New Records of the Psychrophilic Tetracladium Species Isolated from Freshwater Environments in Korea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Fungal Strains
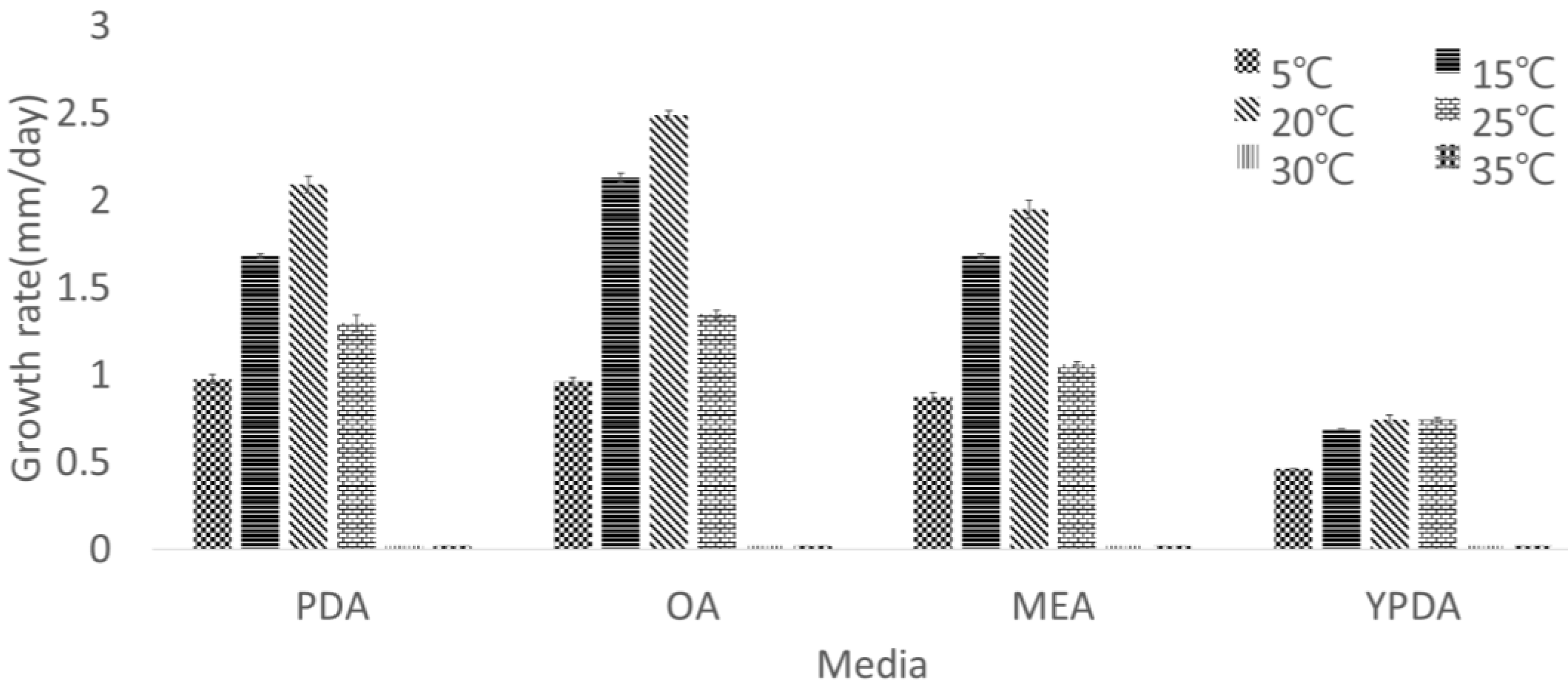
2.2. Induction of Sporulation and Investigation of Culture Conditions
2.3. DNA Extraction and Phylogenetic Analysis
3. Results
3.1. Phylogeny
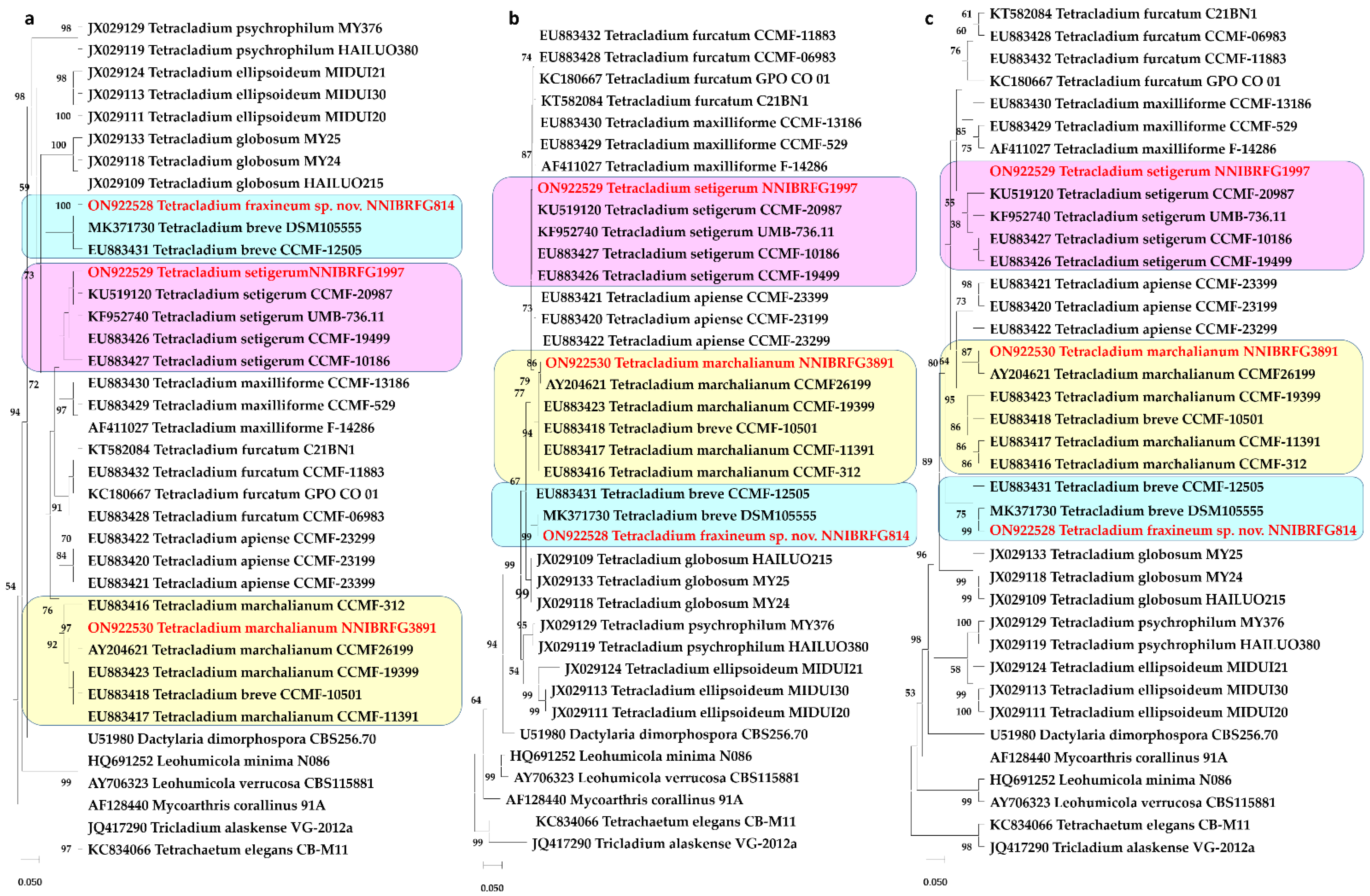
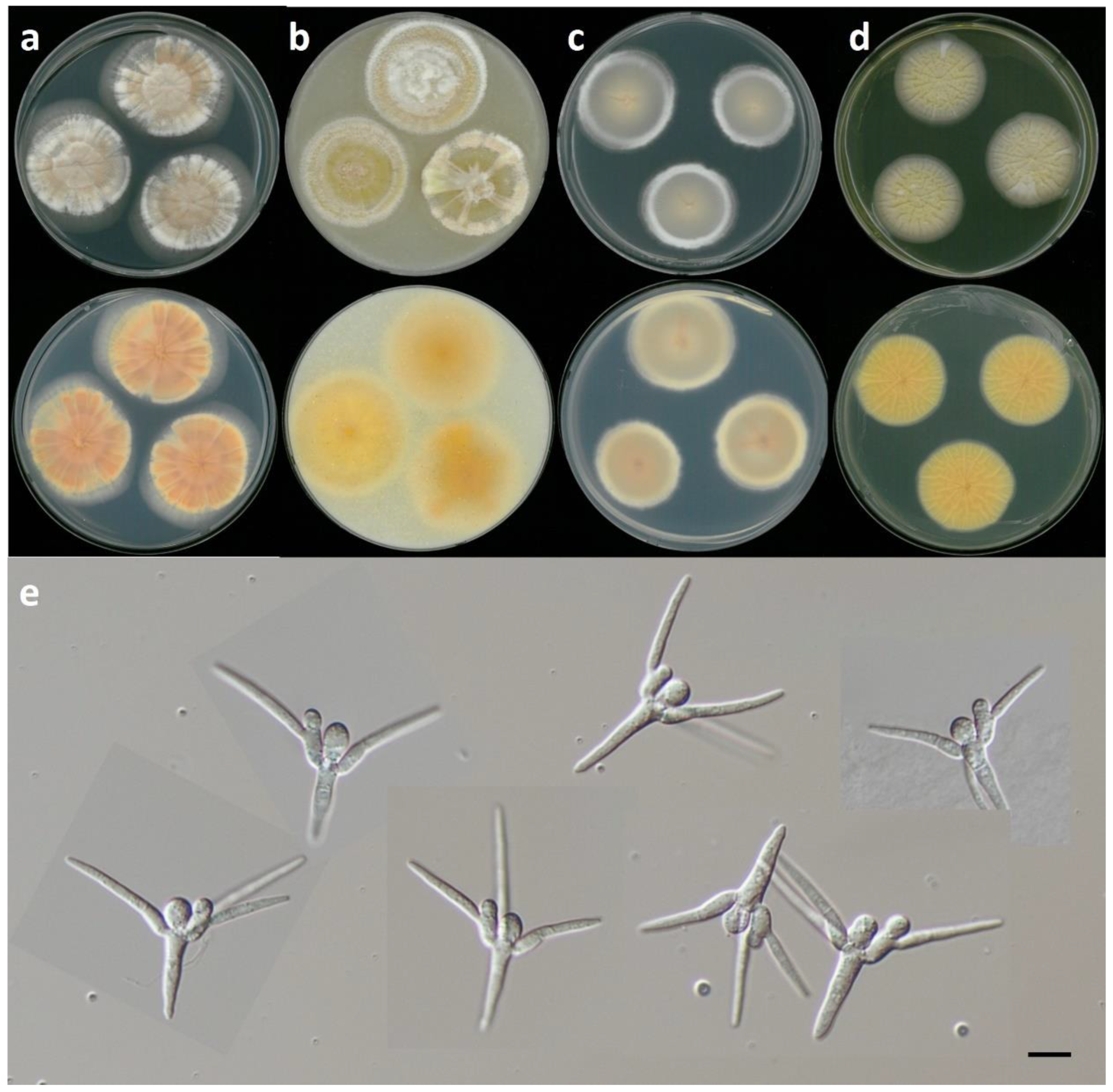
3.2. Taxonomy
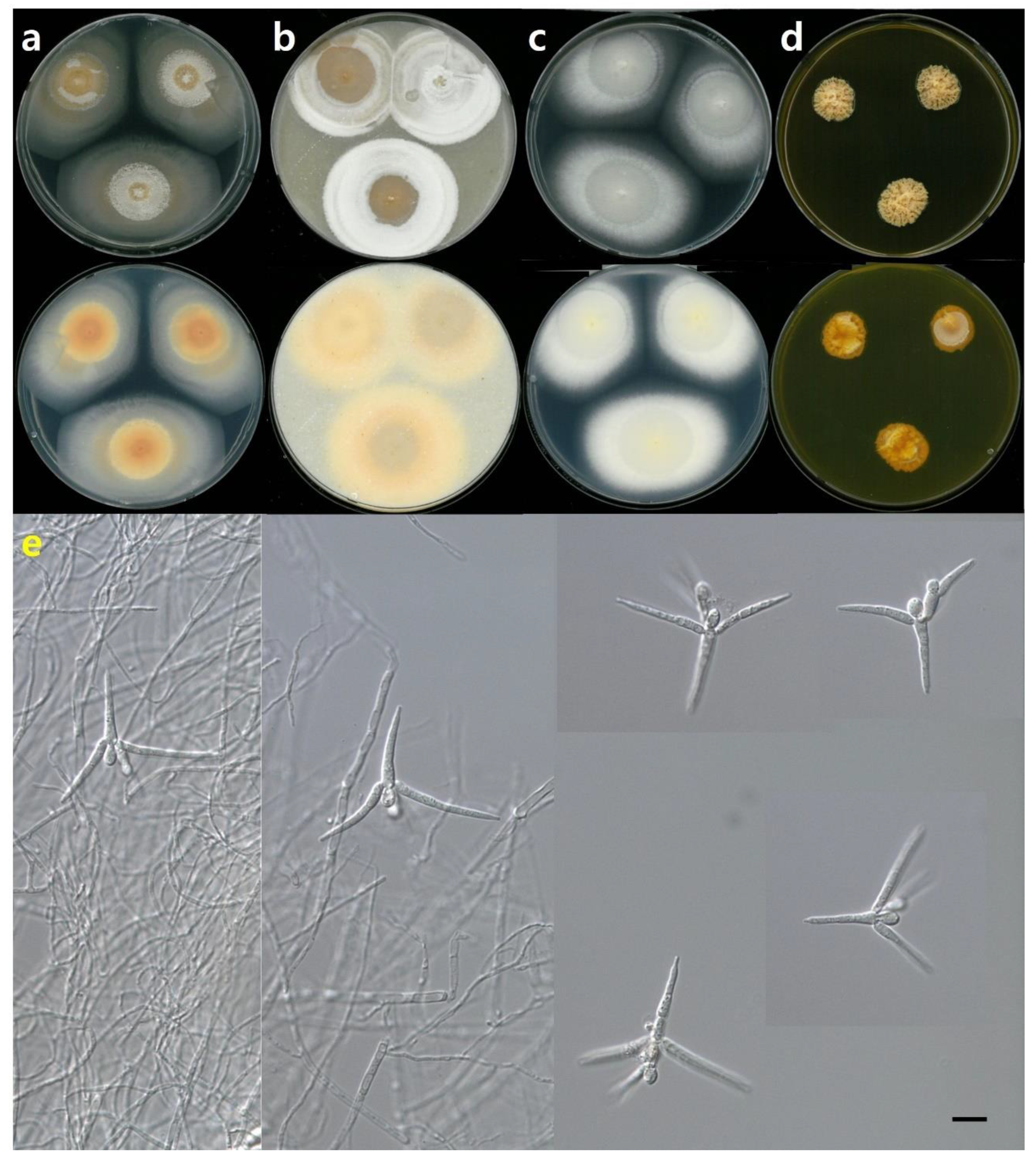
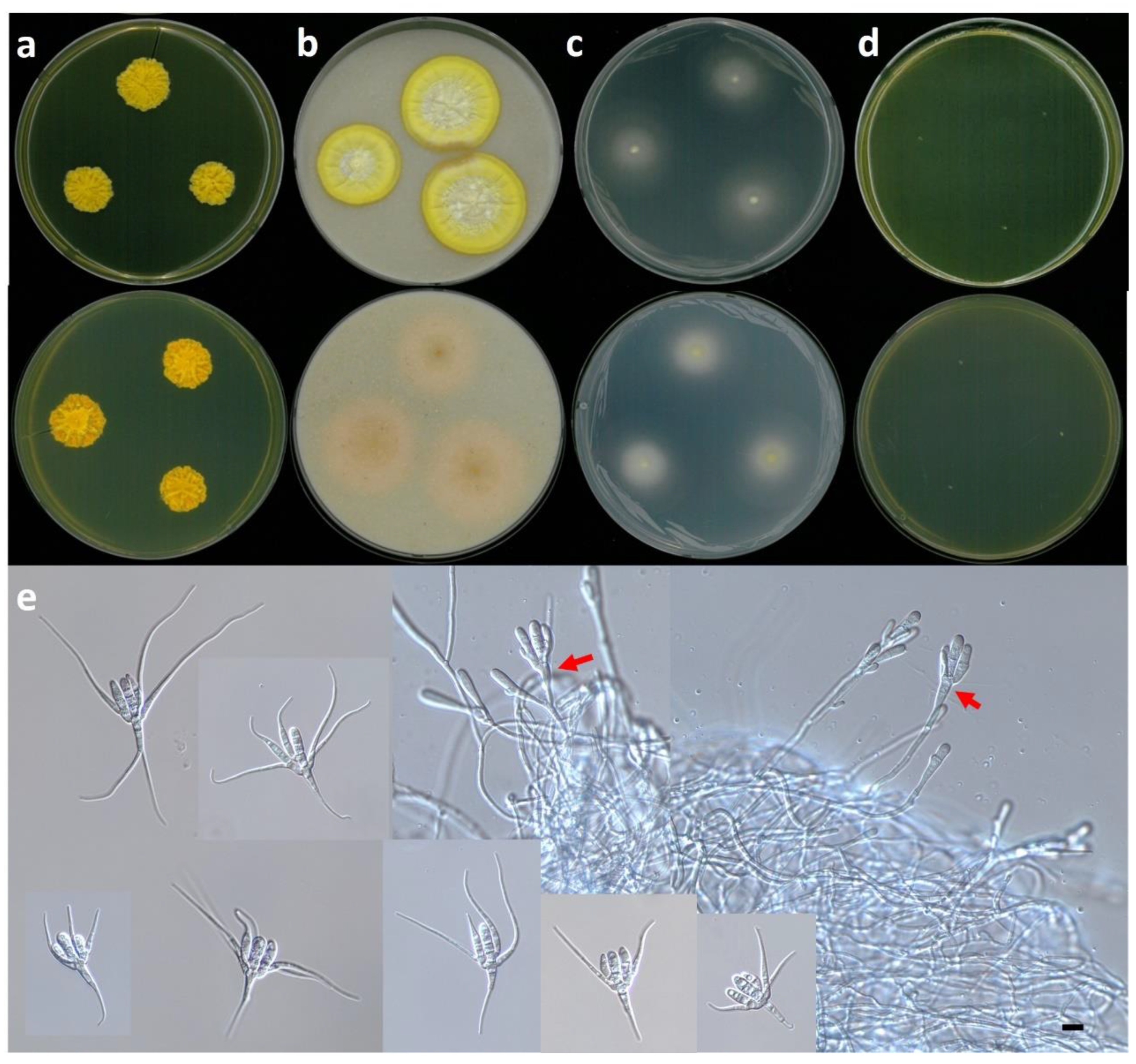
3.2.1. Tetracladium fraxineum sp. nov. J. Goh & H.Y. Mun (2022)
3.2.2. Tetracladium setigerum (Grove) Ingold
3.2.3. Tetracladium marchalianum De Wild
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Krauss, G.J.; Solé, M.; Krauss, G.; Schlosser, D.; Wesenberg, D.; Bärlocher, F. Fungi in freshwaters: Ecology, physiology and biochemical potential. FEMS Microbiol. Rev. 2011, 35, 620–651. [Google Scholar] [CrossRef] [PubMed]
