Abstract
Marine macroalgae are foundation species that play a critical ecological role in coastal communities as primary producers. The macroalgal genus Ulva is vital in intertidal communities, serving as a food source and shelter for organisms, but these species also form environment-damaging nuisance blooms. This project aimed to demonstrate the utility of DNA barcoding for determining the diversity of Ulva species in the San Juan Islands (Washington, DC, USA). Blade-form Ulva (Ulvophyceae) specimens were collected from the lower, mid, and upper intertidal zones at three sites experiencing different levels of wave exposure. Sequences of plastid-encoded tufA were generated for each specimen and cluster analyses revealed the presence of four species at the collection sites. Two species were positively identified as Ulva expansa and Ulva fenestrata based on their sharing identical tufA sequences with those of the holotype specimens. Sequences of plastid-encoded rbcL and the nuclear-encoded ribosomal ITS regions of representative specimens were used to identify the other two species as Ulva prolifera and Ulva californica based on their similarity to epitype and topotype specimen sequences, respectively. Additional types of specimen sequencing efforts are needed to increase the number of Ulva species that can be accurately identified and realize their true biodiversity.
Keywords:
ITS; macroalgae; rbcL; tufA; Ulva californica; Ulva expansa; Ulva fenestrata; Ulva prolifera 1. Introduction
The northeast Pacific Ocean, from the coasts of Southeast Alaska to Oregon, is characterized by a diverse community of marine algae, including 671 taxa and 284 genera [1]. The San Juan Islands within the Salish Sea are a particularly rich area within this region that experience mixed semidiurnal tides that cause intense tidal flows with vigorous vertical mixing, especially at sills [2,3]. The characteristics of channels through the islands are highly influenced by the Fraser River from the Strait of Georgia [4], and the succession of spring and neap tides modulates the mixing over the sills, regulating the estuarine exchange of water [3]. The mixture of cold ocean waters of high salinity with brackish surface waters, seasonality, and physical factors further supports the diversity of the marine community and affect the interaction among resident organisms [5]. This is especially true for marine macroalgae, which have highly diverse intertidal and subtidal communities in this region. The diversity of these organisms can be masked by the high frequency of cryptic and phenotypically plastic species [6,7].
Marine macroalgae are foundation species that play a critical ecological role in coastal communities as primary producers and habitat-defining organisms [8]. Ulva Linnaeus species are important components of biodiversity and bioindicators [9] However, they have also been associated with the majority of blooms of free-floating green algae responsible for ‘green tides’ because Ulva species can rapidly grow in nutrient-rich habitats and have a high tolerance range for abiotic factors such as temperature and salinity [6,10,11,12,13]. Eutrophication-driven green tides in shallow waters have a direct economic impact on coastal communities, making it essential to identify the species involved for bloom characterization and control [10,14]. In addition, it is important to understand their potential uses in pharmaceutical applications for drug development [12], as well as in biotechnological and industrial processes as bioremediators, biofuels, and food sources [14]. However, their simple morphology and phenotypic plasticity means that diversity assessments and identifications of Ulva species based on morphological characters range from challenging to impossible e.g., [15,16].
The genus Ulva is constituted of nearly 100 taxonomically accepted species [17] including those species previously placed in Enteromorpha Link [18]. This green algal genus is present in both freshwater and marine environments. In the latter, it is ubiquitous along coasts, rocky shores, and protected bays and estuaries, growing attached to substrata or found as drift. The morphological characterization of Ulva species has traditionally included both macro- and microscopic features. Macroscopic features include having distromatic blade-form or monostromatic tubular thalli, thallus shape, size, extent of branching and presence or absence of marginal dentation. Cellular features considered key to identification include cellular shape and dimensions, number of pyrenoids, arrangement of the cells in regular or irregular patterns, and thallus thickness e.g., [19,20,21,22]. Although previous studies used these characters for identification, they have been found to vary within species depending on thallus age, reproductive state, wave exposure, tidal factors, temperature, salinity, light, life-history stage, and biological factors such as herbivory and associated microbiome e.g., [9,23,24]. In addition, the morphological plasticity of Ulva species results in a variety of forms and ecotypes. Therefore, the taxonomic status of species in this genus remains uncertain and difficult to assess [9,11,16,24,25].
Molecular analysis of Ulva spp. is greatly expanding our understanding of their taxonomic and phylogenetic status [25]. Studies utilizing DNA sequence data have defined many molecular-based species e.g., [11,16,26,27], but sequences from type specimens have been generated for relatively few historical species [25,28,29,30] and only recently have type sequences been included in new species descriptions [16,31,32]. The historical types that have been sequenced demonstrate that very few of the specimen identifications for sequences in public databases are correct [30,33]. Accordingly, while Ulva species can be easily delimited with DNA sequence data, the identification of most species remains problematic.
Up to 17 species and varieties of Ulva (including taxa formerly classified as Enteromorpha) have been reported in the northeast Pacific [34]. Hayden and Waaland [6] reported 12 species based on molecular and morphological analyses in the most recent treatment of the genus from this region. Little is known about Ulva species in the San Juan Islands; however, multiple studies have focused on the surrounding Salish Sea ecosystem [6,35,36,37,38]. Ulva species within this area proliferate into blooms comprised of multiple species in the intertidal zone, similar to many other anthropogenically influenced coastal ecosystems [39,40,41,42]. They were found to outcompete other macroalgae within these zones, exhibiting harmful characteristics that alter species interactions [42,43]. A better understanding of the species involved is needed for these reasons.
