F.O.; Santiago, L.R.; Mizusawa, Y.M.; Oldroyd, B.P.; Arias, M.C. Genetic Structure of Island and Mainland Populations of a Neotropical Bumble Bee Species. J. Insect Conserv. 2016, 20, 383–394. [Google Scholar] [CrossRef]
- Ehlers, T.A.; Poulsen, C.J. Influence of Andean Uplift on Climate and Paleoaltimetry Estimates. Earth Planet. Sci. Lett. 2009, 281, 238–248. [Google Scholar] [CrossRef]
- Ortiz-Jaureguizar, E.; Cladera, G.A. Paleoenvironmental Evolution of Southern South America during the Cenozoic. J. Arid Environ. 2006, 66, 498–532. [Google Scholar] [CrossRef]
- Abrahamovich, A.H.; Díaz, N.B.; Morrone, J.J. Distributional Patterns of the Neotropical and Andean Species of the Genus Bombus (Hymenoptera: Apidae). Acta Zoológica Mex. 2004, 20, 99–117. [Google Scholar]
- Moure, J.S.; Urban, D.; Melo, G.A.R. Catalogue of Bees (Hymenoptera, Apoidea) in the Neotropical Region—Online Version. Available online: http://www.moure.cria.org.br/catalogue (accessed on 2 November 2019).
- Olesen, J.M. Behaviour and Nest Structure of the Amazonian Bombus Transversalis in Ecuador. J. Trop. Ecol. 1989, 5, 243–246. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).