- Ingold, C.T. Aquatic Hyphomycetes of decaying alder leaves. Trans. Br. Mycol. Soc. 1942, 25, 339–417, IN1–IN6. [Google Scholar] [CrossRef]
- Seena, S.; Pascoal, C.; Marvanová, L.; Cássio, F. DNA barcoding of fungi: A case study using ITS sequences for identifying aquatic hyphomycete species. Fungal Divers. 2010, 44, 77–87. [Google Scholar] [CrossRef]
- Bärlocher, F.; Seena, S.; Wilson, K.P.; Dudley Williams, D.D. Raised water temperature lowers diversity of hyporheic aquatic Hyphomycetes. Freshw. Biol. 2008, 53, 368–379. [Google Scholar] [CrossRef]
- Belliveau, M.J.-R.; Bärlocher, F. Molecular evidence confirms multiple origins of aquatic Hyphomycetes. Mycol. Res. 2005, 109, 1407–1417. [Google Scholar] [CrossRef] [PubMed]
- Mun, H.Y.; Goh, J.; Oh, Y.; Chung, N. New records of three aquatic fungi isolated from freshwater in Samcheok and Yeongju, Korea. Korean J. Mycol. 2016, 44, 247–251. [Google Scholar]
- Mun, H.Y.; Oh, Y.; Goh, J.; Chung, N. First report of three Filosporella species isolated from freshwater ecosystem in Korea. Korean J. Mycol. 2019, 47, 165–172. [Google Scholar]
- Ranzoni, F.V. Aquatic Hyphomycetes from Hawaii. Mycologia 1979, 71, 786–795. [Google Scholar] [CrossRef]
- Sinclair, R.C.; Eicker, A. Tetracladium apiense, a new aquatic species from South Africa. Trans. Br. Mycol. Soc. 1981, 76, 515–517. [Google Scholar] [CrossRef]
- Descals, E.; Webster, J. Four new staurosporous Hyphomycetes from mountain streams. Trans. Br. Mycol. Soc. 1983, 80, 67–75. [Google Scholar] [CrossRef]
- Roldán, A.; Descals, E.; Honrubia, M. Pure culture studies on Tetracladium. Mycol. Res. 1989, 93, 452–465. [Google Scholar] [CrossRef]
- Sati, S.C.; Arya, P.; Belwal, M. Tetracladium nainitalense sp. nov., a root endophyte from Kumaun Himalaya, India. Mycologia 2009, 101, 692–695. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Jiang, X.; Wu, W.; Hao, Y.; Su, Y.; Cai, L.; Xiang, M.; Liu, X. Psychrophilic fungi from the world’s roof. Pers. Mol. Phylogeny Evol. Fungi 2015, 34, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Bråthen, I. The aquatic, stauroconidial Hyphomycetes of Norway with notes on the Nordic species. Nord. J. Bot. 1984, 4, 375–392, IN12–IN13. [Google Scholar] [CrossRef]
- Letourneau, A.; Seena, S.; Marvanová, L.; Bärlocher, F. Potential use of barcoding to identify aquatic Hyphomycetes. Fungal Divers. 2010, 40, 51–64. [Google Scholar] [CrossRef]
- Chauvet, E.; Suberkropp, K. Temperature and sporulation of aquatic Hyphomycetes. Appl. Environ. Microbiol. 1998, 64, 1522–1525. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Czeczuga, B.; Orlowska, M. Hyphomycetes in rain water, melting, snow and ice. Acta Mycol. 1999, 34, 181–200. [Google Scholar] [CrossRef]
| Species | Strain | Accession Number |
|---|---|---|
| ITS | ||
| Dactylaria dimorphospora | CBS 256.70 | U51980 |
| Leohumicola minima | N086 | HQ691252 |
| Leohumicola verrucosa | CBS 115881 | AY706323 |
| Mycoarthris corallinus | 91A | AF128440 |
| Tetrachaetum elegans | CB-M11 | KC834066 |
| Tetracladium apiense | CCM F-23199 | EU883420 |
| Tetracladium apiense | CCM F-23299 | EU883422 |
| Tetracladium apiense | CCM F-23399 | EU883421 |
| Tetracladium breve | CCM F-12505 | EU883431 |
| Tetracladium ellipsoideum | MIDUI20 | JX029111 |
| Tetracladium ellipsoideum | MIDUI21 | JX029124 |
| Tetracladium ellipsoideum | MIDUI30 | JX029113 |
| Tetracladium fraxineum sp. nov. | NNIBRFG814 | ON922528 |
| Tetracladium furcatum | C21BN1 | KT582084 |
| Tetracladium furcatum | CCM F-06983 | EU883428 |
| Tetracladium furcatum | CCM F-11883 | EU883432 |
| Tetracladium furcatum | GPO CO 01 | KC180667 |