DNA barcoding was originally envisioned as a utilitarian method that could be simply applied for the identification of species by a non-specialist using a single universal marker, the mitochondria-encoded cytochrome c oxidase subunit 1 gene 5′ region [44,45]. While this vision has been realized for many groups of organisms, others have been found to require different and multiple markers [46,47]. Ulva and other green algae are part of the latter group, but studies have shown that plastid-encoded rbcL and tufA, as well as nuclear-encoded ITS, are useful singly or in combination for barcoding these algae [48,49,50]. The objectives of this study were to demonstrate the utility of DNA barcoding in its simplest application to determine the number and, if possible, identity of Ulva species. This was achieved by exploring the diversity of blade-form Ulva species present at three environmentally different study sites in the San Juan Islands, Washington.
2. Materials and Methods
2.1. Sample Collection
Thirty-five blade-form Ulva specimens were collected from the intertidal zone at three sites of differing relative wave exposure within the San Juan Islands, Washington (Table 1). Collections were made within the low, mid, and high intertidal zones at each location. Specimens were chosen at each location based on observed macromorphological variation, and two algal specimens of representative morphologies identified in each intertidal zone were collected at each sampled site. Specimens were only collected if attached and not as drift, and transported on ice back to the lab, where they were placed into a running seawater table until processed. Each specimen was morphologically identified using the Gabrielson and Lindstrom [1] key and vouchers were made and deposited in the University of Washington herbarium (WTU). All specimen data, including photographic images, are available from the Barcode of Life Database system website (dx.doi.org/10.5883/DS-MASJI08).

Table 1.
Ulva specimen collection site information.
2.2. DNA Extraction, Amplification, and Sequencing
Total DNA was extracted from specimens using a Bioline Extract-PCR Kit (Bioline, Taunton, MA, USA) following the protocol of Taylor et al. [50], with small modifications as follows. Approximately 0.5 cm2 of healthy blade tissue was chopped into small pieces, and incubated at 75 °C in 50 µL of Extract-PCR kit enzymatic solution for 1–20 h before enzyme deactivation by heating at 95 °C for 10 min. Cellular debris was pelleted by centrifugation and samples were diluted 1:10 and stored at −20 °C.
The plastid-encoded tufA locus was amplified for each Ulva specimen using MyTaq HS Red Mix following the manufacturer’s protocol (Bioline) with primers described in Fama et al. [51]. Cycling conditions were as follows: an initial denaturing step of 95 °C for 2:45 min, followed by 35 cycles of 95 °C for 15 s, 45 °C for 15 s, and 72 °C for 1 min, with a final extension at 72 °C for an additional 4 min. PCR products were enzymatically cleaned using Exo-Sap (Thermo Fisher Scientific, Waltham, MA, USA) and sent to Genewiz for DNA sequencing (Azenta Life Sciences, South Plainfield, NJ, USA). Based on initial analyses of tufA sequences, nuclear-encoded ITS and plastid-encoded rbcL sequences were generated from representative specimens of the detected species. ITS and rbcL were amplified and sequenced following the protocols of Freshwater et al. [52] but using a MyTaq HS Red DNA Polymerase Kit (Bioline), and the ITS and rbcL primers described by Shimada et al. [53]. Individual sequence reactions were compiled and edited using Sequencher (v. 5.4, Gene Codes Corporation, Ann Arbor, MI, USA).
2.3. DNA Sequence Analyses and Species Identifications
Alignments of DNA sequences were generated using MUSCLE [54] as implemented in MEGA (v. 7.0.26, [55]) or Geneious (v. 9, Biomatters Limited, Aukland, New Zealand). Species were molecularly delineated through barcode sequence clustering. Initially a UPGMA cluster diagram was generated from an alignment of the 35 tufA sequences for the newly collected San Juan Islands Ulva specimens to establish specimen clusters. Inter- and intra-cluster sequence divergence values were then assessed to determine if there were barcode gaps, as defined by Meier et al. [56] between clusters and whether these barcode gaps fit the tufA species divergence threshold ranges of Saunders and Kucera [49] and Kirkendale et al. [57]. GenBank BLAST analyses [58] of the tufA and, where needed, ITS and rbcL sequences were used to explore the identifications of the resulting molecularly defined species.
3. Results
The 35 Ulva specimens were grouped into four species based on UPGMA cluster analysis of tufA sequences (Figure 1). Intraspecific variation in tufA sequences was only seen in Species-1 (0.0–0.7%; 3 haplotypes) and interspecific variation among the four species ranged from 3.2–3.3% to 7.5% (Table 2). BLAST searches revealed that tufA sequences of Species-3 and Species-4 were exact matches to those of the U. expansa (Setchell) Setchell & N.L. Gardner (GenBank # MH731007) and U. fenestrata Postels & Ruprecht (GenBank # MK456404) type specimens, respectively. BLAST searches of the Species-1 and Species-2 tufA sequences returned close matches to specimens within the U. linza-procera-prolifera (LPP) complex clade for Species-1 and specimens predominantly identified as U. californica Wille for Species-2.

Figure 1.
UPGMA tufA cluster analysis for 35 blade-form Ulva specimens collected at different intertidal zones from three sites in the San Juan Islands. Specimen labels include the collection number (e.g., ‘03UA’) followed by the morphological identification of the specimen based on the Gabrielson and Lindstrom [1] key. The 1% sequence divergence level is indicated by the grey vertical line and example images of specimens are shown on the right.

Table 2.
Intra- and interspecific divergences among tufA sequences from 35 specimens of four blade-form Ulva species collected in the San Juan Islands, WA. Gray background = intraspecific divergences; white background = interspecific divergences.
The ITS sequences of Species-1 specimens representative of tufA haplotype 1 (specimen 03UA) and haplotype 2 (specimen 20UA) were identical. There was only a single base-pair difference between the ITS-2 region of this sequence and the ITS-2 sequences of 17 U. prolifera O.F. Müller topotype specimens (GenBank# AJ012276, but see discussion), including the epitype designated by Cui et al. [59]. The ITS-2 region sequence of the Species-1 specimen representative of tufA haplotype 3 (specimen 14UB) is two base pairs different from that of the U. prolifera epitype.
The rbcL sequences of two representative specimens of Species-2 (09UB; 23UB) were identical to each other and a topotype specimen identified by Hayden et al. [18] as U. californica (GenBank #AY255866). Similarly, ITS sequences of specimens 09UB and 23UB were identical and only 0.7% different from the ITS sequence of the Hayden et al. [18] topotype specimen identified as U. californica (GenBank #AY260560).
4. Discussion
Distinguishing Ulva species is a well-known problem in phycology. They have a very simple morphology and the few morphological characters that have been used to describe species exhibit intraspecific variation e.g., [15,16,21,60,61]. Analyses of DNA sequences are currently popular for delineating Ulva species e.g., [9,62,63]. However, as clearly demonstrated in a series of papers by Hughey et al. [25,29,30], Ulva specimens can only confidently be identified to species if DNA sequence data from those specimens can be matched to that of type specimens. These papers, as well as the overall analysis of Ulva sequences in GenBank by Fort et al. [33], demonstrated that many of the names assigned to Ulva sequences in public databases are incorrect. As an extreme example, all sequences in GenBank assigned to U. rigida were incorrectly identified [30]. Fort et al. [33] identified accessions that could be used for species identifications when a query sequence was homologous, and Hughey et al. [30] provide a table with all sequenced historical types and sequence determined synonyms.
Analyses of tufA sequences for blade-form Ulva specimens from different tidal heights at three different locations in the San Juan Islands revealed the presence of four species (Figure 2). Two of these species could be positively identified through the homology of their tufA sequences to that from type specimens. One was identified based on the holotype sequences published by Hughey et al. [28] as U. expansa (type locality: Monterey, CA, USA), a species reported in an older floristic survey of nearby Whidbey Island [35] and by Scagel [34] in his flora of British Columbia and northern Washington. Although reported as far north as British Columbia in these and other treatments of Northeast Pacific macroalgae e.g., [64,65,66], Tanner [36] synonymized U. expansa with U. fenestrata based on the morphological variation observed in herbarium specimens, field collections and culture studies. This synonymy was followed in the recent keys to the marine algae from southeastern Alaska to Oregon that have functioned as de facto floras for this region in recent years [1,67,68]. However, Hughey et al. [25,28] demonstrated the distinction of these two species. and U. expansa was once again recognized in the Northeast Pacific flora e.g., [69].
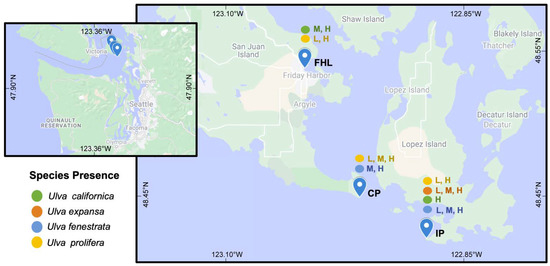
Figure 2.
Map showing the distribution of blade-form Ulva species collected from different intertidal heights at three San Juan Island sites that experience different wave exposures. Species are indicated by color and intertidal zones by letters: L = low; M = middle; H = high. Locations: FHL = Friday harbor Laboratory beach front; CP = Cattle Point; IP = Iceberg Point.
The second positively identified species was determined to be U. fenestrata (type locality: Kamchatka, Russia) based on the tufA sequence for its holotype published by Hughey et al. [25]. Similar to U. expansa, U. fenestrata was included in the Scagel [34] flora and Tanner’s [36] treatment of Northeast Pacific Ulva, but not in the more recent comprehensive keys [1,67,68]. Ulva fenestrata has generally been identified based upon the presence of perforations in the blade e.g., [65,70], but whether the presence or absence of perforations is a true developmental characteristic of species has been questioned, and both perforated and non-perforated specimens have been included in this species [36,70]. Gabrielson et al. [68] included U. fenestrata-type perforated blades within an unidentified Ulva species given the place-holder name U. “lactuca”. Further unpublished observations of San Juan Islands Ulva specimens led to this species being considered to represent U. fenestrata [71], and the current study verifies this identification.