| Tetracladium globosum | HAILUO215 | JX029109 |
| Tetracladium globosum | MY24 | JX029118 |
| Tetracladium globosum | MY25 | JX029133 |
| Tetracladium marchalianum | CCM F-11391 | EU883417 |
| Tetracladium marchalianum | CCM F-19399 | EU883423 |
| Tetracladium marchalianum | CCM F-26199 | AY204621 |
| Tetracladium marchalianum | CCM F-312 | EU883416 |
| Tetracladium marchalianum | NNIBRFG3891 | ON922530 |
| Tetracladium maxilliforme | CCM F-13186 | EU883430 |
| Tetracladium maxilliforme | CCM F-529 | EU883429 |
| Tetracladium maxilliforme | F-14286 | AF411027 |
| Tetracladium psychrophilum | HAILUO380 | JX029119 |
| Tetracladium psychrophilum | MY376 | JX029129 |
| Tetracladium setigerum | CCM F-10186 | EU883427 |
| Tetracladium setigerum | CCM F-19499 | EU883426 |
| Tetracladium setigerum | CCM F-20987 | KU519120 |
| Tetracladium setigerum | NNIBRFG1997 | ON922529 |
| Tetracladium setigerum | UMB 736.11 | KF952740 |
| Tricladium alaskense | VG-2012a | JQ417290 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goh, J.; Jeon, Y.J.; Mun, H.Y. New Records of the Psychrophilic Tetracladium Species Isolated from Freshwater Environments in Korea. Diversity 2022, 14, 789. https://doi.org/10.3390/d14100789
Goh J, Jeon YJ, Mun HY. New Records of the Psychrophilic Tetracladium Species Isolated from Freshwater Environments in Korea. Diversity. 2022; 14(10):789. https://doi.org/10.3390/d14100789
Chicago/Turabian StyleGoh, Jaeduk, Yu Jeong Jeon, and Hye Yeon Mun. 2022. "New Records of the Psychrophilic Tetracladium Species Isolated from Freshwater Environments in Korea" Diversity 14, no. 10: 789. https://doi.org/10.3390/d14100789
APA StyleGoh, J., Jeon, Y. J., & Mun, H. Y. (2022). New Records of the Psychrophilic Tetracladium Species Isolated from Freshwater Environments in Korea. Diversity, 14(10), 789. https://doi.org/10.3390/d14100789