The remaining two species revealed by the tufA analysis did not have close homology to any currently available tufA sequences from an Ulva type or topotype specimen. The tufA sequences from specimens of one of these species included three different haplotypes that had close homology to GenBank sequences from specimens placed in the Ulva linza-procera-prolifera (LPP) complex clade, a group composed of specimens variously identified as U. linza Linnaeus, U. procera (K.Ahlner) H.S.Hayden, Blomster, Maggs, P.C.Silva, Stanhope & Waaland, and U. prolifera O.F.Müller. Cui et al. [59] used morphological, molecular and crossing studies to examine the status of LPP complex specimens collected from Lolland Island, Denmark, the type locality of U. prolifera. Combining their results with those of previous LPP-complex-related studies e.g., [62,72,73], it was determined that U. prolifera was best represented by tubular branched specimens with sexual or asexual life histories [59]. The lectotype of U. prolifera is a drawing in Müller [74], thus an epitype was designated and sequences for the ITS-2 and 5S rDNA spacer regions generated. The ITS-2 sequence of this specimen was not made publicly available, but was reported to be identical to a previously published sequence with GenBank accession number AJ012276. The AJ012276 sequence includes not only ITS-2 but also the ITS-1 and 5.8S rRNA regions and, therefore, the epitype ITS-2 sequence is only the 215 bp portion of the AJ012276 sequence between the annealing sites of the two primers used by Cui et al. [59] to amplify and sequence this region in their specimens. ITS-2 sequences for representative specimens of the LPP complex species collected in the San Juan Islands were only 1–2 base pairs different (0.47–0.93%) from that of the U. prolifera epitype. This divergence from the epitype ITS-2 sequence is less than or equal to that of any LPP complex specimens included in the Cui et al. [59] study.
The San Juan Islands specimens were identified as U. prolifera based on these results. However, an unquestioned identification will require an understanding of whether any taxa within the LPP complex clade represent U. linza. Interestingly, the San Juan Islands specimens molecularly identified in this study as U. prolifera, were not branched tubes, the characteristic morphology of the species, but distromatic blades that became tubular where they were basally narrow near the point of attachment. This latter morphology has been identified as U. linza in the Northeast Pacific e.g., [1,64,68], and all these specimens were morphologically identified as U. linza (Figure 1). Similar to U. prolifera the lectotype of U. linza is an illustration [75] (pl. 9, Figure 6), but there is an epitype in OXF. Unfortunately, requests for the minimal type specimen material needed for current DNA sequence generation techniques have not been fulfilled, and the status of U. linza remains unresolved [76]. Regardless, the findings herein indicate that the concept of U. prolifera needs to be expanded to include blade-form thalli.
Representative specimens of the fourth species resolved in this study by the tufA analysis had rbcL sequences that were identical to that generated by Hayden et al. [18] from a La Jolla, California specimen of U. californica, the type locality for this species. The ITS sequences of San Juan Island specimens and that of this topotype specimen were also closely homologous and varied by only four base pairs (0.7%). The Hayden et al. [18] topotype specimen (WTU 344798) agrees with the type specimen (US 57108) in being ca. 2 cm or less and having turfy blades, and provides a basis for molecularly identifying specimens as U. californica in lieu of sequence data from the type specimen.
Tanner [60] conducted field, herbarium and culture studies of U. californica, U. angusta Setchell & N.L. Gardner, and U. scagelii Chihara, three morphologically similar Northeast Pacific species that differed in size, habit and distribution. The results of these studies led to the synonymy of U. angusta and U. scagelii with U. californica, increasing the size range and geographic distribution of U. californica. Specimens of U. californica sequenced in this study were also variable in size and distribution (Figure 2; dx.doi.org/10.5883/DS-MASJI08). The close homology of the topotype DNA sequences with those of specimens from this and other studies, e.g., [6,49,77], verifies the wider Northeast Pacific distribution and the environmentally determined morphological variation found in the study of Tanner [60].
As demonstrated in this study, simple analyses of DNA barcode sequences can be a useful tool for quickly distinguishing Ulva species, and only with an understanding of the number and extent of these species can applied questions concerning their diversity, ecology and physiology be addressed. For example, determining the species composition of blooms, or establishing monocultures or mixed cultures for industrial applications. However, this in no way diminishes the importance of extensive specimen collection combined with thorough phylogenetic and species delimitation analyses, e.g., [16,26], to establish the species and sequence characteristics upon which utilitarian DNA barcoding methods are based. The application of names to barcode-defined species, however, remains problematic. Two of the four species included in this study could be positively identified because DNA sequences were publicly available for their holotype specimens, and the best current identifications were possible for the other two species based on DNA sequences of epitype and topotype specimens. This fortuitous result is unusual because so few historical Ulva types have been sequenced, and only additional type specimen sequencing efforts and cooperation of the herbaria housing Ulva types can ensure that the application of additional species names is accurate.
Author Contributions
Conceptualization, G.M.K., B.C.-A. and D.W.F.; methodology, G.M.K., B.C.-A. and D.W.F.; validation, G.M.K., B.C.-A., J.E. and D.W.F.; formal analysis, G.M.K., B.C.-A., J.E. and D.W.F.; investigation, G.M.K., B.C.-A., J.E. and D.W.F.; resources, D.W.F.; data curation, G.M.K., B.C.-A., J.E. and D.W.F.; writing—original draft preparation, G.M.K., B.C.-A. and J.E.; writing—review and editing, G.M.K., B.C.-A. and D.W.F.; visualization, G.M.K. and D.W.F.; supervision, D.W.F.; project administration, G.M.K., B.C.-A., J.E. and D.W.F.; funding acquisition, D.W.F. All authors have read and agreed to the published version of the manuscript.
Funding
Research conducted at the University of Washington’s Friday Harbor Marine Laboratory was funded by the Marine Botany class budget and research conducted the University of North Carolina at Wilmington’s Center for Marine Science was supported by the CMS DNA-Algal Trust.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in the BOLD system database at dx.doi.org/10.5883/DS-MASJI08 and GenBank accessions OP347101-OP347108; OP347119-OP347153; OP347156-OP347160.
Acknowledgments
We would like to thank Tom Mumford for his guidance and mentorship during the Friday Harbor Laboratory Marine Botany class. We would also like to thank the Friday Harbor Laboratory for their facilities, resources, and accessibility during this study. Multiple reviewers provided valuable guidance for this publication.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gabrielson, P.W.; Lindstrom, S.C. Keys to the Seaweeds and Seagrasses of Southeast Alaska, British Columbia, Washington and Oregon; Phycological Contribution Number 9; Island Blue/Printorium Bookworks: Victoria, BC, Canada, 2018; 180p. [Google Scholar]
- Klinger, T.; Fluharty, D.; Evans, K.; Byron, C. Assessment of Coastal Water Resources and Watershed Conditions at San Juan Island National Historical Park; Technical Report NPS/NRWRD/NRTR-2006/360; U.S. Department of the Interior: Washington, DC, USA, 2006; p. 144.
- Masson, D.; Cummins, P.F. Fortnightly modulation of the estuarine circulation in Juan de Fuca Strait. J. Mar. Res. 2000, 58, 439–463. [Google Scholar] [CrossRef]
- Zamon, J.E. Tidal changes in copepod abundance and maintenance of a summer Coscinodiscus bloom in the southern San Juan Channel, San Juan Islands, USA. Mar. Ecol. Prog. Ser. 2002, 226, 193–210. [Google Scholar] [CrossRef][Green Version]
- Burnaford, J.L. Habitat modification and refuge from sublethal stress drive a marine plant-herbivore association. Ecology 2004, 85, 2837–2849. [Google Scholar] [CrossRef]
- Hayden, H.S.; Waaland, J.R. A molecular systematic study of Ulva (Ulvaceae, Ulvales) from the northeast Pacific. Phycologia 2004, 43, 364–382. [Google Scholar] [CrossRef]
- Steffensen, D.A. Morphological variation of Ulva in the Avon-Heathcote Estuary, Christchurch. N. Z. J. Mar. Fresh. Res. 2010, 10, 329–341. [Google Scholar] [CrossRef]
- Burke, C.; Thomas, T.; Lewis, M.; Steinberg, P.; Kjelleberg, S. Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 2011, 5, 590–600. [Google Scholar] [CrossRef]
- Wolf, M.A.; Sciuto, K.; Andreoli, C.; Moro, I. Ulva (Chlorophyta, Ulvales) biodiversity in the North Adriatic Sea (Mediterranean, Italy): Cryptic species and new introductions. J. Phycol. 2012, 48, 1510–1521. [Google Scholar] [CrossRef]
- Duan, W.; Guo, L.; Sun, D.; Zhu, S.; Chen, X.; Zhu, W.; Xu, T.; Chen, C. Morphological and molecular characterization of free-floating and attached green macroalgae Ulva spp. in the Yellow Sea of China. J. Appl. Phycol. 2012, 24, 97–108. [Google Scholar] [CrossRef]
- Guidon, M.; Thornber, C.; Wysor, B.; O’Kelly, C. Molecular and morphological diversity of Narragansett Bay (RI, USA) Ulva (Ulvales: Chlorophyta) populations. J. Phycol. 2013, 49, 979–995. [Google Scholar] [CrossRef]
- Ismail, M.M.; Mohamed, S.E. Differentiation between some Ulva spp. by morphological, genetic and biochemical analyses. Vavilovskii Zh. Genet. Sel. 2017, 21, 360–367. [Google Scholar] [CrossRef]
- Rybak, A.S. Species of Ulva (Ulvophyceae, Chlorophyta) as indicators of salinity. Ecol. Indic. 2018, 85, 253–261. [Google Scholar] [CrossRef]
- Wichard, T.; Charrier, B.; Mineur, F.; Bothwell, J.H.; De Clerck, O.; Coates, J.C. The green seaweed Ulva: A model system to study morphogenesis. Front. Plant Sci. 2015, 6, 72. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, L.C.; Nettleton, J.C.; Neefus, C.D.; Mathieson, A.C. Cryptic diversity of Ulva (Ulvales, Chlorophyta) in the Great Bay Estuarine System (Atlantic, USA): Introduced and indigenous distromatic species. Eur. J. Phycol. 2010, 45, 230–239. [Google Scholar] [CrossRef]
- Lagourgue, L.; Gobin, S.; Brisset, M.; Vandenberghe, S.; Bonneville, C.; Jauffrais, T.; Van Wynsberge, S.; Payri, C.E. Ten new species of Ulva (Ulvophyceae, Chlorophyta) discovered in New Caledonia: Genetic and morphological diversity, and bloom potential. Eur. J. Phycol. 2022, 1–21. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M. AlgaeBase. World-Wide Electronic Publication, National University of Ireland, Galway. Available online: https://www.algaebase.org (accessed on 29 June 2021).
- Hayden, H.S.; Blomster, J.; Maggs, C.A.; Silva, P.C.; Stanhope, M.J.; Waaland, J.R. Linnaeus was right all along: Ulva and Enteromorpha are not distinct evolutionary entities. Eur. J. Phycol. 2003, 38, 277–294. [Google Scholar] [CrossRef]
- Bliding, C. A critical survey of European taxa in Ulvales. Part I: Capsosiphon, Percursaria, Blidingia, Enteromorpha. Opera Bot. 1963, 8, 1–160. [Google Scholar]
- Bliding, C. A critical survey of European taxa in Ulvales. Part II: Ulva, Ulvaria, Monostroma, Kornmannia. Bot. Not. 1968, 121, 535–629. [Google Scholar]
- Kapraun, D.F. Field and cultural studies of Ulva and Enteromorpha in the vicinity of Port Aransas, Texas. Contrib. Mar. Sci. 1970, 15, 205–285. [Google Scholar]
- Kapraun, D.F. An illustrated guide to the benthic marine algae of coastal North Carolina. II. Chlorophyta and Phaeophyta. Bibloth. Phycol. 1984, 58, 1–173. [Google Scholar]
- Matsuo, Y.; Imagawa, H.; Nishizawa, M.; Shizuri, Y. Isolation of an algal morphogenesis inducer from a marine bacterium. Science 2005, 307, 1598. [Google Scholar] [CrossRef]
- Kazi, M.A.; Kavale, M.G.; Singh, V. Morphological and molecular characterization of Ulva chaugulii sp. nov. U. lactuca and U. ohnoi (Ulvophyceae, Chlorophyta) from India. Phycologia 2016, 55, 45–54. [Google Scholar]
- Hughey, J.R.; Maggs, C.A.; Mineur, F.; Jarvis, C.; Miller, K.A.; Shabaka, S.H.; Gabrielson, P.W. Genetic analysis of the Linnaean Ulva lactuca (Ulvales, Chlorophyta) holotype and related type specimens reveals name misapplications, unexpected origins, and new synonymies. J. Phycol. 2019, 55, 503–508. [Google Scholar] [CrossRef]
- Fort, A.; McHale, M.; Cascella, K.; Potin, P.; Usadel, B.; Guiry, M.D.; Sulpice, R. Foliose Ulva species show considerable inter-specific genetic diversity, low intra-specific genetic variation, and the rare occurrence of inter-specific hybrids in the wild. J. Phycol. 2021, 57, 219–233. [Google Scholar] [CrossRef]
- Melton, J.T.; Lopez-Bautista, J.M. Diversity of the green macroalgal genus Ulva (Ulvophyceae, Chlorophyta) from the east and gulf coast of the United States based on molecular data. J. Phycol. 2021, 57, 551–568. [Google Scholar] [CrossRef]
- Hughey, J.R.; Miller, K.A.; Gabrielson, P.W. Mitogenome analysis of a green tide forming Ulva from California, USA confirms its identity as Ulva expansa (Ulvaceae, Chlorophyta). Mitochondrial DNA B Resour. 2018, 3, 1302–1303. [Google Scholar] [CrossRef]
- Hughey, J.R.; Gabrielson, P.W.; Maggs, C.A.; Mineur, F.; Miller, K.A. Taxonomic revisions based on genetic analysis of type specimens of Ulva conglobata, U. laetevirens, U. pertusa and U. spathulata (Ulvales, Chlorophyta). Phycol. Res. 2021, 69, 148–153. [Google Scholar] [CrossRef]
- Hughey, J.R.; Gabrielson, P.W.; Maggs, C.A.; Mineur, F. Genomic analysis of the lectotype specimens of. European Ulva rigida and Ulva lacinulata (Ulvaceae, Chlorophyta) reveals the ongoing misapplication of names. Eur. J. Phycol. 2021, 57, 143–153. [Google Scholar] [CrossRef]
- Hiraoka, M.; Shimada, S.; Uenosono, M.; Masuda, M. A new green-tide-forming alga, Ulva ohnoi Hiraoka et Shimada sp. nov. (Ulvales, Ulvophyceae) from Japan. Phycol. Res. 2003, 51, 17–29. [Google Scholar] [CrossRef]
- Spalding, H.L.; Conklin, K.Y.; Smith, C.M.; O’Kelly, C.J.; Sherwood, A.R. New Ulvaceae (Ulvophyceae, Chlorophyta) from mesophotic ecosystems across the Hawaiian archipelago. J. Phycol. 2016, 52, 40–53. [Google Scholar] [CrossRef]
- Fort, A.; McHale, M.; Cascella, K.; Potin, P.; Perrineau, M.-M.; Kerrison, P.D.; da Costa, E.; Calado, R.; Domingues, M.R.; Azevedo, I.C.; et al. Exhaustive reanalysis of barcode sequences from public repositories highlights ongoing misidentifications and impacts taxa diversity and distribution. Mol. Ecol. Resour. 2021, 22, 86–101. [Google Scholar] [CrossRef]
- Scagel, R.F. Marine Algae of British Columbia and Northern Washington, Part I: Chlorophyceae (Green Algae); National Museum of Canada Bulletin 207; National Museum: Ottawa, ON, Canada, 1966; 257p.
- Phillips, R.C.; Vadas, R.L. Marine algae of Whidbey Island, Washington. J. Inst. Res. Ser. A 1967, 7, 2–81. [Google Scholar]
- Tanner, C.E. The Taxonomy and Morphological Variation of Distromatic Ulvaceous Algae (Chlorophyta) from the Northeast Pacific. Ph.D. Thesis, Department of Botany, University of British Columbia, Vancouver, BC, Canada, 1979. [Google Scholar]
- Nelson, T.A.; Olson, J.; Imhoff, L.; Nelson, A.V. Aerial exposure and desiccation tolerances are correlated to species composition in “green tides” of the Salish Sea (northeastern Pacific). Bot. Mar. 2010, 53, 103–111. [Google Scholar] [CrossRef]
- Van Alstyne, K.L. Seasonal changes in nutrient limitation and nitrate sources in the green macroalga Ulva lactuca at sites with and without green tides in a northeastern Pacific embayment. Mar. Pollut. Bull. 2016, 103, 186–194. [Google Scholar] [CrossRef]
- Shelford, V.E.; Weese, A.O.; Rice, L.A.; Rasmussen, D.I.; Maclean, A. Some marine biotic communities of the Pacific coast of North America. Part I. General survey of the communities. Ecol. Monogr. 1935, 5, 249–329. [Google Scholar]
- Hylleberg, J.; Henriksen, K. The central role of bioturbation in sediment mineralization and element re-cycling. Ophelia Suppl. 1980, 1, 1–16. [Google Scholar]
- Nelson, T.A.; Nelson, A.V.; Tjoelker, M. Seasonal and spatial patterns of “green tides” (ulvoid algal blooms) and related water quality parameters in the coastal waters of Washington State, USA. Bot. Mar. 2003, 46, 263–275. [Google Scholar] [CrossRef]
- Nelson, T.A.; Haberlin, K.; Nelson, A.V.; Ribarich, H.; Hotchkiss, R.; Van Alstyne, K.L.; Buckingham, L.; Simunds, D.J.; Fredrickson, K. Ecological and physiological controls of species composition in green macroalgal blooms. Ecology 2008, 89, 1287–1298. [Google Scholar] [CrossRef]
- Nelson, T.A.; Lee, D.J.; Smith, B.C. Are “green tides” harmful algal blooms? Toxic properties of water-soluble extracts from two bloom-forming macroalgae, Ulva fenestrata and Ulvaria obscura (Ulvophyceae). J. Phycol. 2003, 39, 874–879. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; Dewaard, J.R. Biological identification through DNA barcodes. Proc. Royal Soc. B 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Ratnasingham, S.; Dewaard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B 2003, 270, 596–599. [Google Scholar] [CrossRef]
- Hollingsworth, M.L.; Clark, A.A.; Forrest, L.L.; Richardson, J.; Pennington, R.T.; Long, D.G.; Cowan, R.; Chase, M.W.; Gaudeul, M.; Hollingsworth, P.M. Selecting barcoding loci for plants: Evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol. Ecol. Resour. 2009, 9, 439–457. [Google Scholar] [CrossRef] [PubMed]
- McFadden, C.S.; Benayahu, Y.; Pante, E.; Thoma, J.N.; Nevarez, P.A.; France, S.C. Limitations of mitochondrial gene barcoding in Octocorallia. Mol. Ecol. Resour. 2011, 11, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.D.; Fucikova, K.; Lo, C.; Lewis, L.A.; Karol, K.G. An assessment of proposed DNA barcodes in freshwater green algae. Crypt. Algol. 2010, 31, 529–555. [Google Scholar]
- Saunders, G.W.; Kucera, H. An evaluation of rbcL, tufA, UPA, LSU and ITS as DNA barcode markers for the marine green macroalgae. Crypt. Algol. 2010, 31, 487–528. [Google Scholar]
- Taylor, R.L.; Bailey, J.C.; Freshwater, D.W. Systematics of Cladophora spp. (Chlorophyta) from North Carolina, USA, based upon morphology and DNA sequence data with a description of Cladophora subtilissima sp. nov. J. Phycol. 2017, 53, 541–556. [Google Scholar] [CrossRef]
- Fama, P.; Wysor, B.; Kooistra, W.; Zuccarello, G.C. Molecular phylogeny of the genus Caulerpa (Caulerpales, Chlorophyta) inferred from chloroplast tufA gene. J. Phycol. 2002, 38, 1040–1050. [Google Scholar] [CrossRef]
- Freshwater, D.W.; Miller, C.E.; Fankovich, T.A.; Wynne, M.J. DNA sequence analyses reveal two new species of Caloglossa (Delesserieaceae, Rhodophyta) from the skin of West Indian Manatees. J. Mar. Sci. Eng. 2021, 9, 163. [Google Scholar] [CrossRef]
- Shimada, S.; Hiraoka, M.; Nabata, S.; Iima, M.; Masuda, M. Molecular phylogenetic analyses of the Japanese Ulva and Enteromorpha (Ulvales, Ulvophyceae), with special reference to the free-floating Ulva. Phycol. Res. 2003, 51, 99–108. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Meier, R.; Zhang, G.; Ali, F. The use of mean instead of smallest interspecific distances exaggerates the size of the “barcoding gap” and leads to misidentification. Syst. Biol. 2008, 57, 809–813. [Google Scholar] [CrossRef] [PubMed]
- Kirkendale, L.; Saunders, G.W.; Winberg, P. A molecular survey of Ulva (Chlorophyta) in temperate Australia reveals enhanced levels of cosmopolitanism. J. Phycol. 2013, 49, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Karlin, S.; Altschul, S.F. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl. Acad. Sci. USA 1990, 87, 2264–2268. [Google Scholar] [CrossRef]
- Cui, J.; Monotilla, A.P.; Zhu, W.; Takano, Y.; Shimada, S.; Ichihara, K.; Matsui, T.; He, P.; Hiraoka, M. Taxonomic reassessment of Ulva prolifera (Ulvophyceae, Chlorophyta) based on specimens from the type locality and Yellow Sea green tides. Phycologia 2018, 57, 692–704. [Google Scholar] [CrossRef]
- Tanner, C.E. Investigations of the taxonomy and morphological variation of Ulva (Chlorophyta): Ulva californica Wille. Phycologia 1986, 25, 510–520. [Google Scholar] [CrossRef]
- Bloomster, J.; Maggs, C.A.; Stanhope, M.J. Molecular and morphological analysis of Enteromorpha intestinales and E. compressa (Chlorophyta) in the British Isles. J. Phycol. 1998, 34, 319–340. [Google Scholar] [CrossRef]
- Shimada, S.; Yokoyama, N.; Arai, S.; Hiraoka, M. Phylogeography of the genus Ulva (Ulvophyceae, Chlorophyta), with special reference to the Japanese freshwater and brackish taxa. J. Appl. Phycol. 2008, 20, 979–989. [Google Scholar] [CrossRef]
- Phillips, J.A.; Lawton, R.J.; Denys, R.; Paul, N.A.; Carl, C. Ulva sapora sp. nov., an abundant tubular species of Ulva (Ulvales) from the tropical Pacific Ocean. Phycologia 2016, 55, 55–64. [Google Scholar] [CrossRef]
- Smith, G.M. Marine Algae of the Monterey Peninsula California; Stanford University Press: Stanford, CA, USA, 1944; p. 622. [Google Scholar]
- Doty, M.S. The marine algae of Oregon, Part I. Chlorophyta and Phaeophyta. Farlowia 1947, 3, 1–65. [Google Scholar] [CrossRef]
- Abbott, I.A.; Hollenberg, G.J. Marine Algae of California; Stanford University Press: Stanford, CA, USA, 1976; 827p. [Google Scholar]
- Gabrielson, P.W.; Widdowson, T.B.; Lindstrom, S.C. Keys to the Seaweeds and Seagrasses of Southeast Alaska, British Columbia, Washington and Oregon; Phycological Contribution Number 7; PhycoID: Hillsborough, NC, USA, 2006; 209p. [Google Scholar]
- Gabrielson, P.W.; Lindstrom, S.C.; O’Kelly, C.J. Keys to the Seaweeds and Seagrasses of Southeast Alaska, British Columbia, Washington and Oregon; Phycological Contribution Number 8; Island Blue/Printorium Bookworks: Victoria, BC, Canada, 2012; 192p. [Google Scholar]
- Lindstrom, S.C.; Lemay, M.A.; Starko, S.; Hind, K.R.; Martone, P. New and interesting seaweed records from the Hakai area of the central coast of British Columbia, Canada: Chlorophyta. Bot. Mar. 2021, 64, 343–361. [Google Scholar] [CrossRef]
- Setchell, W.A.; Gardner, N.L. The marine algae of the Pacific coast of North America. II. Chlorophyceae. Univ. Calf. Publ. Bot. 1920, 8, 139–374. [Google Scholar]
- O’Kelly, C.J. (University of Hawai’i, Honolulu, HI, USA). Personal communication, 2021.
- Hiraoka, M.; Ichihara, K.; Zuh, W.; Ma, J.; Shimada, S. Culture and hybridization experiments on an Ulva clade including the Qingdao strain blooming in the Yellow Sea. PLoS ONE 2011, 6, e19371. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Ohki, K.; Kamiya, M. High heterozygosity and phenotypic variation of zoids in apomictic Ulva prolifera (Ulvophyceae) from brackish environments. Aquat. Bot. 2014, 120, 185–192. [Google Scholar] [CrossRef]
- Müller, O.F. Florae Danicae, fasc. 13; Havniae: Copenhagen, Denmark, 1778; Volume 5, 8p. [Google Scholar]
- Dillenius, J.J. Historia Muscorum in Qua Circiter Sexcentae Species Veteres et Novae ad Sua Genera Relatae Describuntur et Iconibus Genuinis Illustrantur: Cum Appendice et Indice Synonymorum; e Theatro Sheldoniano: Oxford, UK, 1742; 576p. [Google Scholar]
- Gabrielson, P.W. (University of North Carolina at Chapel Hill, Chapel Hill, NC, USA). Personal communication, 2022.
- O’Kelly, C.J.; Wysor, B.; Bellows, W.K. Gene sequence diversity and the phylogenetic position of algae assigned to the genera Phaeophila and Ochlochaete (Ulvophyceae, Chlorophyta). J. Phycol. 2004, 40, 789–799. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).